Abstract
Tris(1,3-dichloro-2-propyl) phosphate (TDCIPP) has been frequently detected in environmental media and has adverse health effect on wildlife and humans. It has been implicated to have hepatotoxicity, but its molecular mechanisms remain unclear. In the present study, adult male zebrafish were exposed to TDCIPP and global hepatic gene expression was examined by RNA-Seq and RT-qPCR in order to understand the molecular mechanisms of TDCIPP-induced hepatotoxicity. Our results indicated that TDCIPP exposure significantly up-regulated the expression of genes involved in endoplasmic reticulum stress and Toll-like receptor (TLR) pathway, implying an inflammatory response, which was supported by up-regulation of inflammation-related biomaker genes. Hepatic inflammation was further confirmed by histological observation of increase of infiltrated neutrophils and direct observation of liver recruitment of neutrophils labeled with Ds-Red fluorescent protein of Tg(lysC:DsRed) zebrafish upon TDCIPP exposure. To further characterize the hepatotoxicity of TDCIPP, the expression of hepatotoxicity biomarker genes, liver histopathology and morphology were examined. The exposure to TDCIPP significantly up-regulated the expression of several biomarker genes for hepatotoxicity (gck, gsr and nqo1) and caused hepatic vacuolization and apoptosis as well as increase of the liver size. Collectively, our results suggest that exposure to TDCIPP induces hepatic inflammation and leads to hepatotoxicity in zebrafish.
Similar content being viewed by others
Introduction
Tris(1,3-dichloro-2-propyl) phosphate (TDCIPP) has been used as flame retardants and plasticizers in various products (e.g. plastics, foams, textiles, varnishes, electronics equipment and furniture) for decades and its annual production is estimated to be in the range of 4500 to 22,700 tons between 1998 and 2006 in the United States1. In recent years, TDCIPP has been increasingly used as the primary replacement of the phased-out flame retardant polybrominated diphenyl ether (PBDE)1.
Like PBDEs, TDCIPP is not chemically bonded to the related products and it is released to the environment easily2. TDCIPP is frequently detected in indoor air, dust, surface water, drinking water, influents, effluents, sediments, wildlife and human body1,3,4,5,6,7,8,9,10. For example, it has been documented that TDCIPP is present in more than 96% of the indoor dust samples in the United States and the concentrations range from <90 ng/g to 56,000 ng/g11. The concentrations of TDCIPP in surface water and effluent of sewage treatment plants in Germany and Norway have been reported to be up to 50 ng/L and 740 ng/L, respectively1,3. In the wildlife, TDCIPP has been detected at 36–140 μg/kg lipid weight in freshwater perches10. Recently, TDCIPP has also been detected in human milk and urine of office workers10,12,13.
Despite of its high volume of use and frequent detection in the environment, to date only limited information is available about the toxic effects of TDCIPP. For example, it has been reported that exposure to TDCIPP inhibits DNA synthesis and promotes neuron differentiation in PC12 cells14. In zebrafish, TDCIPP also causes developmental toxicity and endocrine disruption15,16,17,18,19,20. Exposure to TDCIPP in primary cultured avian hepatocytes also causes cytotoxicity with deregulation of genes involved in phase I and II metabolism, thyroid hormone pathway, lipid regulation and growth21. Furthermore, injection of TDCIPP to chicken eggs has resulted in a significant accumulation of TDCIPP in the liver and changes of expression of hepatic genes related to xenobiotic metabolism, thyroid hormone pathway and immune responses22,23. These studies suggest that liver is a target for TDCIPP exposure, however to support reliable risk assessment, underlying molecular mechanisms for hepatotoxicity need to be further explored.
The recently developed RNA-Seq technology provides a powerful tool to determine molecular mechanisms in organisms after chemical exposure, especially for emerging environmental pollutants with limited toxicological information since it allows a global examination of biological responses through gene expression. In this study, the effect of TDCIPP on zebrafish hepatic transcriptome was evaluated by RNA-Seq and we found an apparent inflammatory response based on TDCIPP-induced transcriptomic changes. The inflammatory response was confirmed by up-regulation of some biomarker genes and migration of neutrophils to the liver following TDCIPP treatment. Finally, the hepatotoxicity of TDCIPP was further characterized by measuring the expression of biomarker genes for hepatotoxicity and the change of liver histology and morphology.
Materials and Methods
Animals and Chemical Exposure
TDCIPP was purchased from Sigma (St. Louis, MO, USA) and dissolved in dimethyl sulfoxide (DMSO) as a stock solution. Briefly, wild-type male zebrafish (Singapore strain, 5 months old) were acclimated for 2 weeks in our aquarium at 28 °C, with a 14:10 light/dark cycle. Zebrafish were then exposed to 0.01% DMSO (vehicle control) or 1 mg/L TDCIPP for 4 days in 15 L glass tanks with 10 L exposure solution and half of the water with the same chemical of the same concentration was daily renewed. During semi-static exposure period, water pH (7.0–7.3), hardness (3-4 dGH) and dissolved oxygen (6.4–6.8 mg/L) were routinely monitored. The exposure concentrations were selected based on the information from a previous study16, where exposure to 1 mg/L TDCIPP caused reproductive toxicity and endocrine disruption in adult zebrafish. There were two replicated tanks for each group and each tank contained 12 fish. No mortality was observed in any of the treatment and control groups during the exposure period. After the exposure, the fish were anesthetized and livers were collected for RNA extraction, RNA-Seq and Western blotting analyses. All experimental procedures in this study were carried out following the approved protocol by Institutional Animal Care and Use Committee of National University of Singapore (Protocol 079/07). All experiments of this study were performed in accordance with relevant guidelines and regulations in Singapore.
