Abstract
DNA methyltransferase inhibitors (DNMT inhibitors) are administered for high-risk MDS, but their action mechanisms are not fully understood. Hence, we performed a genome-wide DNA methylation assay and focused on cholesterol 25-hydroxylase (CH25H) among the genes whose expression was up-regulated and whose promoter region was hypomethylated after decitabine (DAC) treatment in vitro. CH25H catalyzes hydroxylation of cholesterol and produces 25-hydroxycholesterol (25-OHC). Although CH25H mRNA expression level was originally low in MDS/leukemia cell lines, exposure to DNMT inhibitors enhanced CH25H mRNA expression. The promoter region of CH25H was originally hypermethylated in HL-60 and MDS-L cells, but DAC treatment induced their hypomethylation together with increased CH25H mRNA expression, activation of CH25H-oxysterol pathway, 25-OHC production and apoptotic cell death. We further confirmed that normal CD34-positive cells revealed hypomethylated status of the promoter region of CH25H gene. CH25H-knockdown by transfection of shRNA lentiviral vector into the cell lines partially protected the cells from DAC-induced cell death. Exogenous addition of 25-OHC suppressed leukemic cell growth. The present study raises a possibility that DNMT inhibitors activate CH25H-oxysterol pathway by their hypomethylating mechanism and induce leukemic cell death. Further investigations of the promoter analysis of CH25H gene and therapeutic effects of DNMT inhibitors on MDS/leukemia will be warranted.
Similar content being viewed by others
Introduction
The myelodysplastic syndromes (MDS) are a group of acquired hematopoietic disorders characterized by cytopenia(s) and dysplasia as a result of clonal growth of pathological stem cells showing ineffective hematopoiesis and by the increased risk of progression to acute myeloid leukemia (AML)1,2. Hematopoietic stem cell transplantation (HSCT) is the only potentially curative treatment, but HSCT is restricted to a small population of MDS patients owing to the factors such as advanced ages, concomitant comorbidities and donor availability3. DNA methyltransferase(DNMT) inhibitors, 5-azacytidine (azacitidine; AZA) and 5-aza-2′-deoxycytidine (decitabine; DAC) have recently been used as chemotherapeutic agents for MDS patients who are not eligible for HSCT4,5,6,7,8,9. Notably, the treatment with AZA significantly increased overall survival in patients with high-risk MDS as compared with conventional treatments (clinical trial: AZA-001)10. However, the mechanism of action of DNMT inhibitors has not been clearly defined11,12,13,14,15,16.
We previously investigated the effects of DNMT inhibitors on the MDS cell lines established in our laboratory and demonstrated that DAC-induced cell death was preceded by a DNA damage response via a p53-independent pathway17. Furthermore, we researched some genes involved in the mechanism of action of DAC by a gene expression profiling.
In this study, we performed a genome-wide DNA methylation assay and consequently focused on cholesterol 25-hydroxylase (CH25H) among the genes whose expression was up-regulated and whose promoter region was hypomethylated after DAC treatment in vitro. CH25H is an important enzyme which catalyzes the hydroxylation of cholesterol and produces 25-hydroxycholesterol(25-OHC)18. Twenty-five-OHC, one of the endogenous oxysterols, has been reported to inhibit cell growth in various tumor cells19,20,21,22. We confirmed that the promoter region of CH25H in the two cell lines was originally hypermethylated and DAC treatment induced their hypomethylation which was associated with increased CH25H mRNA expression, activation of CH25H-oxysterol pathway, 25-OHC production and apoptotic cell death. We further confirmed that normal CD34-positive cells present hypomethylated status of the promoter region of CH25H. Therefore, we propose a hypothesis that CH25H is one of the candidate genes whose methylation status is related to myeloid neoplasms and one of the target genes of DNMT inhibitors.
Materials and Methods
Reagents
Five-aza-2′-deoxycytidine (decitabine; DAC, Sigma-Aldrich Co, St. Louis, MO, USA), 5-azacytidine (azacitidine; AZA, Wako Pure Chemical Industries, Ltd, Osaka, Japan) and cytosine arabinoside (cytarabine; ara-C, Nippon Shinyaku Co., Ltd, Kyoto, Japan) were dissolved in distilled water and stored at −20 °C. For the in vitro studies, we used each agent at the concentrations of 1 to 104 nM. They were added to the cultured cells daily without changing the culture medium.
Twenty-five-hydroxycholesterol (cholest-5-ene-3β,25-diol; 25-OHC) and a CH25H enzyme inhibitor, desmosterol (5,24-cholestadien-3β-ol)19 were purchased from Sigma Aldrich. Co (St. Louis, MO, USA). Twenty-four-hydroxycholesterol (cholest-5-ene-3ß,24(S)-diol; 24-OHC) and 27-hydroxycholesterol (cholest-5-ene-3β,27-diol; 27-OHC) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). They were dissolved in ethanol and stored at −20 °C.
Cell lines and culture
A myelodysplastic cell line, MDS92 was established from the bone marrow of a patient with MDS23. This cell line proliferated in the presence of interleukin-3 (IL-3) or granulocyte-macrophage colony-stimulating factor (GM-CSF) with a tendency to mature gradually24,25.
MDS-L and MDS92T cell lines were established independently each other from the long-term culture of parental MDS92. MDS-L cells showed blastic morphology and were positive for CD34, c-Kit, HLA-DR, CD13 and CD3326. MDS92T cell line consisted of immature myeloid cells with indented nucleus and was negative for CD34 exclusively.
MDS92, MDS-L and MDS92T cells were maintained in RPMI1640 medium supplemented with 10% fetal bovine serum, 50 μM 2-mercaptoethanol, 2.0 mM L-glutamine and 100 U/ml IL-3. A human myeloid leukemia cell line, HL-60, a blastic cell line from chronic myelogenous leukemia, K562 and a diffuse histiocytic lymphoma cell line, U937 were also used.
