Abstract
The full-length cDNAs of amh and dax1 in the hermaphrodite, rice-field eel (Monopterus albus), were cloned and characterized in this study. Multiple sequence alignment revealed Dax1 was well conserved among vertebrates, whereas Amh had a low degree of similarity between different vertebrates. Their expression profiles in gonads during the course of sex inversion and tissues were investigated. The tissue distribution indicated amh was expressed mostly in gonads and was scarcely detectable in other tissues, whereas the expression of dax1 was widespread among the different tissues, especially liver and gonads. amh was scarcely detectable in ovaries whereas it was abundantly expressed in both ovotestis and testis. By contrast, dax1 was highly expressed in ovaries, especially in ♀IV (ovaries in IV stage), but it was decreased significantly in ♀/♂I (ovotestis in I stage). Its expression was increased again in ♀/♂III (ovotestis in III stage) and then decreased to a low level in testis. These significant different expression patterns of amh and dax1 suggest the increase of amh expression and the decline of dax1 expression are important for the activation of testis development and the high level of amh and a low level of dax1 expression are necessary for maintenance of testis function.
Similar content being viewed by others
Introduction
Rice-field eel (Monopterus albus) is a species of the teleost family Synbranchidae of the order Synbranchiformes (Neoteleostei, Teleostei, Vertebrata). This freshwater fish, which is an important species for fishery production, is a good model for comparative genomic studies of vertebrates, especially for sexual development owing to its special characteristics, including a relatively small genome compared to other teleosts and natural sex inversion (from female to male via intersex) during its life cycle1,2.
Anti-Müllerian hormone (AMH), also called Müllerian-inhibiting substance (MIS), which encodes a glycoprotein member of the transforming growth factor-β (TGF-β) superfamily3,4, is a gonadal hormone that initializes the regression of Müllerian ducts. Müllerian ducts in mammals are characteristic reproductive structures of the female that would differentiate into fallopian tubes and uterus5,6. In males, Amh is one of the first genes to be expressed strongly in Sertoli cells and causes regression of the Müllerian ducts during testicular differentiation7. AMH prevents the FSH-stimulated aromatase activity in Sertoli cell. It also inhibits the testosterone production stimulated by LH in cultured fetal Leydig cells in immature testis8. AMH dramatically decreases the conversion rate of testosterone to estradiol through reducing the activity of aromatase when ovine fetal ovaries exposed to AMH9. AMH acts mainly through the Amh/AmhrII system in gonad differentiation and development10,11. Müllerian ducts develop in only a few species of fish, such as sturgeon (Acipenser spp.), without undergoing regression in males12. amh orthologs, however, have been identified in various teleost species, including Paralichthys olivaceus (Japanese flounder)13, Danio rerio (Zebrafish)14, Oryzias latipes (Medaka)15, Dicentrarchus labrax (European seabass)16, Squalius alburnoides (Bordalo)17, Acanthopagrus schlegeli (Black Porgy)18 and Carassius auratus var. Pengze (Pengze crucian carp)19. It was also involved in sex differentiation and development in these species.
DAX1 (dosage-sensitive sex reversal, adrenal hypoplasia congenita, critical region on the X-chromosome, gene 1), also called NR0B1, is a member of an unusual orphan nuclear receptor superfamily20. In general, it contains a typical ligand-binding domain (LDB) in the C-terminal region and a characteristic DNA-binding domain (DBD) in the N-terminal region21. DAX1 would be able to modulate the transcription of several genes involved in the development of the adrenal and gonadal tissues and steroidogenic activity, such as estrogen receptors α and β22, androgen receptor23, Amh24, Cyp1925 and CYP1726. Over-expression of Dax1 induced male to female sex inversion in a normal male mouse20,27. Dax1 was also shown to be essential for normal testicular development and influences male fertility in mice28. dax1 is a conservative gene and its orthologs have been cloned in fish, including Oreochromis niloticus (Nile tilapia)29, D. labrax30 and O. latipes31. Recent studies showed dax1 might participate in gonad differentiation and development.
In the hermaphrodite fish M. albus, only a few genes potentially participating in sex inversion have ever been isolated and there is a paucity of knowledge about the mechanisms of sex inversion. amh is one of the genes reported to participate actively in the pathway of male sex determination. dax1 might be involved in gonad differentiation and development in both sexes of several teleost species. Taking into account the function of amh and dax1 in sex inversion of M. albus is not fully understood, we cloned the full-length cDNA of amh and dax1 and analyzed their patterns of expression in different periods during sex inversion, as well as their tissue distributions. With this approach, we aimed to elucidate the role of these genes in gonad development and provide insight into the mechanism underlying sex inversion in M. albus.
Results
Cloning and sequence analysis of amh and dax1
The full-length cDNAs of M. albus amh (KF770790.1) and dax1 (KF770791.1) are 1985 bp and 1366 bp, respectively and the corresponding open reading frames (ORFs) are 1647 bp and 882 bp, which encode 548 and 293 amino acids, respectively. The 5′ untranslated regions (5′-UTRs) of amh and dax1 are 68 bp and 148 bp and the 3′-UTRs are 270 bp and 336 bp, respectively. All of them contain a putative polyadenylation signal (AATAAA) closely upstream of the poly(A) tail (Fig. 1).