In order to confirm our findings in RNA-Seq, the second set of TDCCP exposure experiments, including time- and dose-dependent exposure experiments, were conducted. Wild-type male zebrafish (5 months old) were acclimated for 2 weeks, followed by exposure to 0.01% DMSO (vehicle control) or different concentrations of TDCIPP (0.1, 0.3 or 1 mg/L) as above described. There were three replicated tanks for each group and each tank contained 12 fish. For 1 mg/L exposure group, fish were sampled after 1, 2 and 4 days of exposure; for 0.1 and 0.3 mg/L exposure groups, fish were sampled only after 4 days of exposure. Liver tissues were preserved in TRIzol reagent (Invitrogen, New Jersey, NJ, USA) for RT-qPCR validation or collected for histological examination. No mortality was observed in any of the treatment and control groups during the exposure period. For dose-dependent exposure experiment, one mL exposure solution from each tank was collected before and after renewal of the water solutions on the last day of exposure and concentrations of TDCIPP were quantified.
Quantification of TDCIPP in Exposure Solutions
Exposure solutions were sampled before and after renewal of the water solutions on the last day of exposure and concentrations of TDCIPP were quantified according to previously published protocols24,25. Briefly, exposure solutions were filtered through 0.22 μm membrane filters and then diluted using Milli-Q water. Water samples were analyzed using a Waters ACQUITY UPLC® I-Class system (UHPLC) coupled to Waters® XevoTM TQ-S mass spectrometer (TQ-S/MS) (Milford, MA, USA) using electrospray ionization (ESI(+)) in the multiple reaction monitoring (MRM) mode. LC separation was performed on a CortecsTM UHPLC C18 column (2.1 mm × 50 mm, 1.6 μm particle size) (Waters, Mississauga, ON, Canada). Mobile phases for LC were water (A) and methanol (B) and flow rate was set to 0.5 mL/min. Gradient was as follows: 0 min, 5% B; 0−5 min, 95% B (linear); hold for 1 min; 6−6.1 min, 5% B (linear) and hold for 4.9 min. The capillary voltage was 0.5 KV. The source and desolvation temperatures were 150 and 600 °C, respectively. The desolvation and cone gas flow rates were 800 and 150 L/h, respectively. Method limits of quantification were 0.01 ng TDCIPP /mL water. Triplicates of beakers were conducted for each concentration.
RNA Isolation and Sequencing
Total RNA was isolated using TRIzol reagent and treated with DNase I (Invitrogen, New Jersey, NJ, USA) to remove genomic DNA contamination. RNA concentrations, ratios of 28S/18S and RNA integrity were determined using Agilent Bioanalyser 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA). RNA concentrations ranged from 1 to 2 μg/μL and ratios of 28S/18S ranged from 1.85 to 2.05. Six livers were pooled to generate one biological replicate for RNA preparation and two biological replicates from two individual tanks were included for each treatment group. Magnetic beads with Oligo(dT) were used to isolate poly A + RNA, which was then fragmented into short fragments in fragmentation solution. cDNA was synthesized using the mRNA fragments as templates and short fragments were purified and ligated with adapters. After agarose gel electrophoresis, suitable fragments were selected for PCR amplification and sequenced as 2 × 90 bp paired-end reads on Illumina HiSeqTM 2000 sequencer (Illumina, San Diego, CA, USA). RNA sequencing was performed by BGI Tech Solutions (Hong Kong) Co., Limited using the Illumina’s Solexa platform.
Sequence Tag Preprocessing, Mapping and Statistical Analyses
The original image data were transferred into sequence data and saved as FASTQ files. The quality control of alignment was performed to determine if resequencing was necessary. Briefly, the raw reads were cleaned by removing reads with adaptors, low sequence quality (>30%) or high-proportion unknown bases (>5%) in a read. The clean data were then mapped to the zebrafish Reference Sequence database (http://www.ncbi.nlm.nih.gov/RefSeq) using the SOAPaligner/SOAP2 software, with allowance of maximum 5 nucleotide mismatches. Finally, the alignment data was utilized to calculate the distribution of gene coverage. Gene expression level was normalized as RPKM (reads per kilobase transcriptome per million mapped reads). In this study, we selected differentially expressed genes based on fold change >2 (statistical power >0.8) and P value < 0.05.
Gene Ontology and Pathway Analyses
Gene ontology and pathway analyses were conducted using DAVID (The Database for Annotation, Visualization and Integrated Discovery) with the total zebrafish genome information as the background26. Gene Ontology Fat and KEGG-pathway categories were used in this study and P value (modified Fisher’s exact t-test) cut-off was set at 0.05.
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was isolated using TRIzol reagent and treated with DNase I to remove genomic DNA contamination as previously described27. The synthesis of first-strand cDNA and RT-qPCR were performed by use of Maxima® First Strand cDNA Synthesis kit (Fermentas, St Leon-Rot, Germany) and SYBR® Green PCR kit (Toyobo, Osaka, Japan), respectively according to the manufacturer’s instructions. PCR primers were designed using Primer 3 software (http://frodo.wi.mit.edu/as) (Table S1, see Supporting Information) and glyceraldehyde-3-phosphate dehydrogenase (gapdh), whose cycle threshold (Ct) values were not changed upon TDCIPP exposure in this study (Figure S1), was used as an internal reference. The mRNA levels were expressed as fold change using the 2-ΔΔCt method. There were 3 replicated tanks for each concentration and three fish from each tank were used and thus totally 9 fish were analyzed in each treatment.