Primary cells
Five human normal bone marrow CD34-positive progenitor cells were purchased from LONZA Group Ltd, Basel, Switzerland and cultured with the serum-free medium with recombinant cytokines for myelopoiesis of hematopoietic progenitor cells, STEM ALPHA, AG (FUNAKOSHI, Tokyo, Japan). Pathological bone marrow samples were obtained from untreated patients with MDS or AML at the Department of Hematology, Kawasaki Medical School Hospital after obtaining informed consent from each patient. We performed all experiments in accordance with the Declaration of Helsinki and approved guidelines. The usage of patient samples was approved by the Ethical Committee of Kawasaki Medical School. The mononuclear cell fraction was isolated from the bone marrow samples by Ficoll-Hypaque density centrifugation as manufacturer’s protocols and CD34-positive fraction was purified by the magnetic beads method with anti-CD34 monoclonal antibody. The purity of this fraction was more than 95%.
Cell growth assay and MTT assay
Cell growth was assessed by counting the number of living cells after trypan blue staining. The morphological assessment was performed with May-Gruenwald Giemsa-stained cytospin slides. Cell suspensions were plated into 96-well plates in the presence of the drug or solvent alone, incubated as above at 37 °C for 3–7days and analyzed by the 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay as previously described17.
Apoptosis assay
Apoptosis was examined using Annexin V Apoptosis Detection Kit (BD Pharmingen, San Diego, CA, USA) and all samples were analyzed with FACS Calibur flow cytometer and CellQuest software (Becton Dickinson, Franklin Lakes, NJ, USA)27,28.
Genome-wide methylation sequencing analysis (MethylC-seq analysis)
The detailed information of this analysis is described in Supplementary Materials and methods (Ref. 29-32).
Quantitative real-time reverse transcription PCR
Quantitative real-time reverse transcription PCR (q-PCR) was performed with Applied Biosystems StepOne PlusTM Real time PCR system (Life Technologies Japan Ltd, Tokyo). Total cellular RNA (3–5 μg) was used to synthesize cDNA by using Ready-To-Go T primed First-Strand Kit (GE Healthcare UK Ltd, Amersham Place, Little Chalfont, Buckinghamshire HP7 9NA, UK) in a final volume of 33 μl. Five μl of 1:10 diluted cDNA reactions was used as input for each of the real-time quantitative PCR reactions by using SYBR Premix Ex TaqTM (Takara Bio Inc, Shiga, Japan). Initial denaturation at 95 °C for 20 s was followed by 40 cycles of a denaturation step at 95 °C for 3 s and an annealing and extension step at 60 °C for 30 s. Primers were designed by using Primer 3 v 4.0.0 software (Initial development of Primer3 was funded by Howard Hughes Medical Institute and by the National Institutes of Health, National Human Genome Research Institute). Since human CH25H gene spans one exon, we used total RNA treated with DNase I (Qiagen, Tokyo, Japan) so that the experimental results are not compromised through genomic DNA contamination33.
CH25H (NM_003956) forward primer was 5′-TGGCAACGCAGTATATGAGC-3′ and its reverse primer was 5′-AGGGAAGTTGTAGCCGGAGT-3′. Human housekeeping gene RPL27 (NM_000988) was used as endogenous control30. Primers for RPL27 were as follows: forward: 5′-CTCTGGTGGCTGGAATTGAC-3′ and reverse: 5′-AAACCGCAGTTTCTGGAAGA-3′.
The copy ratio of CH25H to RPL27 was calculated and indicated as a quantification of CH25H expression in several experiments.
Gene expression profiling and the gene set enrichment analysis (GSEA)
The detailed information of these experiments is described in Supplementary Materials and methods (Ref.35-36).
Methylation analysis of the promoter region with bisulfite sequencing
Genomic DNA was extracted using SepaGene DNA extraction Kit (Sanko Junyaku, Tokyo, Japan) and chemically modified with sodium bisulfite using MethylEasyTM Xceed Rapid DNA Bisulfite Modification Kit (Human Genetic Signatures Pty Ltd, North Ryde, Australia), which converts all unmethylated cytosines to uracils, whereas methylated cytosines remain unchanged. Analysis of the obtained sequences confirmed a complete bisulfite reaction in all samples33. Then, the 1700 bp upstream sequence of the transcription initiation site which might include the promoter region was divided into six regions and they were defined as F1 to F6, respectively.
Bisulfite-treated DNA was amplified in PCR with cholesterol 25-hydroxylase (CH25H)-specific primers F1-6. Primers for each region of CH25H and the PCR product sizes are shown in supplementary Table S1.
PCR was performed in 50 μl of reaction mixture with 0.4 μM of each primer, 5 ng of genomic DNA, 1 × EpiTaqPCR buffer (Mg2 + free), 0.3 mM of each deoxynucleotide triphosphate, 2.5 mM MgCl2 and 1.25 U of EpiTaq HS (Takara, Tokyo, Japan). Initial denaturation at 98 °C for 3 min was followed by 40 cycles of a denaturation step at 98 °C for 10 s, an annealing step at 55 °C for 30 s, an extension step at 72 °C for 1 min and a final extension step of 72 °C for 7 min was added. The products were separated by electrophoresis on 1.5% agarose gel. PCR products were gel-purified using WizardRSV Gel and PCR Clean-up System (Promega Co, Madison, USA) and ligated into the pCRTMII-TOPOR vector (Invitrogen by Life Technologies, Tokyo, Japan) according to the manufacturer’s instructions. Independent plasmid clones were purified from several bacterial colonies. Five to ten clones per sample were sequenced using the Big Dye terminator ver 3.1 Cycle Sequencing Kit (Applied Biosystems) and ABI Prism 3130 Genetic Analyzer (Applied Biosystems, Tokyo, Japan).