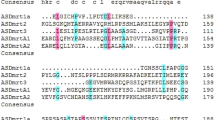
Multiple sequence alignment of Amh with all deduced protein sequences between M. albus and other vertebrates showed the level of conservation of Amh among different species was low; e.g. 64.3% between M. albus and P. olivaceus. A low level of similarity has been also detected between M. albus and Mus musculus (Mouse) (18.2%) and between M. albus and Homo sapiens (Human) (18.1%) (Fig. 2A). The C-terminal region shared a higher degree of identity among different species compared to the N-terminal regions. The amino acid sequence of Amh contains two domains characteristic of the AMH protein family, the AMH-N domain in the middle of this protein and the TGF-β domain at the C terminus. It showed the presence of two consecutive putative plasmin cleavage sites (R/K-XX-R/K) between these two domains (arrow in Fig. 2A). Finally, analysis of the cysteine distribution showed there are nine cysteine residues conserved among different vertebrates (asterisks in Fig. 2A).
Amino acid sequence comparison of M. albus Amh (A) and Dax1 (B) with known orthologs.(A) The AMH domain (AMH-N in solid line box) that characterize this protein family are labeled and the TGF-β domain at the C-terminus (TGF-β in dotted line box). The asterisks indicated the conservative cysteines. Mon: Monopterus albus; Par: Paralichthys olivaceus, BAD37138.1; Sal: Salmo salar, AAU85130.1; Ang: Anguilla japonica, BAB93107.1; Dan: Danio rerio, AAX81416.1; Hom: Homo sapiens, AAH49194.1; Mus: Mus musculus, NP_031471.2; Gal: Gallus gallus, NP_990361.1. (B) Four LXXLL-like motifs were found in DAX1s (in the fine line boxes). Three of them are located in the DNA/RNA/nuclear receptor-binding domain (in unbroken line box). The fourth LXXLL-like motif and AF-2 core are located in the ligand-binding domain (in dotted line box). Mon: Monopterus albus; Epi: Epinephelus coioides, ADR80691.1; Dic: Dicentrarchus labrax, CAG17628.1; May: Maylandia zebra, XP_004551298.1; Ore: Oreochromis niloticus, AAN17672.1; Tak: Takifugu rubripes, XP_003961802.1; Cyn: Cynoglossus semilaevis, ACZ51263.1; Kry: Kryptolebias marmoratus, ACL00866.1; Ory: Oryzias latipes, BAF74811.1; Onc: Oncorhynchus mykiss, ACM80361.1; Dan: Danio rerio, AAI33943.1; Hom: Homo sapiens, ADZ17343.1; Mus: Mus musculus, NP_031456.1.
Multiple alignment of the Dax1 deduced sequence showed the highest degree of identity between M. albus and Epinephelus coioides (Orange-spotted grouper) (89.8%). Furthermore, a higher level of conservation was found in the C-terminal region compared to the N-terminal regions. Four LXXLL-like motifs were found in mammalian DAX1 proteins; three were located in the DNA/RNA/nuclear receptor-binding domain and one was located in the ligand-binding domain. By contrast to mammals, teleost Dax1 has only two LXXLL-like motifs, one located in the DNA/RNA/nuclear receptor-binding domain and the other in the ligand-binding domain. Finally, an AF-2 core was found in the ligand-binding domain of mammals and fish (Fig. 2B).
Phylogenetic analysis of amh nucleotide sequences showed M. albus clusters within the clade of P. olivaceus with high bootstrap support (100), well separated from mammals and Gallus gallus (chicken) as an outgroup (Fig. 3A). A phylogenetic tree of dax1 nucleotide sequences showed M. albus was related most closely to D. labrax and E. coioides (bootstrap 87) and was well separated from mammals as an outgroup (Fig. 3B).
Phylogenetic tree based on amh (A) and dax1 (B) nucleotide sequences. The abbreviations of species are the same as those used in Fig. 2. The numbers are the percentage of bootstrap values supporting each node from 1000 replicas on the basis of the neighbor-joining method.
Tissue-specific distribution of amh and dax1
amh and dax1 were detected in brain, muscle, intestine, kidney, spleen, heart, liver, skin, blood, eye and gonads by RT-PCR, with the highest level in the gonads (Fig. 4). The transcript levels of amh were scarcely detectable in tissues other than gonads. By contrast, the level of dax1 expression was highest in liver and gonads followed by heart, spleen, blood, muscle, brain and eye (Fig. 4A). Among the different development stages of gonads, the highest level of amh mRNA was observed in ovotestis and testis, whereas the highest level of dax1 mRNA was detected in ovaries (Fig. 4B).