Western Blotting Analyses
Western blotting analyses were performed as previously described with some modifications28. After exposure, liver tissues were sampled and homogenized. Homogenates were centrifuged for 5 min at 4 °C and protein contents were determined using commercial BCA (bicinchoninic acid) kit from Sigma (St. Louis, MO, USA). Equal quantity of proteins (50 μg) from control and exposure groups were denatured, electrophoresed and transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were cut into stripes (Figure S2) and blocked by 5% non-fat dry milk for 1 h. The blots were probed with primary antibodies from Cell Signaling Technology, Inc. (MA, USA) for 12 h at 4 °C, followed by three washes and incubation with corresponding secondary antibodies (Cell Signaling Technology, Inc., MA, USA) for 30 min at room temperature. ECLTM reaction solution was prepared according to the manufacturer’s instructions (PerkinElmer Inc., SC, USA). Membranes were washed three times and exposed to Kodak film for two min at room temperature. Films were developed and Grp78, Chop, Fos and Il6 were detected by chemiluminescence.
Zebrafish Larva Assays
Two transgenic zebrafish lines were used in the present study: Tg(lysC:DsRed)29 and Tg(fabp10a:DsRed; elaA:egfp)30, named Tg(fabp10a:DsRed) in the following text. Tg(lysC:DsRed) line has red fluorescent protein (DsRed) expression in neutrophils and it is feasible to observe recruitment of neutrophils in the liver31 and other organs29. Tg(fabp10a:DsRed) line has liver-specific DsRed expression under the gene fabp10a promoter and allows an easy measurement of liver size30. It has been confirmed to be useful in screening hepatotoxin (e.g., acetaminophen, aspirin, isoniazid and phenylbutazone) which can induce hepatic damages, oxidative stress and cellular necrosis32. The embryos from the two transgenic lines were collected and cultured in egg water as described in a previous study33. At 96 hour postfertilization (hpf), larvae from the two transgenic lines were exposed to 0.01% DMSO or different concentrations of TDCIPP (0.1, 0.3 or 1 mg/L) in 6-well plates. Half of the exposure media was daily replaced. There were three replicated wells for each exposure concentration at each sampling time point and each well contained 10 larvae. Both TDCIPP and control groups received 0.01% DMSO. For 1 mg/L exposure group, fish were sampled after 1, 2 and 4 days of exposure; for 0.1 and 0.3 mg/L exposure groups, fish were sampled only after 4 days of exposure. The number of neutrophils in the liver area in Tg(lysC:DsRed) larvae was determined using Carl Zeiss Axiovert 200M fluorescent and Carl Zeiss LSM 510 Meta fluorescent microscopes. Liver size was measured for Tg(fabp10a:DsRed) larvae using ImageJ software (http://rsbweb.nih.gov/ij/) based on 2D liver images as previously described32.
Histological Examination
After 4 days of exposure, the liver tissues from males were sampled for histological examination as previously described34. Briefly, the livers were fixed in Bouin’s solution and dehydrated in ethanol. The samples were then embedded in paraffin wax, sectioned at 5 μm and stained with haematoxylin and eosin.
Statistical Analysis
Statistical analyses of the data for the number of neutrophils in the liver, liver size and gene expression were conducted using Kyplot Demo 3.0 software (Tokyo, Japan). Normality and homogeneity of data were evaluated by the Kolmogorov-Smirnow and Levene’s tests, respectively. ANOVA (one-way analysis of variance) was adopted to determine significant differences between the control and TDCIPP exposure groups. A level of significance for type I error was set at P value < 0.05.
Results
Measured Concentrations of TDCIPP in Exposure Solutions
The nominal concentrations of TDCIPP in the exposure solutions were 0.1, 0.3 and 1 mg TDCPP/L. The analytical measured and actual TDCIPP concentrations in the same three solutions were 0.12 ± 0.00, 0.41 ± 0.01 and 1.13 ± 0.05 mg/L before water renewing and 0.12 ± 0.00, 0.43 ± 0.02 and 1.17 ± 0.02 mg/L after water renewing, respectively (Figure S3, see Supporting Information).
Transcriptomic Responses to TDCIPP in Adult Zebrafish Liver
No mortalities were observed in any of the treatment groups during exposure period. To analyse transcriptomic responses in the liver following acute TDCIPP exposure, four RNA libraries were constructed for RNA-Seq: two replicates from the TDCIPP treatment group (1 mg/L, 4 days) and two replicates from the 0.01% DMSO vehicle treatment group. Representative images for the composition and quality distribution of bases are showed in Figures S4 and S5 (see Supporting Information), where the T and C curves were in accordance with the A and G curves, respectively (Figure S4, see Supporting Information). The percentage of the bases with low quality (<20) was very low in all the samples tested (Figure S5, see Supporting Information), indicating good-quality of sequencing data without the need of resequencing. After filtering out reads with adaptors, low sequence quality (>30%) or high-proportion unknown bases (>5%), over 50 million clean reads were obtained from each library (Table S2) and over 70% of these reads (or at least 35 million from each library) were mappable to the zebrafish Reference Sequence database (http://www.ncbi.nlm.nih.gov/RefSeq), representing a total of 16,631 genes (Table S2). Using fold change >2 and P value < 0.05 as selection criteria, 583 differentially expressed genes (306 up-regulated and 277 down-regulated) were identified between control and TDCIPP groups (Figure S6 and excel data, see Supporting Information). Furthermore, using relative (log 2) RPKM values of the 583 differentially expressed genes, regression analysis (standard line assay) was conducted among the four RNA-Seq groups. As shown in Figure S7 (see Supporting Information), the adjusted correlation coefficients (R2) were very high within the same treatment group (DMSO control, 0.9181; TDCIPP, 0.9612), but low between the two groups (0.6155–0.6842), indicating good repeatability and reliability of our data.