Plasmids and transfection
To inhibit CH25H gene expression, a vector for short-hairpin RNA (shRNA) against CH25H incorporated in pLKO.1-puro (Sigma-Aldrich, Mission shRNA code: TRCN115963) was transfected into HL-60 and MDS-L cells by ViraPower lentiviral Directional TOPO Expression kit (Invitrogen)37 and positive clones were selected with 2 μg/ml puromycin. Likewise, we introduced the control vector (Sigma-Aldrich, Mission TurboGFPTM shRNA Control Vector) into HL-60 and MDS-L cells.
Ultraperformance liquid chromatography coupled with Q-TOF mass spectrometry (UPLC/Q-TOF MS) and gas chromatography-mass spectrometry (GC-MS)
The detailed information of this analysis is described in Supplementary Materials and methods (Ref.38-39).
Statistical analyses
All results are shown as the mean values with ranges. Comparisons between the groups were done using the Dunnett’s and Scheffe tests. Differences were considered statistically significant if p-values were less than 0.05. These analyses were carried out using SPSS for Windows version 14.0.
Results
Decitabine (DAC) treatment leads to up-regulation of cholesterol 25-hydroxylase (CH25H) gene expression in MDS-L cell line
We previously explored the molecular pathways affected by DAC using the gene expression profiling of MDS-L cells treated with 4 nM DAC for 7 days17. Genes whose expression changed by more than 2-fold after DAC treatment were defined as regulated genes. 956 genes were up-regulated and 461 genes were down-regulated in MDS-L cells treated with DAC (4 nM) as compared with the control. In the present study, we further performed the genome-wide DNA methylation assay (MethylC-seq)29,30,31,32 and identified 13 genes whose mRNA expression was up-regulated more than 20-fold and whose CpG islands at the promotor region (around 1000 bp upstream sequence of the transcription initiation site) were hypomethylated more than 10% after DAC treatment as compared with the control (Table 1).
Next, we surveyed the genes involved in cell growth suppression, differentiation and apoptosis among these 13 genes and focused on cholesterol 25-hydroxylase (CH25H) gene. In DNA microarray analysis, the expression of CH25H in DAC-treated MDS-L cells increased more than 180-fold as compared with that of control cells and the 1000 bp upstream sequence of the transcription initiation site in CH25H was dominantly demethylated 45% by DAC treatment (Table 1).
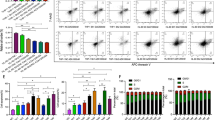
We further applied the data of DNA microarray analysis to the gene set enrichment analysis (GSEA) and found that two gene sets were strongly up-regulated by DAC treatment: ‘metabolism of lipids and lipoproteins’ and ‘bile acid and bile salt metabolism’. Both gene sets contained CH25H as the most activated gene by DAC treatment and its downstream genes, CYP7B1 and HSD3B7 (Fig. 1 and Supplementary Figure S1). CH25H catalyzes the hydroxylation of cholesterol and alters it to 25-hydroxycholeterol (25-OHC). Twenty-five-OHC is one of the natural oxysterols and is reported to inhibit the growth of several tumor cells19,20,21,22. The present data indicated that CH25H-oxysterol pathway is activated by DAC treatment in MDS-L cells.
Activation of CH25H-related pathways in the gene set enrichment analysis (GSEA).
DAC-treated or untreated MDS-L cells were harvested on day 7 in triplicate assays and the gene expression profiling data were used for GSEA by handling the GSEA software and the Molecular Signatures Database according to the references (35,36). Two gene sets strongly up-regulated by DAC treatment are presented: (a) metabolism of lipids and lipoproteins and (b) bile acid and bile salt metabolism. Both gene sets contain CH25H as the most activated gene by DAC treatment (the heat map data are shown in Supplementary Figure S1). FDR q-value: false discovery rate; FWER p-value: family-wise error rate. The accumulation of the gene set is considered as statistically significant when both values are less than 0.25.
The treatment with DNMT inhibitors increases the expression of CH25H gene in MDS/leukemia cell lines
We examined CH25H mRNA expression level in five MDS/leukemia cell lines (HL-60, MDS-L, MDS92T, U937 and K562) by quantitative real-time PCR (q-PCR). The cells were treated with DAC or AZA at different concentrations as indicated for 4 or 7 days. Whereas CH25H mRNA expression level was originally low in all five cell lines, exposure of each cell line to DNMT inhibitors significantly increased the expression of CH25H (Fig. 2a–f).
Induction of CH25H expression in MDS/leukemia cell lines treated with DNMT inhibitors.
Cells were treated with DAC, AZA and cytarabine at various concentrations for 4 days or 7 days. Relative mRNA expression was analyzed by quantitative real-time PCR (described in Materials and Methods). Induction of CH25H mRNA expression in MDS-L treated with different concentrations of DAC or AZA for 4 days (a) and for 7 days (b), in HL-60 treated with different concentrations of DAC or AZA for 4 days (c), in MDS92T treated with different concentrations of DAC for 7 days (d) in U937 treated with different concentrations of DAC for 4 days (e) and in K562 treated with different concentrations of DAC for 4 days (f,g) Induction of CH25H mRNA expression in MDS-L treated with cytarabine for 4 days is shown. The data shown are the average ± SD of three independent experiments. *p < 0.05; **p < 0.01.
CH25H mRNA expression in MDS-L cells treated with DAC for 4 or 7 days was increased in a dose-dependent and time-dependent manner (Fig. 2a,b). Likewise, the treatment with AZA for 4 or 7 days in MDS-L cells induced CH25H mRNA expression at high concentrations as compared with DAC (Fig. 2a,b). CH25H mRNA expression was also increased in HL-60 and the other cell lines treated with DAC or AZA (Fig. 2c–f). To confirm that up-regulation of CH25H mRNA in DAC-treated cell lines actually causes the production of 25-OHC, we tried to quantify 25-OHC in cell lysates or culture supernatants of DAC-treated cell lines by UPLC/Q-TOF MS and GC-MS. The analysis using the samples from less than several millions cells did not show any detectable amount of oxysterols, but we finally found a detectable amount of 25-OHC (46 ng/gram cell pellet) in a larger-scale sample of DAC-treated cells (109 cells) in our assay system (detection sensitivity was 0.1 ng/g). In contrast, neither of the other oxysterols (24-OHC and 27-OHC) was detected in the same sample.
