Abstract
Arginine methylation is a post-translational modification required for the maintenance of genomic integrity. Cells deficient in protein arginine methyltransferase 1 (PRMT1) have DNA damage signaling defects, defective checkpoint activation and extensive genomic instability. Herein we identify the DNA damage protein and RNA binding protein, hnRNPUL1, to be a substrate of PRMT1. We identify the dimethylation of R584, R618, R620, R645 and R656, as well as the monomethylation of R661 R685 and R690 within hnRNPUL1 in U2OS cells by mass spectrometry. Moreover, we define the arginines within the RGG/RG motifs as the site of methylation by PRMT1 both in vitro and in vivo. The arginines 612, 618, 620, 639, 645, 656 and 661 within the human hnRNPUL1 RGG/RG motifs were substituted with lysines to generate hnRNPUL1RK. hnRNPUL1RK was hypomethylated and lacked the ability to interact with PRMT1, unlike wild type hnRNPUL1. Co-immunoprecipitation studies showed that hnRNPUL1RK had impaired ability to associate with the DNA damage protein NBS1. Moreover, hnRNPUL1RK was not recruited to sites of DNA damage, unlike wild type hnRNPUL1, in the presence of transcriptional inhibitors. These findings define a role for arginine methylation during the DNA damage response to regulate protein-protein interactions for the recruitment at sites of damage.
Similar content being viewed by others
Introduction
Arginine methylation is a common post-translational modification that takes place in eukaryotic cells1,2,3,4. The link between arginine methylation and the DNA damage response pathway was initially observed during a proteomic analysis, where the meiotic recombination 11 homolog (MRE11) protein was identified in immunoprecipitations of methyl-specific antibodies5,6. MRE11 is a DNA damage sensing protein part of trimeric complex termed the MRN complex with NBS1 and RAD507. The key roles of the RGG/RG motif of MRE11 in double strand break (DSB) repair was determined biochemically to alter the processivity of its exonuclease activity8,9. A knockin strategy where the MRE11 RGG/RG motif was replaced by a KGG/KG motif showed that the mice are hyper-sensitive to ionizing radiation (IR), as predicted for a hypomorphic allele of MRE1110. It is now known that several DNA repair proteins including 53BP111,12, p5313, FEN114, BRCA115, RAD916 and DNA polymerase β are regulated by arginine methylation17.
There are 3 main forms of methylated arginine identified in eukaryotes: ω-NG-monomethylarginine (MMA), ω-NG,NG-asymmetric dimethylarginine (aDMA) and ω-NG,N’G-symmetric dimethylarginine (sDMA). Protein arginine methyltransferases (PRMTs) are conserved from yeast to mammals and there are 9 human PRMTs that have been identified that catalyze the transfer of a methyl group from S-adenosylmethionine to a guanidino nitrogen of arginine2. Type I PRMTs (PRMT1, 3, 4, 6 and 8) lead to the production of aDMA, while type II PRMTs (PRMT5 and PRMT9) catalyze the formation of sDMA PRMT7 is a type III enzyme known to generate MMAs18. The discovery of lysine demethylases demonstrates that protein methylation is reversible19, however, an arginine demethylase remains to be identified3.
PRMT1 is the main type I mammalian enzyme with a preference for RGG/RG motifs4 and embryos from PRMT1 knockout mice die shortly after implantation at E6.520,21. The generation of conditional PRMT1 knockout mouse embryo fibroblasts (MEFs) revealed that the inducible loss of PRMT1 leads to spontaneous DNA damage, checkpoint defects, hypersensitivity to DNA-damaging agents, chromosomal instability, aneuploidy and polyploidy, suggesting that PRMT1 is a key player in the DDR pathway21. PRMT1 substrates in the DDR pathway include MRE11, 53BP1 and BRCA1 (for review6. An additional type I enzyme, PRMT6 functions to repress gene activation by methylating histones22,23,24. PRMT6 null mice are viable, but the mouse embryo fibroblasts display premature senescence25.