The up- and down-regulated transcripts were further subjected to Gene Ontology (GO) and KEGG pathway analyses. The most significantly enriched GO term for biological process was Innate immunity response, where five toll-like receptor (tlr) genes were included (Table 1). Other relevant enriched terms in the biological process include Defense response, Response to inorganic substance, Transmembrane transport, Metal ion transport, Cation transport, Ion transport, Response to xenobotic stimulus and Response to virus (Table 1). Only one enriched GO term with 14 deregulated genes, endoplasmic reticulum (ER), was observed for cellular component (Table 1). The enriched terms for molecular function included nucletidyltransferase activity, oxidoreductase activity (acting on the CH-NH2 group of donors, oxygen as receptor) and dipeptidyl-peptidase activity. In KEGG pathway analysis, two enriched terms, toll-like receptor signaling pathway and steroid hormone biosynthesis, were obtained (Table 1). For toll-like receptor signaling pathway, 6 genes were involved, including 5 up-regulated and 1 down-regulated genes (Table 1).
Time- and Dose-Dependent Response of Biomarker Genes Involved in ER Stress and Inflammation
To validate our findings in RNA-Seq, time- and dose-dependent exposure experiment was conducted and expression of 19 selected genes involved in Innate immune response (GO)/Toll-like receptor signaling pathway (KEGG), ER response and Inflammation response, were determined by RT-qPCR (Table 2 and Table 3). TDCIPP exposure caused a time-dependent up-regulation of genes enriched in Innate immunity response/Toll-like receptor signaling pathway. While the expression of these genes (tlr18, tlr8a, tlr8b, tlr20a, tlr9, fos, stat1b and irf7) was not significantly altered after 1 day of TDCIPP treatment, their expression was all significantly up-regulated by 2 days of exposure and by 4 days of TDCIPP treatment. The two ER stress biomarker genes, grp78 (glucose-regulated protein 78) and chop (CCAAT/enhancer-binding protein-homologous protein), also showed similar up-regulation by TDCIPP, with 3.2- and 9.9-fold up-regulation for grp78 and 1.9- and 10.6-fold up-regulation for chop after 2 and 4 days of exposure, respectively. Finally, abundances of some marker genes for inflammation were also examined after TDCIPP exposure for 1, 2 and 4 days. At least seven of them (il1b, il6, il10, il12a, il13, il15 and il26) also showed time-dependent increase following TDCIPP. The highest up-regulated genes were two interleukin genes, il13 and il26, with over 23 fold of increase of expression after 4 days of TDCIPP exposure. Exposure to lower concentrations of TDCIPP (0.1 or 0.3 mg/L) for 4 days only up-regulated the expression of stat1b, irf7, grp78, il13, il22 and il26, while the expression of other genes was not significantly changed.
To further confirm the up-regulation of some of these genes at protein level, four proteins from Toll-like receptor signaling pathway (Fos), ER stress (Grp78 and Chop) and Inflammation response (Il6) were selected for Western blot analysis because of the availability of their antibodies. Their expression in liver of male zebrafish exposed to 0 or 1 mg TDCIPP/L was examined. As shown in Fig. 1, exposure to 1 mg TDCIPP/L for 4 days significantly up-regulated expressions of Grp78, Chop, Fos and Il6 by 2.05, 4.77, 1.71 and 2.13 fold, respectively.
Effects on expressions of four proteins selected (Grp78, Chop, Fos and Il6) in response to 1 mg/L TDCIPP.
(A) Western blots of four proteins selected from control and TDCIPP groups; (B) Quantification of the relative expressions of four proteins selected in control and treatment groups. C: control group; T: treatment group. Values represent mean ± SEM (n = 3). Asterisks indicate significant differences from matched control samples (P < 0.05).
Up-regulation of Hepatotoxicity Biomarker Genes by TDCIPP
To further confirm the hepatotoxicity caused by TCDPP exposure, a panel of 14 hepatotoxicity biomarker genes included in “generalized hepatotoxicity” based on Qiagen Hepatotoxicity RT2 Profiler PCR array (http://www.qiagen.com/) were selected for RT-qPCR analysis. We found that exposure to 1 mg/L TDCIPP for 2 or 4 days up-regulated the expression of three of these biomarker genes, gclc (glutamate-cysteine ligase catalytic subunit), gsr (glutathione reductase) and nqo1 (NAD(P)H dehydrogenase [quinone] 1 isoform 1) (Fig. 2A), while expression of other genes (krt8, plazg12a, hmox1, krt18, gadd45ab, cryl1, ccng1, casp3b, casp3a, apex1 and aldoaa) was not significantly changed (data not shown). Furthermore, out of the 14 genes selected, thirteen genes were detected in RNA-seq and only plazg12a was not detected possibly due to low expression abundance. The fold changes of the 13 genes were consistent in qRT-PCR with those in RNA-seq although up-regulation of gclc (1.52) and nqo1 (1.77) was not statistically significant in RNA-Seq data and thus the two genes were not initially selected by out cutoff criteria for differentially expressed genes. No significant changes were observed after exposure to lower concentrations of TDCIPP (0.1 or 0.3 mg/L) for 4 days (Fig. 2B).
Dose- (A) and time-dependent (B) effects on the expression of selected hepatotoxicity biomarker genes (gclc, gsr and nqo1) in response to TDCIPP.(C): control group; T: treatment group. Values represent mean ± SEM (n = 9). Asterisks indicate significant differences from matched control samples (P < 0.05).
Validation of Inflammatory Response and Hepatotoxicity by Histological Examination and Transgenic Zebrafish Larvae
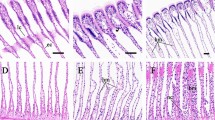
As RNA-Seq data indicated a potential inflammatory response caused by TDCIPP exposure, histological examination was also carried out for the effects of TDCIPP on liver cells. In this experiment, adult zebrafish were exposed to 1 mg/L TDCIPP for 4 days. Histological examination revealed that the TDCIPP exposure caused an increase of infiltrated neutrophils, hepatic vacuolization and apoptosis (Fig. 3). Examples of moderately and severely affected liver sections are shown in Fig. 3B,C, respectively.