Next, we evaluated whether the increase of CH25H mRNA expression is specific to DNMT inhibitors or whether cytotoxic drugs such as cytarabine also induce it and found that CH25H mRNA expression in MDS-L cells was increased after cytarabine treatment (Fig. 2g).
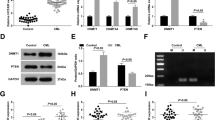
The promoter region of CH25H is originally hypermethylated in both MDS-L and HL-60 cells
To further investigate the methylation status at the promoter region of CH25H in MDS-L and HL-60 cells, we performed the methylation analysis by bisulfite sequencing assay focusing on 1700 bp of the CH25H promoter region, by dividing the area into six regions, which spans from 1,647 bp-upstream of transcription initiation site to 32 codons of coding region in exon 1 (F1 to F6 regions in Fig. 3a and in Materials and methods). The DNA sequence in each region was amplified by PCR reaction using the primer sequences in the promoter region of CH25H (Supplementary Table S1). Each PCR product was ligated into the pCRTMII-TOPOR vector and more than five clones per sample were sequenced. All primers do not contain any CpG sites. F6 region contains 32 codons of coding region in exon 1.
The methylation analysis in the promoter region of CH25H in the cell lines and normal and MDS/leukemia CD34-positive progenitor cells.
(a) The schematic presentation of CH25H promoter sequence divided into 6 regions. Methylation status of independent plasmid clones derived from MDS-L (b), HL-60 (c) treated without or with DAC, CD34-positive cells of MDS92 (d) and a representative case (n1) of five normal CD34-positive progenitor cells (e). (● methylated cytosine, ○ unmethylated cytosine, △ not defined). (f) The expression of CH25H mRNA in normal CD34-positive progenitor cells (n = 5), six cell lines and CD34-positive or blastic cells from primary MDS/leukemia cells was compared. (g) Induction of CH25H mRNA expression is indicated in primary MDS/leukemia cells treated with 1 μM DAC for 4 days. In (f,g), The primary cases were categorized by FAB or WHO classification as follows: MDS (Refractory anemia: UPN1), MDS (Refractory cytopenia with multilineage dysplasia: UPN2), ALL(L2: UPN3), AML (M2: UPN4), AML (M7: UPN5), AML (M2: UPN6), AML (AML with myelodysplasia-related changes: UPN7), AML (M5b: UPN8). Purified CD34-positive cells in cases except UPN8 were used. Leukemia cells occupied more than 90% of bone marrow cells in case with UPN8. The data shown are the average ± SD of three independent experiments (*p < 0.05; **p < 0.01).
In untreated MDS-L and HL-60 cells, most CpG sites at F1, F2, F5 and F6 were methylated, whereas those of F3 and F4 remained unmethylated (Fig. 3b,c). In DAC-treated MDS-L and HL-60 cells, a considerable part of CpG sites at F2, F5 and F6 were demethylated (Fig. 3b,c). MDS92 cell line grows slowly with a tendency of maturation and appears to be less malignant than its descendant blastic subline, MDS-L and MDS92T.
Hence, we separated CD34-positive fraction from MDS92 for methylation analysis. In CD34-positive fraction of MDS92 cells, CpG sites at F5 of CH25H was hypermethylated, but not at F6 (Fig. 3d). CH25H mRNA expression in CD34-positive fraction of MDS92 cells was increased as compared with that of MDS-L and MDS92T (Fig. 3f). We further evaluated the methylation status of CH25H in CD34-positive progenitor cells derived from bone marrow of five healthy donors as a control. Strikingly, CpG sites at F5 and F6 in normal CD34-positive cells were hardly methylated (Fig. 3e). The mRNA expression in normal CD34-positive progenitor cells was generally increased as compared with the vigorous cell lines MDS-L, MDS92T, HL-60, U937 and K562 (p < 0.05) (Fig. 3f).
Next, we investigated the methylation status of CH25H F6 region in bone marrow CD34-positive cells derived from 3 MDS patients (UPN1, 2, 9) and one AML patient (UPN10) and found one case (UPN9) whose CH25H F6 region was hypermethylated out of 4 patients studied (Supplementary Figure S2). We could examine the expression of CH25H in two out of the above 4 cases and found that the expression of CH25H was increased in these cases (low-risk MDS: UPN1 and UPN2) (Fig. 3f). As both UPN1 and UPN2 revealed hypomethylation of CH25H F6 region (Supplementary Figure S2), it was consistent with increased CH25H mRNA expression. Regarding primary MDS/leukemia cells, CD34-positive or blastic cells indicated various levels of CH25H mRNA expression including two above described MDS cases showing marked CH25H expression (Fig. 3f). DAC treatment led to enhanced CH25H mRNA expression in most of primary leukemia cells studied (Fig. 3g).
CH25H-knockdown by transfection of shRNA lentiviral vector in HL-60 and MDS-L cells partially protects the cells from DAC-induced cell death
To further examine the role of CH25H, we established CH25H-knockdown HL-60 clones (HL-60-CH25HshRNA) and CH25H-knockdown MDS-L clones (MDS-L-CH25HshRNA) by transfecting CH25H-shRNA lentiviral vector into each cell line and simultaneously established the control clones transfected with the control vector (HL-60-CV and MDS-L-CV). CH25H-knockdown clones revealed a drastic decrease in CH25H mRNA expression (the copy ratio of CH25H to RPL27 after DAC treatment was 410 ± 40 in HL-60-CV and 0.5 ± 0.1 in HL-60-CH25HshRNA: p < 0.01) (Fig. 4a.).