Non-coding RNAs, RNA helicases and RNA binding proteins (RBP) have recently been shown to participate in DNA damage signaling. RBPs including hnRNPK26, p54nrb/NONO27, hnRNPUL128, RBMX and DDX1729 have been identified as participants in the DDR pathway. hnRNPUL1 was shown to bind with NBS1 and CtIP28, whereas the mechanism of action of RBMX and DDX17 is unknown29. The identification of non-coding RNAs at DNA breaks30 and its requirement for DNA repair31 and 53BP1 recruitment32, defines a role for RNA and RBPs during DNA repair. Thus, the role and regulation of RNA, RBPs and ribonucleoprotein complexes at DSBs is unknown.
hnRNPUL1 is known as an hnRNPU-like protein and belongs to the hnRNP family. It was first identified as an adenoviral early region 1B-associated protein 5 (E1B-AP5), since it was known to associate with the adenovirus early protein E1B-55 kDa (Ad5EE1B55K) during lytic infection33. hnRNPUL1 binds to the MRN complex and is recruited to the damage site to participate in DSB repair28. Specifically, hnRNPUL1 has a RGG/RG motif at its C-terminus that is required to associate with NBS1 and recruit it to DNA damage sites28. hnRNPUL1 was shown to function downstream of MRN and CtIP in the DNA resection pathway and induce DNA resection with the recruitment of the BLM helicase28. It has been demonstrated that hnRNPUL1 is methylated34,35, however, the precise methylated arginine residues and the functional implication of the methylation have remained undefined. Herein, we demonstrate that arginine methylation of hnRNPUL1 is required for its association with NBS1 and recruitment to the DNA damage sites.
Materials and Methods
Antibodies, immunoprecipitations and immunoblotting
Rabbit anti-hnRNPUL1 antibody was purchased from Proteintech (Chicago, IL). Mouse anti-FLAG (M2) antibody, anti-β-tubulin antibody and protein-A-Sepharose beads were purchased from Sigma (St. Louis, MO). Anti-GFP antibody was purchased from Novus Biologicals (Littleton, CO). Rabbit anti-PRMT1 antibody and ASYM25b were described previously21,36. Immunoprecipitations and immunoblotting were performed as previously described37. Briefly, cells were lysed in 50 mM HEPES pH 7.4, 150 mM NaCl and 1% Triton X-100 on ice for 15 min. After removal of the Triton insoluble matter by centrifugation, the supernatant was incubated with the indicated antibodies on ice for 2 h. The bound proteins were immunopurified using protein A Sepharose beads tumbled at 4 °C for 1 h and separated by SDS-PAGE, transferred to nitrocellulose membranes and immunoblotted with the indicated antibodies, as previously described37.
Plasmids
The human hnRNPUL1 cDNA purchased from ORIGENE (Rockville, MD) was FLAG epitope-tagged and subcloned into pcDNA3.1 with the following primers 5′-GGG GGA TCC GAT GTG CGC CGT CTG AAG GTG-3′ and 5′-GGG GTC GAC CTA CTG TGT ACT TGT GCC ACC-3. FLAG-hnRNPUL1RK (R612K, R618K, R620K, R639K, R645K, R656K and R661K) in pcDNA3.1 was generated from FLAG-hnRNPUL1 by Mutagenex Inc (Hillsborough, NJ). GFP-hnRNPUL1 and GFP-hnRNPUL1RK were also generated by Mutagenex Inc. DNA constructs were entirely sequenced. Oligonucleotide sequences used to generate the GST-hnRNPUL1 fragments are as follows:
Di-RG: 5′- GAT CTA TGA AGA AAA CCG GGG ACG GGG GTA CTT TGA GCA CTGA-3′ and 5′-TCG ATC AGT GCT CAA AGT ACC CCC GTC CCC GGT TTT CTT CAT A-3′. RRGR: 5′-GAT CCA CCG AGA GGA TAG GAG GGG CCG CTC TCC TCA GCC TTG A-3′ and 5′-TCG ATC AAG GCT GAG GAG AGC GGC CCC TCC TAT CCT CTC GGT G-3′. RIRG: 5′-GAT CCC CCT TAG TGA GCG TAT CCG GGG CAC CGT TGG ACC ATG A-3′ and 5′-TCG ATC ATG GTC CAA CGG TGC CCC GGA TAC GCT CAC TAA GGG G-3′.