Expression of amh and dax1 during sex inversion of M. albus
The level of amh expression was down-regulated significantly when the gonad developed to ♀IV (ovaries in IV stage) from ♀III (ovaries in III stage). But it was up-regulated dramatically when the gonad inversed to ovotestis from ovaries and the level of expression was very high throughout the ♀/♂II (ovotestis in II stage), ♀/♂III (ovotestis in III stage) and ♂ (testis) stages (Friedman ANOVA, P < 0.05; Fig. 5A). The expression of dax1 appeared to be more stable compared to amh during the development stages of gonads. It was increased significantly during oocyte maturation (♀IV) (Friedman ANOVA, P < 0.05; Fig. 5B). Thereafter, dax1 expression declined dramatically as the oocytes degenerate, paralleling with the initiation of spermatogonial proliferation (♀/♂I, ovotestis in I stage) (Friedman ANOVA, P < 0.05; Fig. 5B). The dax1 expression was increased again (♀/♂III) and then decreased to a low level in testis (♂) (Friedman ANOVA, P < 0.05; Fig. 5B).
The copies of amh (A) and dax1 (B) in different development stages of gonads in M. albus. ♀III: ovaries in III stage, ♀IV: ovaries in IV stage, ♀V: ovaries in V stage, ♀/♂I: ovotestis in I stage, ♀/♂II: ovotestis in II stage, ♀/♂III: ovotestis in III stage, ♂: testis. Two biological replicates have been used in each stage and each samples being measured three times. Bars with the different letter indicate significantly difference from one another (Friedman ANOVA, P < 0.05). Data are given as mean ± SE. (A) amh expression significantly decreased when ovary developed from ♀III to ♀IV and then significantly increased when the gonadal development changed into the ovotestis (Friedman ANOVA, P < 0.05). (B) The expression of dax1 significantly increased when gonadal development entered into ♀IV from ♀III and then significantly decreased when the gonadal developed from ♀V to ♀/♂I (Friedman ANOVA, P < 0.05).
Discussion
In this study, we isolated the full-length cDNAs of amh and dax1 from M. albus and then analyzed and characterized their deduced amino acid sequences. Multiple sequence alignment of Amh showed the C-terminal region shared a high degree of homology among different species owing to the conservatism of the TGF-β family. In most members of this family, the N-terminal region does not have its own biological activity but it is critical to enhance the biological function of the C-terminal region32. Sequence analysis of the region between TGF-β and the AMH-N domain suggest that there are two putative cleavage sites (R/K-XX-R/K) in most fish species, including M. albus. However, only one cleavage site form in humans and cattle3. Phylogenetic analysis of amh showed a significant difference between fish and mammal, suggesting a high rate of diversification during the evolution of amh16. Multiple sequence alignment of Dax1 showed the protein shared a high level of similarity in fish. Four LXXLL-like motifs were found in mammalian DAX1 but only two such motifs have been found in the M. albus Dax1 protein, which is highly conservative among teleosts19,30. The second and fourth LXXLL-like motifs were located in the DNA/RNA/nuclear receptor-binding domain and the ligand-binding domain, respectively. It is documented these LXXLL-like motifs were capable of interacting with SF-133,34 and other nuclear receptors, including estrogen receptor22, androgen receptor23 and progestin receptor35. Mutation or deletion of these motifs impaired DAX1 repressed activity against SF136. In M. albus, D. labrax and the Takifugu rubripes (Pufferfish), the first LXXLL-like motif in the Dax1 protein was followed by a polyglutamine (poly(Q)) stretch. There were seventeen glutamines in T. rubripes, thirteen in D. labrax and only six in M. albus. Glutamine was the most commonly repeated amino acid in eukaryotic proteins. It has been reported that such repeats are part of a general evolutionary mechanism for adding new amino acid sequence37. As in mammals, fish have the AF-2 core in the ligand-binding domain and mutation of this residue lowered the frequency of nuclear localization of sf1, suggesting an important role of the AF-2 core in nuclear localization processes21.
The expression of M. albus amh and dax1 mRNAs was examined in various tissues by RT-PCR. amh expression was detected in the gonads (both sexes) but no obvious signal was obtained in other tissues, suggesting amh is a related gene in gonadal development in M. albus. The similar pattern of expression was found in O. latipes15 and P. olivaceus13. It has been well described in mammals that the expression of Amh occurs in the Sertoli cells of the testis and the granulosa cells of the ovary38,39. The high level of amh expression was observed in the ovotestis and testis in M. albus and it was also detected in ovary but at a low level. At the same time, in other teleosts, amh was expressed in the gonads of one or both sexes, with the highest level in the testis13,14,16,40. These findings suggested that regulation of spermatogonial proliferation and differentiation might be one of the common functions of amh among vertebrates. In mammals, higher levels of Dax1 were apparent in different tissues encompassing the gonads but not in the liver or intestine41,42. Nevertheless, dax1 mRNA was expressed abundantly in the liver in non-mammalian vertebrates43,44. The expression of dax1 has been found in liver and it is also expressed in brain, muscle, spleen, heart, blood, eye and gonads in M. albus. dax1 expressed in the liver might be involved in the regulation of vitellogenesis by interacting with nuclear receptors in C. auratus var. Pengze19. In D. rerio, dax1 morpholino antisense nucleotide injected embryos exhibited abnormal development in the central nervous systems, indicating it was important for brain development45. Therefore, the wide and different expression pattern among vertebrates suggested the function of dax1 was complex and remained unclear46. In addition, Dax1 has been thought to act as an “anti-testis” gene or a “pro-ovarian” gene during gonadal development27 and it was involved in the regulation of ovarian steroidogenesis via inhibition of the transcription of LRH-1 in a granulosa cell line47. Dax1 was also found to be involved in testis cord organization during development48. In the M. albus gonad, the expression of dax1 has been detected in male and female, revealing dax1 was closely related to gonad development including testis and ovaries.