The role of CH25H in DAC-induced cell death.
To investigate the role of CH25H in detail, we established CH25H-knockdown HL-60 cell clones (HL-60-CH25HshRNA) and MDS-L cell clones (MDS-L-CH25HshRNA) by transfecting shRNA-CH25H into each cell line using a lentivirus vector. (a) Displayed are histograms of CH25H expression level in HL-60-CV (CV) and HL60-CH25HshRNA (shRNA) treated with or without DAC for 4 days, respectively. (b) Shown is the survival (%) from DAC-induced HL-60 cell death by CH25H-knockdown. HL-60-CV cells (CV) and HL-60-CH25HshRNA cells (shRNA) were treated daily with DAC (0–500 nM) for 3 to 4 days and the change in the cell number was evaluated by MTT assay. (c) HL-60-CV cells (CV) and HL-60-CH25HshRNA cells (shRNA) were treated with different concentrations (0 and 500 nM) of DAC for 96 h and apoptosis was assessed by flow cytometry using annexin V and propidium iodide (PI) staining. Positive staining for annexin V or for annexin V and PI show early or late apoptosis, respectively. The value of right area indicates the percentage of the cells in early and late apoptosis. (d) Displayed are the histograms of CH25H expression level in MDS-L-CV (CV) and MDS-L-CH25HshRNA (shRNA) treated with or without DAC for 4 days, respectively. (e) Shown is the survival (%) from DAC-induced MDS-L cell death by CH25H-knockdown. MDS-L-CV cells (CV) and MDS-L-CH25HshRNA cells (shRNA) were treated daily with DAC (0–100 nM) for 4 days and the change in the cell number was evaluated by MTT assay. The data shown are the average ± SD of three (a,d) or five (b,e) independent experiments. *p < 0.05; **p < 0.01.
Both HL-60-CV and HL-60-CH25HshRNA cell clones were treated daily with DAC for 3 or 4 days and the effect of CH25H-knockdown on DAC-induced growth suppression was examined by MTT assay. As shown in Fig. 4b, DAC-induced cell death was partially but significantly rescued by CH25H-knockdown. The percentage of growth inhibition by DAC treatment (100 nM and 500 nM) was 48.9 ± 1.3% and 24.9 ± 2.3% in HL-60-CV clones; 57.9 ± 2.1% and 35.6 ± 0.8% in HL-60-CH25HshRNA clones, respectively on day 3 (p < 0.01); 15.0 ± 0.6% and 4.7 ± 0.6% in HL-60-CV clones; 30.3 ± 1.2% and 13.0 ± 0.7% in HL-60-CH25HshRNA clones, respectively on day 4 (p < 0.01).
Apoptosis was detected using flow cytometry by positive staining for annexin V (early apoptosis) or for annexin V and propidium iodide (late apoptosis). The percentage of annexin V(+) and PI (−) and annexin V(+) and PI (+) in DAC (500 nM)-treated HL-60-CH25HshRNA clones was decreased as compared with that of HL-60-CV (Fig. 4c). DAC-induced cell death in MDS-L was also partially but significantly rescued by CH25H-knockdown (Fig. 4d,e). Taken together, CH25H-knockdown by shRNA lentiviral transfection partially protected these cell lines from DAC-induced cell death.
Exogenous addition of 25-OHC suppresses the growth of MDS/leukemia cell lines in vitro
As above described, we could not detect the obvious production of 25-OHC in the cell lysates nor in the culture supernatants of DAC-treated cells. Therefore, to know whether 25-OHC actually suppresses cell growth in vitro, we added several concentrations of 25-OHC to the MDS/leukemia cell lines in vitro. We also used another oxysterol, 27-OHC as a comparative experiment. MDS-L cells were found to be most sensitive to 25-OHC, with reduced cell viability observed at around 500 nM (Fig. 5a). Half-maximal inhibitory concentration (IC50) values of 25-OHC and 27-OHC on the cell lines are shown in Table 2. Except for U937, these cell lines were susceptible to 25-OHC rather than 27-OHC (Table 2). In addition, to examine whether growth suppression by 25-OHC was due to the apoptosis, we performed annexin V and PI staining with flow cytometer. The percentage of annexin V(+) and PI (−) and annexin V(+) and PI (+) in 25-OHC (1 and 5 μM) -treated MDS-L cells was increased as compared with that of untreated MDS-L cells (Fig. 5b).
Growth suppressive effect of 25-hydroxycholesterol (25-OHC) on MDS/leukemia cell lines.
(a) The indicated cell lines were cultured for 3 days in the presence of various concentrations of 25-OHC or 27-OHC and growth suppression was evaluated by MTT assay. (b) MDS-L cells were treated with 0, 1 or 5 μM of 25-OHC for 72 h and apoptosis was assessed by flow cytometry using annexin V and PI staining. Positive staining for annexin V or for annexin V and PI show early or late apoptosis, respectively. The value of lower right area and upper right area indicate the percentage of cells in early apoptosis and late apoptosis, respectively. (c) MDS-L cells were treated with 5 nM DAC in the presence or absence of 10 μM desmosterol for 4 days. Shown is the survival (%) from DAC-induced MDS-L cell death by desmosterol. The change in the cell number was evaluated by MTT assay. The data shown are the average ± SD of five independent experiments and there was significant difference between DAC-treated and DAC plus desmosterol-treated cells. *p < 0.05.
To examine whether the inhibition of CH25H enzymatic activity modifies its growth suppressive effect, we used a CH25H inhibitor, desmosterol. MDS-L cells were cultivated with 5 nM DAC and/or 10 μM desmosterol for 4 days and cell growth was measured by MTT assay. In the presence of desmosterol, DAC-induced cell growth suppression was rescued to a limited extent but significantly (n = 5, p < 0.05) (Fig. 5c).