Tri-RG: 5′-GAT CTT TGA CAA CCG AGG TGG TGG TGG CTT CCG GGG CCG CGG GGG TGG TGG TGG CTT CCA GTG A-3′ and 5′- TCG ATC ACT GGA AGC CAC CAC CAC CCC CGC GGC CCC GGA AGC CAC CAC CAC CTC GGT TGT CAA A-3′.
Di-RGG: 5′-GAT CCC TGG AGG CAA CCG TGG CGG CTT CCA GAA CCG AGG GGG AGG CAG CGG TGG AGG ATG A-3′ and 5′-TCG ATC ATC CTC CAC CGC TGC CTC CCC CTC GGT TCT GGA AGC CGC CAC GGT TGC CTC CAG G-3′.
Mono-RGG: 5′-GAT CGG AGG AGG CAA CTA CCG AGG AGG TTT CAA CCG CAG CGG AGG TGG TGG CTG A-3′ and 5′-TCG ATC AGC CAC CAC CTC CGC TGC GGT TGA AAC CTC CTC GGT AGT TGC CTC CTC C-3′. All glutathione S-transferase (GST) proteins were isolated, as described previously36.
Cell culture, transfection and drug treatments
HEK293 and U2OS cell lines were purchased from the American Type Culture Collection (Manassas, VA). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; BioSera), 1 mM sodium pyruvate and antibiotics under typical culture conditions. Plasmids were transfected with Lipofectamine 2000 (Invitrogen) and siRNAs were transfected with Lipofectamine RNAiMAX (Invitrogen) as per the manufacturer’s instructions. siRNAs purchased from Dharmacon Inc. (Lafayette, CO) along with their respective target sequences were as follows: siLuciferase (siCTL, 5′-CGU ACG CGG AAU ACU UCG A dTdT- 3′) and sihnRNPUL1 (5′-GCA GUG GAA CCA GUA CUA U dTdT-3′). siPRMT1 (5′-CGT CAA AGC CAA CAA GTT A dTdT- 3′) was as reported previously to be specific for PRMT121. Cells were analyzed 72 h after transfection.
In vitro methylation assays
The methylation of the hnRNPUL1 RGG/RG motifs by PRMT1 was performed using recombinant proteins, as described36. The indicated recombinant GST-hnRNPUL1 protein was incubated with GST-PRMT1 and 0.55 μCi of methyl-3H-S-adenosyl-L-methionine in the presence of 25 mM Tris- HCl at pH 7.4 for 1 h in a final total volume of 30 μl. The reactions were stopped by adding 30 μl of 2X Laemmli protein loading buffer, followed by boiling the samples at 100 °C for 10 min. The methylation of the GST-RGG/RG motifs was assessed by polyacrylamide gel electrophoresis and stained with Coomassie Blue. The destained gel was soaked in EN3HANCE for 1 h, followed by a gentle agitation in cold water for 30 min. The washed gel was dried at 80 °C for 1.5 h and visualized by fluorography.
Laser irradiation and confocal microscopy
Immunofluorescence localization of specific proteins at laser-induced DSBs in vivo was performed as described previously38.
Results
Identification of methylated arginine residues of hnRNPUL1 in U2OS cells
To identify the methylated arginines within hnRNPUL1, an expression vector encoding FLAG-epitope tagged hnRNPUL1 was transfected in U2OS cells. The cells were lysed under denaturing conditions to identify the post-translational modifications of hnRNPUL1 and not its co-immunoprecipitating proteins. Seventy peptides with 62% coverage of hnRNPUL1 and 42 of the total 66 arginines were detected by mass spectrometry (Supplementary Table1 and Supplementary Figure 1). hnRNPUL1 C-terminus residues 732 to 856 do not contain lysines or arginines and, therefore, were not detected in our analysis. We identified arginines 661, 685 and 690 to harbor monomethyls, while arginines 584, 618, 620, 645 and 656 were dimethylated (Table 1). Arginines 618, 620, 645 and 656 are located within the RGG/RG motifs4. Moreover, four out of the five monomethylated arginines contain an N-terminal asparagine (Table 1).