Next, we used absolute quantitative real-time RT-PCR to examine amh and dax1 expression in different development stages of gonads during sex inversion in M. albus. The expression of amh has been detected in both testis and ovaries, agreeing with the expression pattern of amh in D. rerio14. In Anguilla japonica (Japanese eel)40 and P. olivaceus13 testis, amh expression has been detected in Sertoli cells surrounding spermatogonia. In the mammalian fetus, AMH is one of the earliest Sertoli cell-specific proteins expressed by the gonad and it was responsible for the initiation of Müllerian duct regression in the male fetus49. amh in A. schlegeli plays important roles in early testicular18. In M. albus, the expression of amh was up-regulated significantly when gonads developed into the ♀/♂I (ovotestis in I stage), implying amh expression was triggered in differentiating Sertoli cells and early testicular development. amh expression was continuous up-regulated significantly at ♀/♂II (ovotestis in II stage). It was consistent with the maintenance of amh secretion at high levels in the testis, indicating the male development and testis maintained in M. albus required sustained release of amh in testis. In D. rerio14 and D. labrax16, cyp19a is down-regulated by amh. Conversely, AMH is regulated by other transcription factor, such as SF1, SOX9, DAX1 and WT124,50,51. Our previous studies investigated the relationship between miRNA-430 and amh. The parallel expression levels of amh and miRNA-430 were detected in M. albus, suggesting that amh may be the target gene of miRNA-43052. The function of amh during gonadal determination and differentiation is regulated by microRNAs in M. albus52. In addition, the expression of amh was detected in granulosa cells in adult female D. rerio during the stages of oogenesis. It was discovered first in late stage IB, decreased in early stage III and has not been found in the later stages14. Our result in ovaries of M. albus agreed with the expression of amh in D. rerio, which was decreased significantly in ♀IV and hardly detected in ♀V. AMH is an important gene for normal folliculogenesis by regulating the number of follicles53. Recent study in mouse found that AMH and FOXL2 synergistically interact to reserve ovarian follicles54. This finding supported the role of amh in early ovary development. It also approved the important roles of amh in natural sex change fish A. schlegeli in early ovarian development and late ovarian growth (e.g., vitellogenic oocytes)18.
Dax1 was expressed in all regions of the HPG axis during development. In the ovaries, the granulosa and theca cells stained positive for DAX1. Both Sertoli cells and Leydig cells showed expression of DAX1 in the testis55. Dax1-deleted mice might have caused ovarian developmental impairment because duplicated Dax1 caused XY sex inversion, indicating Dax1 might be an ovary-determination gene. Unexpectedly, the Dax1-deficient mice had testis dysgenesis and a spermatogenesis defect in males as the result of disorganized testis cord formation; however, ovarian development was normal48,56. On the other hand, the expression of DAX1 in Sertoli cells and Leydig cells individually was not sufficient to rescue the pathology, suggesting DAX1 function in other somatic cell types was necessary for proper testicular development, as well as in Sertoli and Leydig cell types28,57. In females, DAX1 has been detected in various ovarian cell types, especially in granulosa cells, suggesting DAX1 likely had a role in regulation of gene expression during the specific stage of follicular development21. It has been postulated DAX1 could be involved in the regulation of some key enzymes by inhibiting SF1-mediated transcription in steroid biosynthesis during follicular development58, whereas Dax1-deficient female mice were normal and fertile with a slight ovarian follicular defect59. In this study, M. albus dax1 was detected in the ovaries and in the ovotestis and testis and it was present in significantly greater amounts in the ♀IV stage compared to other development stages. These results implied the function of dax1 in ovary development was complex and not fully understood. Moreover, the expression of dax1 was significantly decreased when the gonad development entered the ♀/♂I stage, indicating down-regulation of dax1 was likely one of the factors stimulating testis development. Our previous study demonstrated that foxl2 may regulate the expression of cyp19a1a according to the correlation expression pattern between foxl2 and cyp19a1a in ovary and brain60. Foxl2 knocked-out led to the up-regulation of Dax1, Foxl2 was also regarded as the negative regulator of Sf161. The direct interaction between DAX1 and SF1 modulates the various target gene expression, such as aromatase25 and MIS62. Furthermore, DAX1 also acts as a transactivator for aromatase25 and regulates the timing of vitellogenic development in protandrous A. schlegeli63. Therefore, the cooperative or antagonistical regulation relationship between DAX-1, FOXL2 and SF-1 are complicated. All of them indirectly or directly regulate aromatase expression during the gonadal determination and differentiation64.