Discussion
In the present study, by gene expression profiling and genome-wide DNA methylation analysis, we identified 13 genes whose mRNA expression was up-regulated more than 20-fold and whose CpG islands at around 1000 bp upstream sequence of the transcription initiation site was hypomethylated more than 10% after DAC treatment as compared with control. Out of these genes we focused on CH25H as the gene which seemed to be associated with cell proliferation, apoptosis and cell death (Table 1).
CH25H is an enzyme to convert cholesterol into 25-OHC, one of the oxysterols and the CH25H-oxysterol pathway is an entrance to the bile acid synthesis. The sterol-regulatory element binding proteins (SREBPs) are transcription factors involved in the synthesis of cholesterol40,41. Twenty-five-OHC inhibits SREBPs and regulates 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR), the rate-limiting enzyme of de novo cholesterol synthesis42,43. In addition, 25-OHC regulates cholesterol homeostasis through a ligand of liver X receptor44,45. It was reported that 25-OHC has a growth inhibitory effects on various cells19,20,21,22. Banker, DE et al. reported that the cholesterol level is abnormally increased in AML cells exposed to chemotherapeutic drugs in vitro and that blocking these acute cholesterol responses facilitates cell death46,47. Zhang F, et al. reported that DAC inhibits leukemic cell growth through modulating endogenous cholesterol biosynthesis48. As shown in Fig. 1 and Supplementary Figure S1, GSEA indicated that lipoprotein and bile acid metabolism pathways are up-regulated by DAC treatment and that these affected gene sets contained CH25H and its downstream genes, CYP7B1 and HSD3B7 as clearly activated genes (Supplementary Figure S1). These data demonstrate the activation of the CH25H-oxysterol pathway by DAC treatment in MDS-L cells.
We hypothesized that up-regulation of CH25H after DAC treatment promotes the accumulation of endogenous 25-OHC and it contributes in part to DAC-induced cell death. DAC promoted the expression of CH25H in various MDS/leukemia cell lines, particularly in MDS-L (Fig. 2). AZA also induced CH25H mRNA expression at high concentrations in HL-60 and MDS-L cells (Fig. 2a–c). To confirm intracellular increase in the enzymatic activity of CH25H, we attempted to detect 25-OHC within cell lysates or culture supernatants of DAC-treated and untreated MDS-L cells with LC/MS and TOF/MS and in addition with GC-MS. Although the production level of 25-OHC seemed to be quite low, we could detect a small but significant amount of 25-OHC in a larger-scale cell sample after DAC treatment.
Unexpectedly, CH25H mRNA expression was induced not only by DNMT inhibitors but also by a similar antimetabolite compound, cytarabine (Fig. 2g). Because it is unlikely that cytarabine acts as a hypomethylating agent, cytarabine might up-regulate CH25H mRNA expression by another mechanism. Hence, induction of CH25H expression itself might not be a specific phenomenon induced by DNMT inhibitors but rather a general intracellular event induced by cytarabine. Anti-cancer agents exert various cytotoxic actions. From our data we speculate that activation of CH25H-mediated pathway is one of their important action mechanisms and that DNMT inhibitors trigger such a universal death signaling as a result of DNA hypomethylation. Further knockdown or overexpression experiments of CH25H gene would serve for elucidating the implication of this gene for the therapeutic targets of cancer chemotherapy.
We paid attention to the promoter region of CH25H gene and performed the methylation analysis on 1700 bp upstream sequence (divided into F1 to F6 regions) of CH25H transcription initiation site. In untreated MDS-L and HL-60 cells, most CpG sites at F1, F2, F5 and F6 were methylated, whereas those of F3 and F4 remained unmethylated (Fig. 3b,c). On the contrary, in DAC-treated cells, a considerable part of CpG sites at F2, F5 and F6 were demethylated (Fig. 3b,c). In particular, release of F6 region from hypermethylated status seems to be important for the recovery of CH25H expression. We next evaluated the methylation status of CH25H in CD34-positive progenitor cells derived from bone marrow of five healthy donors. Strikingly, CpG sites at F5 and F6 in normal bone marrow CD34-positive cells were hardly methylated. These data strongly suggest that: first, the CpG islands at the promoter region of CH25H are hypermethylated and the expression of CH25H is suppressed in MDS/leukemia cell lines; second, the promoter region of CH25H is hardly methylated and CH25H message is expressed to some extent in normal bone marrow CD34-positive cells; and third, DNMT inhibitors promote CH25H expression by altering the hypermethylated status of this gene and induce cell death.
Considering our results that the promoter region of CH25H was hypermethylated in the blastic subline, MDS-L as compared with that of parental MDS92, we speculate that hypermethylation of the promoter region of CH25H is relevant to more aggressive cell growth. To verify this hypothesis on primary MDS/leukemia cells, we investigated the methylation status of CH25H F6 region in 4 MDS/AML patients and found one case (UPN9) whose CH25H F6 region was hypermethylated (Supplementary Figure S2), although CH25H mRNA expression was not examined in this case. About two low-risk MDS cases (UPN1 and UPN2), we verified increased expression of CH25H together with hypomethylation of CH25H F6 region (Fig. 3f and Supplementary Figure S2). Nonetheless, the present data are indeed insufficient to certify the relation of the methylation status of CH25H gene and its expression in primary MDS/leukemia cells. Further information should be accumulated about the data of CH25H gene in MDS/leukemia cases and the effects of DNMT inhibitors on the methylation status of CH25H as well as clinical outcome.