PRMT1 methylates and associates with hnRNPUL1 in vivo
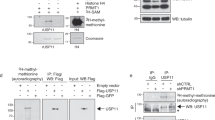
PRMT1 is the major arginine methyltransferase in mammalian cells and it has a preference is for arginines in RG/RGG motifs4. We examined in vitro whether the hnRNPUL1 RGG/RG motifs were indeed substrates of PRMT1. hnRNPUL1 peptide sequences inserted in-frame with glutathione S-transferase (GST, Fig. 1A,B) were incubated with GST-PRMT1 and methyl-3H-S-adenosyl-L-methionine for an in vitro methylation assay. The proteins were stained with Coomassie Blue to confirm their equivalent expression (Fig. 1C, lanes 1 and 3 to 9) and the methylated proteins were visualized by fluorography (Fig. 1C, lanes 10 and 12 to 18). The GST-TriRG, GST-DiRGG and GST-monoRGG proteins were methylated by PRMT1 (Fig. 1C, lanes 16-18). In contrast, the GST-DiRG, GST-RRGR and GST-RIRG proteins were not methylated (Fig. 1C, lanes 13-15). The RGG/RG motif of MRE11 was used as a positive control and GST alone, as a negative control, respectively (Fig. 1C, lanes 10 and 12).
The RGG/RG motif of hnRNPUL1 is methylated by PRMT1 in vitro.
A) SAP designates the SAF- A/B, Acinus and PIAS motif, while BBS denotes BRD7-binding site. The RGG/RG motifs located from 609 to 658 are shown. The RG and RGG repeats are bold and underlined. B) The human hnRNPUL1 peptide sequences fused to recombinant GST used for the methylation assay of panel C. C) PRMT1 in vitro methylation assay using GST-hnRNPUL1 proteins of panel B with H3-SAM. Proteins were resolved by SDS-PAGE, stained with Coomassie blue (left panel), dried and visualized by fluorography (right panel). GST-MRE11:RGG (glycine arginine rich) and GST were used as the positive and negative control, respectively. The indicated sequences (RGG or RG) fused to GST migrate at a similar molecular mass as GST alone because of the short added sequence. GST-MRE11:RGG harbors an extra ~60aa and migrates close to 33 kDa. The migration of GST-PRMT1 is shown with arrows.
We next examined whether PRMT1 was the enzyme responsible for the methylation of endogenous hnRNPUL1. HEK293 cells were transfected with siRNAs targeting luciferase (siCTL) or PRMT1. Cellular extracts were prepared for immunoprecipitation with anti-hnRNPUL1 antibodies and the bound proteins were separated by SDS-PAGE and immunoblotted with ASYM25b, a specific asymmetric dimethylarginine antibody10. hnRNPUL1 was recognized by anti-ASYM25b antibodies in siCTL, but not siPRMT1-transfected cells, suggesting that hnRNPUL1 is hypomethylated in PRMT1-depleted cells (Fig. 2A, α-ASYM25b panel). Equivalent hnRNPUL1 was immunoprecipitated from siCTL and siPRMT1 cells (Fig. 2A, α-UL1 panel). Anti-PRMT1 immunoblotting of whole cell lysates (WCL) confirmed the PRMT1 knockdown and anti-tubulin immunoblotting confirmed equivalent loading (Fig. 2A, lower panels). These findings suggest that PRMT1 is the physiological enzyme responsible for the arginine methylation of endogenous hnRNPUL1.
The RGG/RG motif of hnRNPUL1 is methylated by PRMT1 in vivovia a physical association.
A) Whole cell lysates from HEK293 cells transfected with siControl (CTL) and siPRMT1 were subjected to immunoprecipitation (IP) 48 h post-transfection with the anti-hnRNPUL1 antibody. Whole cell lysates (WCL) and immunoprecipitants were immunoblotted with anti-asymmetrical dimethylarginines (ASYM25b), anti-hnRNPUL1, anti-PRMT1 and anti-Tubulin antibodies. B) HEK293 cells transfected with wild type FLAG-hnRNPUL1 and FLAG-hnRNPUL1RK were lysed and anti-FLAG immunoprecipitations were performed. The bound proteins were separated by SDS-PAGE and immunoblotted with the indicated antibodies. Tubulin was used as loading control.