Changes of liver histology of male zebrafish after exposure to 1 mg/L TDCIPP for 4 days.
Liver sections were stained by hematoxylin and eosin. (A) representative liver section from the DMSO vehicle control group. (B,C) Two representative liver sections from two individual fish of the same TDCIPP group with a moderate effect (B) and a severe effect (C). The left images have a magnification of 200X and the right images have a magnification of 400X. Several features are exampled by arrows of different colors: neutrophils (black); apoptosis (red); hepatic vacuolization (green).
To further confirm the inflammatory response induced by the TDCIPP exposure, Tg(lysC:DsRed) zebrafish larvae, in which neutrophils were labelled by DsRed expression, were treated with different concentrations of TDCIPP from 96 hpf. No mortalities were observed in any of the treatment groups during the exposure period. Exposure to TDCIPP caused time- and dose-dependent increases in infiltration of neutrophils in the liver, while no significant infiltration was observed in other internal organs (Fig. 4A–C). In addition, slight increase of neutrophils was observed in the ventral region of the head and near the mouth (Fig. 4A–C).
Dose- (A) and time-dependent (B) increase of neutrophils in the livers of Tg(lysC:dDsRed) zebrafish larvae in response to TDCIPP exposure.Images were captured with a digital camera attached to a Carl Zeiss LSM 510 Meta fluorescent microscope. (C) Representative images from control group and 4-day TDCIPP (1 mg/L) exposure group. The livers are outlined with white lines. Values represent mean ± SEM. Significant difference between the two groups were observed: P < 0.05.
Transgenic zebrafish Tg(fabp10a:DsRed) was also used to investigate the effect of TDCIPP and this transgenic line has been previously suggested to be a useful model to evaluate the hepatotoxicity of chemicals32. In the present study, no mortalities were observed in any of the treatment groups during TCDPP exposure; the liver size was significantly increased in a time- and dose-dependent manner compared with that of the control (Fig. 5A–C).
Dose- (A) and time-dependent (B) increase of liver size in Tg(fabp10a:DsRed) zebrafish larvae after exposure to TDCIPP.Images were captured with a digital camera attached to a Carl Zeiss LSM 510 Meta fluorescent microscope. Liver sizes were measured based on the 2D image using the ImageJ software. (C) Representative images from control group and 4-day TDCIPP (1 mg/L) exposure group Values represent mean ± SEM. Significant difference between the two groups were observed: P < 0.05.
Discussion
It has been recently demonstrated that TDCIPP is significantly accumulated in the livers of chicken after exposure and the expression of hepatic genes included in xenobiotic metabolism, thyroid hormone pathway, lipid regulation and immune responses are deregulated21,22,23, thus implying an apparent hepatotoxicity. However, these studies only examined responses of certain genes involved in a few pathways and the information provided is rather limited. To devise a reliable risk assessment, further study is needed for a more comprehensive examination of gene responses by using “omic” technologies. In the present study, we have evaluated the effects of TDCIPP on hepatic transcriptome by RNA-Seq in zebrafish. Our data suggest that acute exposure to TDCIPP significantly changes the expression of genes involved in TLR pathway and ER stress in a dose- and time-dependent manner. More interestingly, TDCIPP also leads to the up-regulation of some biomaker genes for inflammation, which has been confirmed by increased neutrophil infiltration in the adult liver and rapid migration of neutrophils to the larval liver. Finally, TDCIPP exposure up-regulates the expression of several biomarker genes for hepatotoxicity (e.g. gclc, gsr and nqo1) and causes hepatocyte vacuolization, apoptosis in liver cells and increase of liver size, further proving the hepatotoxicity of TCDPP.
In this study, transcriptional effect of TDCIPP on the liver has been first evaluated in zebrafish. GO and KEGG pathway analyses indicate that a number of biological processes and pathways are significantly altered. The most significantly enriched GO term for cellular component is Innate immunity response, where five tlr genes are included. KEGG pathway analysis has further confirmed that TDCIPP exposure significantly changes the expression of genes in TLR signaling pathway, including tlr8a, tlr8b, tlr9, ap-1, stat1b and irf7. In mammalian livers, TLR genes are expressed in Kupffer cells, hepatocytes, stellate cells, biliary epithelial cells, sinusoidal endothelial cells, dendritic cells and other types of immune cells35,36. The protein products of these TLR genes are pattern recognition receptors and, once activated, they can interact with a common adaptor, MyD88 (myeloid differentiation factor 88), to activate nuclear transcription factors such as NF-κB, AP-1 and IRFs and to cause the initiation of innate immunity36. In addition, activated IRFs can interact with interferons (IFNs) to activate STAT1 and cause corresponding inflammatory responses37. In this study, exposure to TDCIPP significantly up-regulates the expression of tlr8b, tlr9, ap-1, irf7 and stat1b in a dose- and time-dependent manner. Consistent with this, we have found an up-regulation of eight cytokine genes/proteins in the liver after TDCIPP exposure. Cytokines are a family of secreted and regulatory molecules with molecular masses ranging from 10 to 50 kDa38. Besides classical responses and interactions between immune and neuroendocrine systems, cytokines play a fundamental role in inflammation38 and these genes are considered as main biomarkers of inflammation in fish39. In addition, an increase of neutrophils in the liver of TDCIPP-treated adult fish and Tg(lyzC:DsRed) zebrafish fry are also observed. Although neutrophil infiltration might be also due to liver damage (e.g., fatty liver) induced by TDCIPP, the up-regulation of biomarker genes for inflammation strongly suggests that inflammatory responses do occur.