Although they were cell line-limited studies, we attempted to confirm the close relation of CH25H expression to the suppressive effect on leukemic cell growth by several ways. First, CH25H-knockdown by transfection of shRNA lentiviral vector in HL-60 and MDS-L cells partially protected the cells from DAC-induced cell death (Fig. 4a–e). Second, exogenous addition of 25-OHC suppressed the growth of MDS/leukemia cell lines in vitro (Fig. 5a,b). Third, a CH25H inhibitor, desmosterol rescued the cells from DAC-induced cell growth suppression to a limited extent but significantly (Fig. 5c). These experimental data are of help as evidence that epigenetic silencing of CH25H leads to leukemic cell growth and that hypomethylating agents exert their anti-leukemic action partly via activation of the CH25H-oxysterol pathway. In addition to this hypomethylating effects, there may exist other underlying action mechanisms of DNMT inhibitors, judging from our observation that DAC also exerted some cytotoxic effect on normal CD34-positive cells although their CH25H promoter region was hardly methylated.
The present study raises a possibility that DNMT inhibitors activate the CH25H-oxysterol pathway by their hypomethylating mechanism and induce leukemic cell death. Further investigations of the promoter analysis of CH25H gene and the therapeutic effects of DNMT inhibitors on MDS and AML will be warranted.
Support and Financial Disclosure Declaration
Tohyama K received research funding from Eisai Co., Ltd. The other authors have no competing financial interests to declare
Additional Information
How to cite this article: Tsujioka, T. et al. Five-aza-2'-deoxycytidine-induced hypomethylation of cholesterol 25-hydroxylase gene is responsible for cell death of myelodysplasia/leukemia cells. Sci. Rep. 5, 16709; doi: 10.1038/srep16709 (2015).
References
Bennett, J. M. et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol. 51, 189–199 (1982).
Brunning, R. D. et al. in WHO classification of tumours of Haematopoietic and Lymphoid Tissues, 4th edn (eds Swerdlow, S. H. et al. ) Ch. 5, 88–93 (IARC, 2008).
Cazzola, M. & Malcovati, L. Myelodysplastic syndromes-coping with ineffective hematopoiesis. N. Engl. J. Med. 352, 536–538 (2005).
Issa, J. P. & Kantarjian H. M. Targeting DNA methylation. Clin Cancer Res. 15, 3938–3946 (2009).
Quintás-Cardama, A., Santos, F. P. & Garcia-Manero, G. Therapy with azanucleosides for myelodysplastic syndromes. Nat Rev Clin Oncol. 7, 433–444 (2010).
Silverman, L. R. et al. Randomized controlled trial of azacitidine in patients with the myeloblastic syndrome: a study of the cancer and Leukemia Group B. J Clin Oncol. 20, 2429–2440 (2002).
Kornblith, A. B. et al. Impact of azacytidine on the quality of life of patients with myelodysplastic syndrome treated in a randomized phase III trial: a Cancer and Leukemia Group B study. J Clin Oncol. 20, 2441–2452 (2002).
Kantarjian, H. et al. Decitabine improve patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 106, 1794–1803(2006).
WijerMans, P. et al. Low dose decitabine versus best supportive care in elderly patients with intermediate or high risk MDS not eligible for intensive chemotherapy: final results of the randomized phase III study (06011) of the EORTC Leukemia and German MDS study groups. Blood. 112, 226 (2008).
Fenaux, P. et al. International Vidaza High-Risk MDS Survival Study Group Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 10, 223–232 (2009).
Flotho, C. et al. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 23, 1019–1028 (2009).
Hollenbach, P. W. et al. A comparison of azacitidine and decitabine activities in acute myeloid leukemia cell lines. PLoS One. 5, e9001 (2010).
Qin, T. et al. Effect of cytarabine and decitabine in combination in human leukemic cell lines. Clin Cancer Res. 13, 4225–4232 (2007).
Qin, T., Jelinek, J., Si, J., Shu, J. & Issa, J. P. Mechanisms of resistance to 5-aza-2′-deoxycytidine in human cancer cell lines. Blood. 113, 659–667 (2009).
Qin, T. et al. Mechanisms of resistance to decitabine in the myelodysplastic syndrome. PLoS One. 6, e23372 (2011).
Kimura, S. et al. Antiproliferative and antitumor effects of azacitidine against the human myelodysplastic syndrome cell line SKM-1. Anticancer Res. 32, 795–798 (2012).
Tsujioka, T. et al. Effects of DNA methyltransferase inhibitors (DNMTIs) on MDS-derived cell lines.Exp Hematol. 41, 189–197 (2013).
Lund, E. G., Kerr, T. A., Sakai, J., Li, W. P. & Russell, D. W. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J. Biol. Chem. 273, 34316–34327 (1998).
Wang, J. H. & Tuohimaa, P. Regulation of cholesterol 25-hydroxylase expression by vitamin D3 metabolites in human prostate stromal cells. Biochem Biophys Res Commun. 345, 720–725 (2006).
Larsson, O. & Zetterberg, A. Existence of a commitment program for mitosis in early G1 in tumour cells. Cell Prolif. 28, 33–43 (1995).
Zhou, R. H. et al. Vascular endothelial growth factor activation of sterol regulatory element binding protein: a potential role in angiogenesis. Circ. Res. 95, 471–478 (2004).
Ayala-Torres, S., Zhou, F. & Thompson, E. B. Apoptosis induced by oxysterol in CEM cells is associated with negative regulation of cmyc. Exp. Cell Res. 246, 193–202 (1999).
Tohyama, K., Tsutani, H., Ueda, T., Nakamura, T. & Yoshida, Y. Establishment and characterization of a novel myeloid cell line from the bone marrow of a patient with the myelodysplastic syndrome. Br J Haematol 87, 235–242 (1994).
Tohyama, K. et al. A novel factor-dependent human myelodysplastic cell line, MDS92, contains haemopoietic cells of several lineages. Br J Haematol. 91, 795–799 (1995).
Drexler, H. G., Dirks, W. G. & Macleod, R. A. Many are called MDS cell lines: one is chosen. Leuk Res. 33, 1011–1016 (2009).