We next proceeded to substitute all seven arginines located within the two RGG/RG motifs that are likely methylated by PRMT1 (R612K, R618K, R620K, R639K, R645K, R656K and R661K; hnRNPUL1RK; Fig. 1A). We initially examined whether FLAG-hnRNPUL1RK was hypomethylated in vivo. HEK293 cells were transfected with either FLAG-hnRNPUL1 or FLAG-hnRNPUL1RK. The cells were lysed, immunoprecipitated with anti-FLAG antibodies and the presence of methylation was visualized by immunoblotting with ASYM25b. hnRNPUL1, but not FLAG-hnRNPUL1RK, was arginine methylated (Fig. 2B,α-ASYM25b panel). These findings indicate that the arginines that reside within the RGG/RG motifs are the main methylated residues of hnRNPUL1 recognized by ASYM25b antibodies.
We then tested whether endogenous PRMT1 associated with FLAG-hnRNPUL1 and FLAG-hnRNPUL1RK. HEK293 cells transfected with FLAG-hnRNPUL1 or FLAG-hnRNPUL1RK were lysed and co-immunoprecipitation analyses were performed. We observed that PRMT1 co-immunoprecipitated with FLAG-hnRNPUL1, but not FLAG-hnRNPUL1RK (Fig. 2B, α-PRMT1 upper panel). The fact that PRMT1 did not associate with FLAG-hnRNPUL1RK (Fig. 2B, α-PRMT1 upper panel), likely explains its hypomethylation, as PRMT1 is known to associate with its substrates39. These findings suggest that the arginines located within the RGG/RG motifs of hnRNPUL1 are sites of PRMT1 methylation.
Arginine methylation regulates the interaction of hnRNPUL1 with NBS1
hnRNPUL1 interacts with NBS1, a component of the DNA damage sensing complex MRE11-RAD50-NBS128. To determine whether arginine methylation of hnRNPUL1 affects its interaction with NBS1, U2OS cells were co-transfected with a control pcDNA3.1, FLAG-hnRNPUL1 or FLAG-hnRNPUL1RK and YFP-NBS1 expression vectors. The cells were lysed, anti-FLAG antibody immunoprecipitations were performed and the bound proteins were immunoblotted with anti-GFP antibodies. YFP-NBS1 associated with the FLAG-hnRNPUL1, but had a reduced affinity for FLAG-hnRNPUL1RK (Fig. 3A, upper panel). To further determine whether arginine methylation of hnRNPUL1 regulates its interaction with NBS1, we performed co-immunoprecipitation assays in PRMT1-deficient cells. HEK293 cells were transfected with siRNAs for luciferase (siCTL) or PRMT1 along with expression vectors for YFP-NBS1 and FLAG-hnRNPUL1. The cells were immunoprecipitated with anti-FLAG antibodies and the bound proteins were separated by SDS-PAGE and immunoblotted with anti-GFP antibodies. We observed a reduced interaction between NBS1 and hnRNPUL1 in siPRMT1 transfected cells compared to siCTL cells (Fig. 3B). These findings suggest that arginine methylation by PRMT1 regulates the interaction between hnRNPUL1 and NBS1.
Methylation of hnRNPUL1 is required for its interaction with NBS1.
A) U2OS cells were transfected with empty vector (pcDNA 3.1), FLAG-hnRNPUL1 wild type, FLAG-hnRNPUL1RK mutant and YFP-NBS1. Whole cell lysates (WCL) and FLAG-immunoprecipitants were immunoblotted with the indicated antibodies. B) HEK293 cells were transfected with siControl and siPRMT1 along with YFP-NBS1 and FLAG-hnRNPUL1. The whole cell lysates (WCL) and FLAG immunoprecipitates were subjected to western blot analysis using the antibodies indicated.
The methylation of the hnRNPUL1 RGG/RG motifs regulates its recruitment at sites of DNA damage
hnRNPUL1 exhibits two types of dynamics at DNA damage sites depending on the presence or absence of RNA28. hnRNPUL1 is rapidly excluded from laser microirradiation sites, but in the presence of transcription inhibitors, it is effectively recruited at DNA damage tracks28. To assess the role that the RGG/RG motif in recruitment following DNA damage, we transfected U2OS cells with GFP-hnRNPUL1 or GFP- hnRNPUL1RK. The cells were examined for recruitment to laser microirradiated nuclear regions in the absence and presence of the transcriptional inhibitor, 5,6 dichloro-β-D-ribofuranosylbenzimidazole (DRB). Both GFP-hnRNPUL1 and hnRNPUL1RK were excluded from laser microirradiated nuclear regions in U2OS cells without DRB (Fig. 4). GFP-hnRNPUL1 was effectively recruited at sites of DNA damage, while GFP-hnRNPUL1RK was not recruited at these DNA damage sites with DRB treatment (Fig. 4). These findings suggest that arginine methylation of the RGG/RG motifs is required for the recruitment at sites of DNA damage in the presence of transcription inhibitors, but are not required for the exclusion of hnRNPUL1 from sites of DNA damage.