Recently, it has been reported that some TLRs, such as TLR9, are exclusively sequestered in the ER in unstimulated cells and traffic to endolysosomes upon ligand stimulation40. Moreover, ER stress can directly induce TLRs and synergise with TLRs to cause inflammatory responses or/and related diseases in the liver35,41,42,43,44,45. In this study, ER is also a significantly enriched GO term, where the expression of 14 genes is significantly altered during TDCIPP exposure. For example, the expression of pdia4, hyou1, hsp90b1, hspa8 and pdip5 is significantly down-regulated after TDCIPP exposure. The protein products of these genes are ER chaperones46,47,48,49,50 and the down-regulation of these genes is the evidence for decreased ER function due to stress. Similarly, slc35b1 (solute carrier family 35 member B1) is responsible for sugar transport in ER51 and exposure to 1 mg/L TDCIPP for 4 days significantly down-regulates its expression. Thus, the down-regulation of slc35b1 may be another evidence for the occurrence of ER stress. In addition, treatment with TDCIPP significantly up-regulates the expression of sgk1; this is reminiscent of a previous report that ER stress in PC12 cells induced by overexpression of β-amyloid precursor protein are also accompanied by the up-regulation of sgk1 expression52. Accumulated evidence indicates that when ER stress occurs, cells could initiate an adaptive response called UPR (unfolded protein response) to maintain homeostasis of ER function, such as increase in the expression grp78 and chop53,54,55. Protein coded by grp78 works as a sensor for accumulation of unfolded proteins; once activated it will cause UPR54. Chop plays a key role in the downstream of UPR and works as a chaperone of other proteins (e.g., cleaved Atf6α and Xbp-1) to mediate inflammatory response and cellular apoptosis54. In this study, TDCIPP causes a time-dependent up-regulation of the two genes. Treatment with 1 mg/L TDCIPP for 4 days also increases protein expressions of Grp78 and Chop. However, the expressions of grp78 and chop were not detected in RNA-seq, which might be due to their low abundances of expression. Collectively, our observations suggest that ER stress occurs in zebrafish liver upon TDCIPP exposure, which might be a main reason for the up-regulation of genes included in the Toll-like receptor pathway.
Finally, we have characterized the hepatotoxicity of TDCIPP by measuring the expression of related genes and the change of liver histology and morphology. In this study, a panel of hepatotoxicity biomarker genes based on Qiagen Hepatotoxicity RT2 Profiler PCR array (http://www.qiagen.com/) were selected for RT-qPCR analysis and expressions of three biomarker genes including gclc, gsr and nqo1 were significantly up-regulated. The three genes encode antioxidant and detoxifying enzymes in liver56 and up-regulation of their expression are usually used as biomarker of hepatotoxicity56. Therefore, our results confirm that TDCIPP has potential to induce hepatotoxicity in zebrafish. Histological evidence indicates that exposure to TDCIPP causes hepatocyte apoptosis and vacuolization. Furthermore, we have also used Tg(fabp10a:DsRed) transgenic zebrafish to further evaluate the effect of TDCIPP on liver size. This transgenic line has RFP expression in the liver and it is feasible to measure the size of liver. We have found that treatment with 1 mg/L TDCIPP for 4 days significantly increases liver size. Consistent with this, a previous study also suggested that hepatic inflammation was accompanied by enlarged hepatosomatic index in fish exposed to pollutants57.
Here it should be noted that inflammatory response might be also caused by liver injury and apoptosis induced by TDCIPP, therefore further studies are needed to explore these possibilities. In addition, the concentrations (0.1, 0.3 and 1 mg/L) of TDCIPP that fish have been exposed in this study is several orders of magnitude greater than those reported in the environment, but our study represents an acute toxic experiment. Due to increased use and frequent detection of TDCIPP, further studies to evaluate the potential toxic effects of TDCIPP by chronic exposure with low concentrations are required.
Additional Information
How to cite this article: Liu, C. et al. Acute Exposure to Tris(1,3-dichloro-2-propyl) Phosphate (TDCIPP) Causes Hepatic Inflammation and Leads to Hepatotoxicity in Zebrafish. Sci. Rep. 6, 19045; doi: 10.1038/srep19045 (2016).
References
van der Veen, I. & de Boer, J. Phosphorus flame retardants: Properties, production, environmental occurrence, toxicity and analysis. Chemosphere 88, 1119–1153 (2012).
Betts, K. S. New details on organophosphate flame retardants. Environ. Health Perspect. 121, A168 (2013).
Andresen, J. A., Grundmann, A. & Bester, K. Organophosphorus flame retardants and plasticisers in surface waters. Sci. Total Environ. 332, 155–166 (2004).
Bacaloni, A. et al. Liquid chromatography/tandem mass spectrometry determination of organophosphorus flame retardants and plasticizers in drinking and surface waters. Rapid Commun. Mass Spectrom. 21, 1123–1130 (2007).
Cao, S. et al. Levels and distributions of organophosphate flame retardants and plasticizers in sediment from Taihu Lake, China. Environ. Toxicol. Chem. 31, 1478–1484 (2012).
Wang, X. W., Liu, J. F. & Yin, Y. G. Development of an ultra-high-performance liquid chromatography-tandem mass spectrometry method for high throughput determination of organsphosphorus flame retardants in environmental water. J. Chromatogr. A 1218, 6705–6711 (2011).
Hartmann, P. C., Burgi, D. & Giger, W. Organophosphate flame retardants and plasticizers in indoor air. Chemosphere 57, 781–787 (2004).
Marklund, A., Andersson, B. & Haglund, P. Organophosphorus flame retardants and plasticizers in air from various indoor environment. J. Environ. Monit. 7, 814–819 (2005).