In summary, full length cDNAs of amh and dax1 were isolated and characterized from the hermaphrodite freshwater fish M. albus and the tissue-specific distribution of these two genes was determined. amh is a gonad-related gene expressed mainly in gonads and dax1 is widely distributed among different tissues. The gonad expression patterns of amh and dax1 were investigated during M. albus sex inversion. The amount of amh is increased significantly at the ovotestis and maintained at a high level at testis, whereas dax1 is highly expressed in the ♀IV. It is decreased significantly in the ♀/♂I. Thereafter, its expression was increased again in ♀/♂III and then decreased to a low level in testis. The information provided in this study will broaden our understanding of the molecular mechanisms underlying gonad development in hermaphrodite fish.
Methods
Experimental fish
M. albus were obtained from Wuhan in China and cultured in an aquarium for 7 days (34 to 54 cm body length; 50 to 120 g body weight). The fish have been anesthetized by MS-222. Then, they were sacrificed by spinal amputation and segments of the gonads were fixed in Bouin-Holland fluid for histological assessment. The sexual stages refer to our earlier study65. The different development stages of gonads and tissues (gonad, blood, muscle, skin, liver, eye, spleen, intestine, kidney, heart and brain) of M. albus were frozen in liquid nitrogen and stored at −80 °C. For gonadal analysis: two fishes have been chosen in each stage and each sample was analyzed in triplicate. For tissue distribution analysis: one kind of tissues from random three fishes was pooled as one sample. All experiments were carried out with the approval from the Institutional Animal Care and Use Committee (IACUC) of Huazhong Agricultural University (Wuhan, China) and also performed in accordance with the International Guiding Principles for Biomedical Research Involving Animals as promulgated by the Society for the Study of Reproduction.
RNA extraction and reverse transcription-PCR (RT-PCR)
Total RNA was extracted using TRIzol® reagent (Takara, Dalian) according to the manufacturer’s instructions. The quality was assessed by agarose-gel electrophoresis on the basis of the integrity of the 18 S and 28 S rRNA bands and NANODROP 2000c (Thermo Scientific, USA) with an A260mm/A280mm ratio from 1.8 to 2.1. A sample of total RNA of 1 μg was used for reverse transcription with a PrimeScript® RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara) in a final reaction volume of 20 μL.
Molecular cloning of amh and dax1
Partial cDNA fragments of target genes were amplified using degenerate primers (Table 1) designed on the basis of the conserved regions of other species using Primer Premier 5.0 software. PCR was performed with 2 μL of cDNA template in a 25 μL reaction volume containing 0.5 μL of 10 mM each primer and 12.5 μL of Premix Taq (Takara). PCR products were cloned into the PMD-19T vector (Takara) and sequenced at the Beijing Genomics Institute. Gene-specific primers (Table 1) were designed to obtain the 5′ and 3′ ends of each cDNA on the basis of the sequences of these partial fragments following the manufacturer’s instructions for the SMART RACE Kit (Clontech, America).
Sequence analysis and phylogenetic analyses
The amino acid sequences of Amh and Dax1 were deduced by BioEdit software and aligned with other species using ClustalW. Protein percentage sequence identity was calculated by the MegAlign program of DNAStar software. Phylogenetic trees were constructed using the neighbor-joining (NJ) method of MEGA version 5.05 software and the credibility of each branch was obtained by the use of 1000 bootstrap replicates.
Tissue distribution of amh and dax1 by RT-PCR
The gene-specific primers (Table 1) were designed to analyze the expression patterns of amh and dax1 among different tissues (brain, muscle, intestine, kidney, spleen, heart, liver, skin, blood, eye and gonads). The 25 μL PCR volume contained 2 μL of 10-fold concentrated cDNA template and the protocol was: heat at 94 °C for 3 min followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s and a final extension step at 72 °C for 10 min. The ef1α (KC011266) and rpl-17 (KC011267) reference genes were treated for 28 and 25 cycles, respectively. The stability of reference genes have been analyzed in our previous study65. All PCR products were electrophoresed in 1% (w/v) agarose gel and DNA was stained with GelRed.
amh and dax1 patterns of expression during sex inversion of gonads
Absolute quantitative real-time RT-PCR was used to detect the expression of genes among different development stages of gonads. The gene-specific primers used in this experiment were the same as those described above and the specificity of each pair of primers was verified via the only peak of the melt curve. The 25 μL reactive system containing 2 μL of cDNA, 0.5 μL of 10 mM each primer and 12.5 μL of SYBR® Premix Ex Taq™ II (Perfect Real Time) (Takara) and the protocol was: heat at 95 °C for 30 s, followed by 30 cycles of 95 °C for 5 s, 55 °C for 60 s for amh or 56 °C for 50 s for dax1 and a final extension step at 72 °C for 30 s. A negative control was included in each assay without cDNA and the samples were analyzed in triplicate with a Rotor-Gene Q instrument. The expression of each sample was calculated as copies/ml from the standard curve of serial dilutions.