RGG/RG motif regulates GFP-hnRNPUL1 recruitment at breaks.
A) U2OS cells were transfected with wild type GFP-hnRNPUL1 and GFP-hnRNPUL1RK and subjected to laser micro-irradiation treatment (forms a DNA damage ‘track’; white box). Exclusion of GFP-UL1 proteins from laser damage sites (2 min post-laser scissor damage) was monitored via live imaging (exclusion) or recruitment in the presence of DRB (inclusion) B) Quantification of exclusion and inclusion patterns of GFP-UL1 proteins are indicated as a percentage.
Discussion
In the present manuscript, we identify arginines that are mono- and dimethylated within human hnRNPUL1 in U2OS cells. These methylated arginine residues reside mainly within two separate RGG/RG motifs. In vivo and in vitro experiments confirmed that PRMT1 was the enzyme responsible for the methylation of these RGG/RG sequences. Replacement of these methylated arginines or deficiency of PRMT1 expression significantly inhibited the association of hnRNPUL1 with NBS1. Furthermore, the ability of FLAG-hnRNPUL1RK to localize to sites of DNA damage was impaired. These findings suggest that methylation of the RGG/RG motifs regulates key protein-protein interactions for chromatin recruitment of hnRNPUL1.
Previous studies have shown that upon laser scissor damage, hnRNPUL1 forms unique exclusion and recruitment patterns28. hnRNPUL1 is essentially excluded from sites of DNA damage, however, in the presence of transcription inhibitors, hnRNPUL1 is recruited to them28. Our studies revealed that arginine to lysine substitution in the RG/RGG motifs affected hnRNPUL1 recruitment, but not its exclusion (Fig. 4). We demonstrated that arginine methylation of the RGG/RG motifs plays an important role in regulating the association of hnRNPUL1 with the MRN complex, as hypomethylation of hnRNPUL1 had reduced association with NBS1. This may reflect the requirement of the MRN complex for the recruitment of hnRNPUL1 to DSB damage sites. Moreover, it has been reported that hnRNPUL1 lies between MRN and CtIP and BLM, such that hnRNPUL1 promotes DSB resection by regulating BLM recruitment28. FLAG-hnRNPUL1RK associated normally with BLM and PRMT1 deficiency did not affect the hnRNPUL1/BLM association (data not shown). This suggests that methylation of hnRNPUL1′s RGG/RG motifs is functionally involved in its association with upstream components, but not necessarily with the downstream effectors in the DDR pathway. It is also possible that NBS1-hnRNPUL1-PRMT1 forms a complex and loss of PRMT1 destabilizes this interaction, without involvement of arginine methylation.
hnRNP proteins have been implicated in numerous cellular processes including RNA metabolism, telomere elongation, DNA repair and chromatin reorganization40. hnRNPUL1 has sequence homology to hnRNPU, also known as scaffold attachment factor A (SAF-A), that is a nucleic acid binding ribonucleoprotein which has a characteristic binding preference for scaffold associated DNA regions including areas rich in A/T stretches41,42. hnRNPU contains an RGG/RG motif from residue 778 to 793 which is the preferred site for arginine methylation by PRMT143.
Defects in PRMT1-deficient cells have been previously characterized21. It has been shown that PRMT1 is essential for early development using a conditional null allele of PRMT1 in mice, as null embryos die at embryonic day 6.520,21. PRMT1-deficient MEFs display genomic instability and exhibit spontaneous DNA damage, checkpoint defects and delays in cell cycle progression21. The arginine methylation of the DNA damage response and RNA binding protein hnRNPUL1 further supports a role for PRMT1 in the DDR pathway.