Stackelberg, P. E. et al. Persistence of pharmaceutical compounds and other organic wastewater contaminants in a conventional drinking-water-treatment plant. Sci. Total Environ. 329, 99–113 (2004).
Sundkvist, A. M., Olofsson, U. & Haglund, P. Organophosphorus flame retardants and plasticizers in marine and fresh water biota and in human milk. J. Environ. Monit. 12, 943–951 (2010).
Stapleton, H. M. et al. Detection of organophosphate flame retardants in furniture foam and US house dust. Environ. Sci. Technol. 43, 7490–7495 (2009).
Meeker, J. D., Cooper, E. M., Stapleton, H. M. & Hauser, R. Urinary metabolites of organophosphate flame retardants: Temporal variability and correlations with house dust concentrations. Environ. Health Perspect. 121, 585–585 (2013).
Carignan, C. C. et al. Predictors of tris(1,3-dichloro-2-propyl) phosphate metabolite in the urine of office workers. Environ. Int. 55, 56–61 (2013).
Dishaw, L. V. et al. Is the pentaBDE replacement, tris(1,3-dichloro-2-propyl) phosphate (TDCPP), a developmental neurotoxicant? Studies in PC12 cells. Toxicol. Appl. Pharm. 256, 281–289 (2011).
Liu, X., Ji, K. & Choi, K. Endocrine disruption potentials of organophosphate flame retardants and related mechanisms in H295R and MVLN cell lines and in zebrafish. Aquat. Toxicol. 114–115, 173–181 (2012).
Liu, X. et al. Effects of TDCPP or TPP on gene transcriptions and hormones of HPG axis and their consequences on reproduction in adult zebrafish (Danio rerio). Aquat. Toxicol. 134–135, 104–111 (2013).
Liu, C. et al. Effects of tris(1,3-dichloro-2-propyl) phosphate and triphenyl phosphate on receptor-associated mRNA expression in zebrafish embryos/larvae. Aquat. Toxicol. 128, 147–157 (2013).
Wang, Q. et al. Exposure of zebrafish embryos/larvae to TDCPP alters concentrations of thyroid hormones and transcriptions of genes involved in the hypothalamic–pituitary–thyroid axis. Aquat. Toxicol. 126, 207–213 (2013).
McGee, S. P., Cooper, E. M., Stapleton, H. M. & Volz, D. C. Early zebrafish embryogenesis is susceptible to developmental TDCPP exposure. Environ. Health Perspect. 120, 1585–1591 (2012).
Fu, J. et al. Toxicogenomic responses of zebrafish embryos/larvae to tris(1,3-dichloro-2-propyl) phosphate (TDCPP) reveal possible molecular mechanisms of developmental toxicity. Environ. Sci. Technol. 47, 10574–10582 (2013).
Crump, D., Chiu, S. & Kennedy, S. W. Effects of tris(1,3-dichloro-2-propyl) phosphate and tris(1-chloropropyl) phosphate on cytotoxicity and mRNA expression in primary cultures of avian hepatocytes and neuronal cells. Toxicol. Sci. 126, 140–148 (2012).
Farhat, A. et al. In ovo effects of two organophosphate flame retardants-TCPP and TDCPP-on pipping success, development, mRNA expression and thyroid hormone levels in chicken embryos. Toxicol. Sci. 134, 92–104 (2013).
Farhat, A. et al. Tris(1,3-dichloro-2-propyl) phosphate perturbs the expression of genes involved in immune response and lipid and steroid metabolism in chicken embryos. Toxicol. Appl. Pharm. 275, 104–112 (2014).
Su, G. et al. Liquid chromatography-electrospray-tandem mass spectrometry method for determination of organophosphate diesters in biotic samples including Greate Lakes herring gull plasma. J. Chromatogr. A 1374, 85–92 (2014).
Chen, D., Letcher, R. J. & Chu, S. G. Determination of non-halogenated, chlorinated and brominated organophosphate flame retardants in herring gull eggs based on liquid chromatography-tandem quadrupole mass spectrometry. J. Chromatogr. A 1220, 169–174 (2011).
Huang, da W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Xu, H., Lam, S. H., Shen, Y. & Gong, Z. Genome-wide identification of molecular pathways and biomarkers in response to arsenic exposure in zebrafish liver. PLOS ONE, 8, e68737 (2013).
Liu, C. et al. Effects of fluorotelomer alcohol 8:2 FTOH on steroidogenesis in H295R cells: targeting the cAMP signaling cascade. Toxicol. Appl. Pharm. 247, 222–228 (2010).
Hall, C. et al. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 7, 42 (2007).
Korzh, S. et al. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev. Biol. 8, 84 (2008).
Yan, C. et al. Stimulation of hepatocarcinogenesis by neutrophils upon induction of oncogenic kras expression in transgenic zebrafish. J. Hepatol. 63, 420–428 (2015).
Zhang, X., Li, C. & Gong, Z. Development of a convenient in vivo hepatotoxin assay using a transgenic zebrafish line with liver-specific dsRed expression. PLOS ONE, 9, e91874 (2014).
Zhang, X. & Gong, Z. Fluorescent transgenic zebrafish Tg(nkx2.2a:mEGFP) provides a highly sensitive monitoring tool for neurotoxins. PLOS ONE, 8, e55474 (2013).
Liu, C. et al. Endocrine disruption and reproductive impairment in zebrafish by exposure to 8:2 fluorote lomer alcohol. Aquat. Toxicol. 96, 70–76 (2010).
Lawless, M. W. & Greene, C. M. Toll-like receptor signaling in liver disease: ER stress the missing link? Cytoline 59, 195–202 (2012).
Seki, E. & Brenner, D. A. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology, 48, 322–335 (2008).