Statistical analysis
Two biological replicates were used in each gonadal stage and each sample was measured three times. All experimental data were reported as mean ± standard error and analyzed by the non-parametric Friedman ANOVA. P value less than 0.05 was chosen as the significant level.
Additional Information
How to cite this article: Hu, Q. et al. Molecular cloning and characterization of amh and dax1 genes and their expression during sex inversion in rice-field eel Monopterus albus. Sci. Rep. 5, 16667; doi: 10.1038/srep16667 (2015).
References
Liu, C. Rudimentary hermaphroditism in the symbranchoid eel, Monopterus javanensis. Sinensia 15, 1–8 (1944).
Zhou, R., Cheng, H. & Tiersch, T. R. Differential genome duplication and fish diversity. Rev. Fish Biol. Fisher. 11, 331–337 (2001).
Cate, R. et al. Isolation of the bovine and human genes for Müllerian inhibiting substance and expression of the human gene in animal cells. Cell 45, 685–698 (1986).
Picard, J.-Y., Goulut, C., Bourrillon, R. & Josso, N. Biochemical analysis of bovine testicular anti-Müllerian hormone. FEBS Lett. 195, 73–76 (1986).
Josso, N. et al. Anti-Müllerian hormone in early human development. Early Hum. Dev. 33, 91–99 (1993).
Munsterberg, A. & Lovell-Badge, R. Expression of the mouse anti-mullerian hormone gene suggests a role in both male and female sexual differentiation. Development 113, 613–624 (1991).
Josso, N., Racine, C., di Clemente, N., Rey, R. & Xavier, F. The role of anti-Müllerian hormone in gonadal development. Mol. Cell. Endocrinol. 145, 3–7 (1998).
Rouiller-Fabre, V. et al. Effect of anti-Mullerian hormone on Sertoli and Leydig cell functions in fetal and immature rats. Endocrinology 139, 1213–1220 (1998).
Vigier, B. et al. Anti-Müllerian hormone produces endocrine sex reversal of fetal ovaries. P. Natl. Acad. Sci. USA 86, 3684–3688 (1989).
Baarends, W. M. et al. A novel member of the transmembrane serine/threonine kinase receptor family is specifically expressed in the gonads and in mesenchymal cells adjacent to the mullerian duct. Development 120, 189–197 (1994).
Behringer, R. R., Finegold, M. J. & Cate, R. L. Müllerian-inhibiting substance function during mammalian sexual development. Cell 79, 415–425 (1994).
Wrobel, K. H. The genus Acipenser as a model for vertebrate urogenital development: the müllerian duct. Anat. Embryol. 206, 255–271 (2003).
Yoshinaga, N. et al. Sexually dimorphic expression of a teleost homologue of Müllerian inhibiting substance during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem. Bioph. Res. Co. 322, 508–513 (2004).
Rodríguez-Marí, A. et al. Characterization and expression pattern of zebrafish anti-Müllerian hormone (amh) relative to sox9a, sox9b and cyp19a1a, during gonad development. Gene Expr. Patterns 5, 655–667 (2005).
Klüver, N. et al. Differential expression of anti‐Müllerian hormone (amh) and anti‐Müllerian hormone receptor type II (amhrII) in the teleost medaka. Dev. Dynam. 236, 271–281 (2007).
Halm, S., Rocha, A., Miura, T., Prat, F. & Zanuy, S. Anti-Müllerian hormone (AMH/AMH) in the European sea bass: Its gene structure, regulatory elements and the expression of alternatively-spliced isoforms. Gene 388, 148–158 (2007).
Pala, I., Klüver, N., Thorsteinsdóttir, S., Schartl, M. & Coelho, M. Expression pattern of anti-Müllerian hormone (amh) in the hybrid fish complex of Squalius alburnoides. Gene 410, 249–258 (2008).
Wu, G. C., Chiu, P. C., Lyu, Y. S. & Chang, C. F. The expression of amh and amhr2 is associated with the development of gonadal tissue and sex change in the protandrous Black Porgy, Acanthopagrus schlegeli. Biol. Reprod. 83, 443–453, doi: 10.1095/biolreprod.110.084681 (2010).
Li, M. et al. Molecular cloning and characterization of amh, dax1 and cyp19a1a genes and their response to 17α-methyltestosterone in pengze crucian carp. Comp. Biochem. Phys. C 157, 372–381 (2013).
McCabe, E. Adrenal hypoplasias and aplasias. The Metabolic and Molecular Bases of Inherited Disease, McGraw-Hill, New York, 4263–4274 (2001).
Iyer, A. K. & McCabe, E. R. Molecular mechanisms of DAX1 action. Mol. Genet. Metab. 83, 60–73 (2004).
Zhang, H., Thomsen, J. S., Johansson, L., Gustafsson, J. A. & Treuter, E. DAX-1 functions as an LXXLL-containing corepressor for activated estrogen receptors. J. Biol. Chem. 275, 39855–39859 (2000).