Matsuoka, A. et al. Lenalidomide induces cell death in an MDS-derived cell line with deletion of chromosome 5q by inhibition of cytokinesis. Leukemia. 24, 748–755 (2010).
Ormerod, M. G., Collins, M. K. L., Rodriguez-Tarduchy, G. & Robertson, D. Apoptosis in Interleukin-3 dependent haemopoietic cells: quantification by two flow cytometric methods. J Immunol Meth. 153, 57–65 (1992).
Vermes, I., Haanen, C., Steffens-Nakken, H. & Reutelingsperger, C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Meth. 184, 39–51 (1995).
Lister, R. et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 133, 523–536 (2008).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 462, 315–322 (2009).
Schroeder, D. I., Lott, P., Korf, I. & LaSalle, J. M. Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Res. 21, 1583–91 (2011).
Schillebeeckx, M. et al. Laser capture microdissection-reduced representation bisulfite sequencing (LCM-RRBS) maps changes in DNA methylation associated with gonadectomy-induced adrenocortical neoplasia in the mouse. Nucleic Acids Res. 41, e116 (2013).
Oka, T. et al. Gene silencing of the tyrosine phosphatase SHP1 gene by aberrant methylation in leukemias/lymphomas. Cancer Res. 62, 6390–6394 (2002).
de Jonge, H. J. et al. Evidence based selection of housekeeping genes. PLoS One. 2, e898 (2007)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 102, 15545–50 (2005).
Gene Set Enrichment Analysis (GSEA) Home page in Broad Institute. Available from URL: http://www.broadinstitute.org/gsea/index.jsp. (v5.0 updated April 2015)(Data of access: 22/09/2015).
Tanaka, C. et al. Rab27b regulates c-kit expression by controlling the secretion of stem cell factor. Biochem Biophys Res Commun. 9, 368–373 (2012).
Zhou, M. et al. Identification and comparison of anti-inflammatory ingredients from different organs of Lotus nelumbo by UPLC/Q-TOF and PCA coupled with a NF-κB reporter gene assay. PLoS One. 29, e8197 (2013).
Park, C., Yun, S., Lee, S. Y., Park, K. & Lee, J. Metabolic profiling of Klebsiella oxytoca: evaluation of methods for extraction of intracellular metabolites using UPLC/Q-TOF-MS. Appl Biochem Biotechnol. 167, 425–438 (2012).
Adams, C. M. et al. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 279, 52772–52780 (2004).
Brown, M. S. & Goldstein, J. L. A proteolytic pathway that controls the cholesterol content of membranes, cells and blood. Proc. Natl. Acad. Sci. USA 96, 11041–11048 (1999).
Goldstein, J. L. & Brown, M. S. Regulation of the mevalonate pathway. Nature. 343, 425–30 (1990).
Rodwell, V. W., Nordstrom, J. L. & Mitschelen, J. J. Regulation of HMG-CoA reductase. Adv. Lipid Res. 14, 1–74 (1976).
Espenshade, P. J. & Hughes, A. L. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41, 401–427 (2007).
Song, B. L., Javitt, N. B. & DeBose-Boyd, R. A. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1, 179–189 (2005).
Li, H. Y., Appelbaum, F. R., Willman, C. L., Zager, R. A. & Banker, D. E. Cholesterol-modulating agents kill acute myeloid leukemia cells and sensitize them to therapeutics by blocking adaptive cholesterol responses. Blood. 101, 3628–3634 (2003).
Banker, D. E. et al. Cholesterol synthesis and import contribute to protective cholesterol increments in acute myeloid leukemia cells. Blood. 104, 1816–1824 (2004).
Zhang, F., Dai, X. & Wang, Y. 5-Aza-2′-deoxycytidine induced growth inhibition of leukemia cells through modulating endogenous cholesterol biosynthesis. Mol Cell Proteomics. 11, M111.016915, doi: 10.1074/mcp.M111.016915. (2012).
Acknowledgements
This work was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science; and in part by a Kawasaki Medical School Project Grant and Kurozumi Medical Foundation Grant. The authors thank Prof. Takashi Sugihara and Prof. Hideho Wada, a Division of Hematology, Department of Internal Medicine, Kawasaki Medical School for providing the patient samples and Ms. Aki Kuyama and Ms. Asami Matsuo for editorial assistance.
Author information
Authors and Affiliations
Contributions
T.T. was the principal investigator. T.T., A.Y. and Y.I. performed all the pilot experiments and most of analysis. The study was performed in KaT’s group and A.Y. group under the direction of K.T., K.T. and A.Y. participated in the statistical analysis. T.T., S.O., T.K. and S.S. performed the benchwork. T.T., Y.I., M.O., Y.T., T.K. and K.T. discussed the steps of the study. T.T. and K.T. wrote the manuscript with contributions from the other authors.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Tsujioka, T., Yokoi, A., Itano, Y. et al. Five-aza-2′-deoxycytidine-induced hypomethylation of cholesterol 25-hydroxylase gene is responsible for cell death of myelodysplasia/leukemia cells. Sci Rep 5, 16709 (2015). https://doi.org/10.1038/srep16709
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep16709
This article is cited by
-
Role of reactive oxygen species in regulating 27-hydroxycholesterol-induced apoptosis of hematopoietic progenitor cells and myeloid cell lines
Cell Death & Disease (2022)
-
Novel copper complex CTB regulates methionine cycle induced TERT hypomethylation to promote HCC cells senescence via mitochondrial SLC25A26
Cell Death & Disease (2020)
-
Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer
Journal of Biomedical Science (2018)
-
An MDS-derived cell line and a series of its sublines serve as an in vitro model for the leukemic evolution of MDS
Leukemia (2018)
-
KIF20A, highly expressed in immature hematopoietic cells, supports the growth of HL60 cell line
International Journal of Hematology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.