Peng, G. et al. Toll-like receptor 8-mediated reversal of CD4+ regulatory T cell function. Science 309, 1380–1384 (2005).
Scapigliati, G., Buonocore, F. & Mazzini, M. Biological activity of cytokines: an evolutionary perspective. Curr. Pharm. Design. 12, 1–11 (2006).
Randelli, E., Buonocore, F. & Scapigliati, G. Cell markers and determinants in fish immunology. Fish Shellfish Immun. 25, 326–340 (2008).
Shimasaki, S. et al. Endoplasmic reticulum stress increases the expression and function of toll-like receptor-2 in epithelial cells. Biochem. Biophys. Res. Commun. 402, 235–240 (2010).
Vecchi, C. et al. ER stress controls iron metabolism through induction of hepcidin. Science 5942, 877–880 (2009).
Smith, J. A. et al. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-β induction via X-box binding protein 1. Eur. J. Immunol. 38, 1194–1203 (2008).
Martinon, F. & Glimcher, L. H. Regulation of innate immunity by signaling pathways emerging from the endoplasmic reticulum. Curr. Opin. Immunol. 23, 35–40 (2011).
Guillot, L. et al. Response of human pulmonary epithelial cells to lipopolysaccharide involves toll-like receptor-4 (TLR4)-dependent signaling pathways: evidence for an intracellular compartmentalization of TLR4. J. Biol. Chem. 279, 2712–2718 (2004).
Zeng, L. et al. XBP-1 couples endoplasmic reticulum stress to augmented IFN-β induction via a cis-acting enhancer in macrophages. J. Immunol. 185, 2324–2330 (2010).
Mintz, M. et al. Time series proteome profiling to study endoplasmic reticulum stress response. J. Proteome Res. 7, 2435–2444 (2008).
Sharma, N. K. et al. Endoplasmic reticulum stress markers are associated with obesity in nondiabetic subjects. Endocr. Res. 93, 4532–4541 (2008).
Liu, B. & Li, Z. Endoplasmic reticulum HSP90b1 (gp96, grp94) optimizes B-cell function via chaperoning integrin and TLR but not immunoglobulin. Blood 112, 1223–1330 (2008).
Fu, Y. et al. Pten null prostate tumorigenesis and AKT activation are blocked by targeted knockout of ER chaperone GRP78/BiP in prostate epithelium. P. Natl. Acad. Sci. USA 105, 19444–19449 (2008).
Gupta, S. et al. HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1α-XBP1 signaling through a physical interaction. PLOS Biol. 8, e1000410 (2010).
Csala, M. et al. Transport and transporters in the endoplasmic reticulum. BBA-Biomembranes 1768, 1325–1341 (2007).
Copanaki, E. et al. The amyloid precursor protein potentiates CHOP induction and cell death in response to ER Ca2+ depletion. BBA-Mol. Cell Res. 1773, 157–165 (2007).
Kim, S. R. et al. Endoplasmic reticulum stress influences bronchial asthma pathogenesis by modulating nuclear factor κB activation. J. Allergy Clin. Immunol. 132, 1397–1408 (2013).
Malhi, H. & Kaufman, R. J. Endoplasmic reticulum stress in liver disease. J. Hepatol. 54, 795–809 (2011).
Brenner, C., Galluzzi, L., Kepp, O. & Kroemer, G. Decoding cell death signals in liver inflammation. J. Hepatol. 59, 583–594 (2013).
Soyalan, B. et al. Apple juice intervention modulates expression of ARE-dependent genes in rat colon and liver. Eur. J. Nutr. 50, 135–143 (2011).
van Dyk, J. C., Cochrane, M. J. & Wagenaar, G. M. Liver histopathology of the sharptooth catfish Clarias gariepinus as a biomarker of aquatic pollution. Chemosphere 87, 301–311 (2012).
Acknowledgements
This work was supported by the Singapore National Research Foundation under its Environmental & Water Technologies Strategic Research Programme and administered by the Environment & Water Industry Programme Office (EWI) of the PUB, grant number R-154-000-328-272. This work was also supported by grant from the National Natural Science Foundation of China (21207063), Postdoctoral Science Foundation of China (No. 2013M530054) and Huazhong Agricultural University Scientific & Technological Self-innovation Foundation (2014RC001).
Author information
Authors and Affiliations
Contributions
C.L., G.L., L.Y. and Z.G. conceived and designed the work. C.L., I.A., G.S., J.W. and J.L. performed the experiments. C.L., G.S., J.P.G., R.J.L., G.L., L.Y., J.W. and Z.G. draft the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Liu, C., Su, G., Giesy, J. et al. Acute Exposure to Tris(1,3-dichloro-2-propyl) Phosphate (TDCIPP) Causes Hepatic Inflammation and Leads to Hepatotoxicity in Zebrafish. Sci Rep 6, 19045 (2016). https://doi.org/10.1038/srep19045
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19045
This article is cited by
-
Environmentally relevant concentrations of tris (2-chloroethyl) phosphate (TCEP) induce hepatotoxicity in zebrafish (Danio rerio): a whole life-cycle assessment
Fish Physiology and Biochemistry (2023)
-
Biochemical responses of a freshwater fish Cirrhinus mrigala exposed to tris(2-chloroethyl) phosphate (TCEP)
Environmental Science and Pollution Research (2020)
-
Developing specific molecular biomarkers for thermal stress in salmonids
BMC Genomics (2018)
-
Co-exposure to benzo[a]pyrene and ethanol induces a pathological progression of liver steatosis in vitro and in vivo
Scientific Reports (2018)
-
Toxicity and Transcriptome Sequencing (RNA-seq) Analyses of Adult Zebrafish in Response to Exposure Carboxymethyl Cellulose Stabilized Iron Sulfide Nanoparticles
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.