Holter, E. et al. Inhibition of androgen receptor (AR) function by the reproductive orphan nuclear receptor DAX-1. Mol. Endocrinol. 16, 515–528 (2002).
Nachtigal, M. W. et al. Wilms’ tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell 93, 445–454 (1998).
Wang, Z. J. et al. Aromatase (Cyp19) expression is up-regulated by targeted disruption of Dax1. P. Natl Acad. Sci. USA 98, 7988–7993 (2001).
Hanley, N. A., Rainey, W. E., Wilson, D. I., Ball, S. G. & Parker, K. L. Expression profiles of SF-1, DAX1 and CYP17 in the human fetal adrenal gland: potential interactions in gene regulation. Mol. Endocrinol. 15, 57–68 (2001).
Swain, A., Narvaez, V., Burgoyne, P., Camerino, G. & Lovell-Badge, R. Dax1 antagonizes Sry action in mammalian sex determination. Nature 391, 761–767 (1998).
Jeffs, B. et al. Sertoli cell-specific rescue of fertility, but not testicular pathology, in Dax1 (Ahch)-deficient male mice. Endocrinology 142, 2481–2488 (2001).
Wang, D. S. et al. Molecular cloning of DAX1 and SHP cDNAs and their expression patterns in the Nile tilapia, Oreochromis niloticus. Biochem. Bioph. Res. Co. 297, 632–640 (2002).
Martins, R. S., Deloffre, L. A., Mylonas, C. C., Power, D. M. & Canário, A. V. Developmental expression of DAX1 in the European sea bass, Dicentrarchus labrax: lack of evidence for sexual dimorphism during sex differentiation. Reprod. Biol. Endocrin. 5, 19 (2007).
Nakamoto, M. et al. Dax1 suppresses P450arom expression in medaka ovarian follicles. Mol. Reprod. Dev. 74, 1239–1246 (2007).
Wilson, C. et al. Mullerian inhibiting substance requires its N-terminal domain for maintenance of biological activity, a novel finding within the transforming growth factor-beta superfamily. Mol. Endocrinol. 7, 247–257 (1993).
Ito, M., Yu, R. & Jameson, J. L. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol. Cell. Biol. 17, 1476–1483 (1997).
Kawajiri, K. et al. Role of the LXXLL-motif and activation function 2 domain in subcellular localization of Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1). Mol. Endocrinol. 17, 994–1004 (2003).
Agoulnik, I. U. et al. Repressors of androgen and progesterone receptor action. J. Biol. Chem. 278, 31136–31148 (2003).
Suzuki, T., Kasahara, M., Yoshioka, H., Morohashi, K.-I. & Umesono, K. LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Mol. Cell. Biol. 23, 238–249 (2003).
Kahlem, P., Terre, C., Green, H. & Djian, P. Peptides containing glutamine repeats as substrates for transglutaminase-catalyzed cross-linking: relevance to diseases of the nervous system. P. Natl. Acad. Sci. USA 93, 14580–14585 (1996).
Juengel, J. L. et al. Expression of anti-Müllerian hormone mRNA during gonadal and follicular development in the brushtail possum (Trichosurus vulpecula). Reprod. Fert. Develop. 14, 345–353 (2002).
Pask, A. J. et al. Marsupial anti-Müllerian hormone gene structure, regulatory elements and expression. Biol. Reprod. 70, 160–167 (2004).
Miura, T., Miura, C., Konda, Y. & Yamauchi, K. Spermatogenesis-preventing substance in Japanese eel. Development 129, 2689–2697 (2002).
Bae, D., Schaefer, M., Partan, B. & Muglia, L. Characterization of the mouse DAX-1 gene reveals evolutionary conservation of a unique amino-terminal motif and widespread expression in mouse tissue. Endocrinology 137, 3921–3927 (1996).
Parma, P., Pailhoux, E., Puissant, C. & Cotinot, C. Porcine Dax-1 gene: isolation and expression during gonadal development. Mol. Cell. Endocrinol. 135, 49–58 (1997).
Smith, C. et al. Cloning and expression of a DAX1 homologue in the chicken embryo. J. Mol. Endocrinol. 24, 23–32 (2000).
Sugita, J., Takase, M. & Nakamura, M. Expression of Dax-1 during gonadal development of the frog. Gene 280, 67–74 (2001).
Yajima, Y., Vestergaard, M. D. C. & Takagi, M. Damage in brain development by morpholino knockdown of zebrafish dax1. J. Biosci. Bioeng. 113, 683–688 (2012).
McCabe, E. R. DAX1: increasing complexity in the roles of this novel nuclear receptor. Mol. Cell. Endocrinol. 265, 179–182 (2007).
Peng, N., Kim, J. W., Rainey, W. E., Carr, B. R. & Attia, G. R. The role of the orphan nuclear receptor, liver receptor homologue-1, in the regulation of human corpus luteum 3β-hydroxysteroid dehydrogenase type II. J. Clin. Endocr. Metab. 88, 6020–6028 (2003).
Meeks, J. J. et al. Dax1 regulates testis cord organization during gonadal differentiation. Development 130, 1029–1036 (2003).
Tran, D. & Josso, N. Relationship between avian and mammalian anti-Müllerian hormones. Biol. Reprod. 16, 267–273 (1977).
Giuili, G., Shen, W. H. & Ingraham, H. A. The nuclear receptor SF-1 mediates sexually dimorphic expression of Mullerian Inhibiting Substance, in vivo. Development 124, 1799–1807 (1997).
De Santa Barbara, P. et al. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol. Cell. Biol. 18, 6653–6665 (1998).
Gao, Y. et al. Characterization and differential expression patterns of conserved microRNAs and mRNAs in three genders of the Rice Field Eel (Monopterus albus). Sex. Dev. 8, 387–398 (2014).
Broekmans, F. J. et al. Anti-Müllerian hormone and ovarian dysfunction. Trends Endocrin. Met. 19, 340–347 (2008).
Park, M., Suh, D. S., Lee, K. & Bae, J. Positive cross talk between FOXL2 and antimüllerian hormone regulates ovarian reserve. Fertil. Steril. 102, 847–855. e841 (2014).
Jadhav, U., Harris, R. M. & Jameson, J. L. Hypogonadotropic hypogonadism in subjects with DAX1 mutations. Mol. Cell. Endocrinol. 346, 65–73 (2011).
Meeks, J. J., Weiss, J. & Jameson, J. L. Dax1 is required for testis determination. Nat. Genet. 34, 32 (2003).
Meeks, J. J. et al. Leydig cell-specific expression of DAX1 improves fertility of the Dax1-deficient mouse. Biol. Reprod. 69, 154–160 (2003).
Sato, Y. et al. Immunolocalization of nuclear transcription factors, DAX-1 and COUP-TF II, in the normal human ovary: correlation with adrenal 4 binding protein/steroidogenic factor-1 immunolocalization during the menstrual cycle. J. Clin. Endocr. Metab. 88, 3415–3420 (2003).
Richard, N. Y., Ito, M., Saunders, T. L., Camper, S. A. & Jameson, J. L. Role of Ahch in gonadal development and gametogenesis. Nat. Genet. 20, 353–357 (1998).
Hu, Q., Guo, W., Gao, Y., Tang, R. & Li, D. Molecular cloning and analysis of gonadal expression of Foxl2 in the rice-field eel Monopterus albus. Sci. Rep. 4 (2014), doi: 10.1038/srep06884
Uhlenhaut, N. H. et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139, 1130–1142 (2009).
Tremblay, J. J. & Viger, R. S. Nuclear receptor Dax-1 represses the transcriptional cooperation between GATA-4 and SF-1 in Sertoli cells. Biol. Reprod. 64, 1191–1199 (2001).
Wu, G. C., Tomy, S. & Chang, C. F. The expression of nr0b1 and nr5a4 during gonad development and sex change in protandrous black porgy fish, Acanthopagrus schlegeli [J]. Biol. Reprod., 78, 200–210 (2008)
von Schalburg, K. R. et al. Regulation and expression of sexual differentiation factors in embryonic and extragonadal tissues of Atlantic salmon. BMC genomics 12, 31 (2011).
Hu, Q., Guo, W., Gao, Y., Tang, R. & Li, D. Reference gene selection for real-time RT-PCR normalization in rice field eel (Monopterus albus) during gonad development. Fish physiol. Biochem. 40, 1721–1730 (2014).
Acknowledgements
This work was supported by the Special Fund for Agro-scientific Research in the Public Interest (201003076), the Fundamental Research Funds for the Central Universities (2013PY024) and the National Natural Foundation of China (Project no. 30970529).
Author information
Authors and Affiliations
Contributions
Q.H. wrote this manuscript text; R.T., D.L. and Q.H. designed the experiments; W.G. collected material for study; Q.H., W.G. and Y.G. carried out the experiments and analyzed the data. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Hu, Q., Guo, W., Gao, Y. et al. Molecular cloning and characterization of amh and dax1 genes and their expression during sex inversion in rice-field eel Monopterus albus. Sci Rep 5, 16667 (2015). https://doi.org/10.1038/srep16667
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep16667
This article is cited by
-
Complete Depletion of Primordial Germ Cells Results in Masculinization of Monopterus albus, a Protogynous Hermaphroditic Fish
Marine Biotechnology (2022)
-
A new experimental model for the investigation of sequential hermaphroditism
Scientific Reports (2021)
-
The transcriptome of the newt Cynops orientalis provides new insights into evolution and function of sexual gene networks in sarcopterygians
Scientific Reports (2020)
-
Expression profiles of sex-related genes in gonads of genetic male Takifugu rubripes after 17β-estradiol immersion
Journal of Oceanology and Limnology (2019)
-
Induction of Gonadal Development in Protogynous Grouper with Orally Delivered FSH DNA
Marine Biotechnology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.