The role of arginine methylation and PRMTs in the DDR pathway has not yet been fully characterized. Nevertheless, we observed the methylation status of hnRNPUL1 in PRMT1 knockdown cells using siRNA. hnRNPUL1 was shown to be hypomethylated in the PRMT1-deficient cells, demonstrating that PRMT1 is the physiological enzyme responsible for the methylation of RGG/RG motif of hnRNPUL1. We did not observe an increase in the arginine methylation of hnRNPUL1 with DNA damage, suggesting that the regulation is likely with associated proteins (data not shown). Previous studies have implicated hnRNPUL1 to be required for resection at DSBs and to be required for optimal ATR activation. It was proposed that there are two pools of hnRNPUL1 in the cell, such that one pool is involved in RNA metabolism, while the other participates in DSB repair28 and that perhaps arginine methylation regulates accessibility between these two pools.
Additional Information
How to cite this article: Gurunathan, G. et al. Arginine methylation of hnRNPUL1 regulates interaction with NBS1 and recruitment to sites of DNA damage. Sci. Rep. 5, 10475; doi: 10.1038/srep10475 (2015).
References
Bedford, M. T. & Richard, S. Arginine methylation an emerging regulator of protein function. Mol Cell 18, 263–72 (2005).
Bedford, M. T. & Clarke, S. G. Protein arginine methylation in mammals: who, what and why. Mol Cell 33, 1–13 (2009).
Yang, Y. & Bedford, M. T. Protein arginine methyltransferases and cancer. Nat Rev Cancer 13, 37–50 (2013).
Thandapani, P., O’Connor, T. R., Bailey, T. L. & Richard, S. Defining the RGG/RG motif. Mol Cell 50, 613–23 (2013).
Boisvert, F. M., Côté, J., Boulanger, M. C. & Richard, S. A Proteomic Analysis of Arginine-methylated Protein Complexes. Mol Cell Proteomics 2, 1319–30 (2003).
Auclair, Y. & Richard, S. The role of arginine methylation in the DNA damage response. DNA Repair 12, 459–65 (2013).
Carney, J. P. et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93, 477–86 (1998).
Boisvert, F.-M., Déry, U., Masson, J.-Y. & Richard, S. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes & Dev 19, 671–676 (2005).
Déry, U. et al. A Glycine-Arginine Domain in Control of the Human MRE11 DNA Repair Protein. Mol Cell Biol 28, 3058–3069 (2008).
Yu, Z. et al. The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res 22, 305–320 (2012).
Adams, M. M. et al. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle 4, 1854–61 (2005).
Boisvert, F.-M., Rhie, A., Richard, S. & Doherty, A.J. The GAR Motif of 53BP1 is Arginine Methylated by PRMT1 and is Necessary for 53BP1 DNA Binding Activity. Cell Cycle 4, 1834–1841 (2005).
Jansson, M. et al. Arginine methylation regulates the p53 response. Nat Cell Biol 10, 1431–9 (2008).
Guo, Z. et al. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat Chem Biol 6, 766–73 (2010).
Guendel, I. et al. Methylation of the tumor suppressor protein, BRCA1, influences its transcriptional cofactor function. PLoS One 5, e11379 (2010).
He, W. et al. A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res 39, 4719–27 (2011).
El-Andaloussi, N. et al. Arginine methylation regulates DNA polymerase beta. Mol Cell 22, 51–62 (2006).
Feng, Y. et al. Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J Biol Chem 288, 37010–25 (2013).
Pedersen, M. T. & Helin, K. Histone demethylases in development and disease. Trends Cell Biol 20, 662–71 (2010).
Pawlak, M. R., Scherer, C. A., Chen, J., Roshon, M. J. & Ruley, H. E. Arginine N-Methyltransferase 1 Is Required for Early Postimplantation Mouse Development, but Cells Deficient in the Enzyme Are Viable. Mol. Cell. Biol. 20, 4859–4869 (2000).
Yu, Z., Chen, T., Hébert, J., Li, E. & Richard, S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol Cell Biol 29, 2982–96 (2009).
Guccione, E. et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–7 (2007).
Iberg, A. N. et al. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem 283, 3006–10 (2008).
Michaud-Levesque, J. & Richard, S. Thrombospondin-1 is a transcriptional repression target of PRMT6. J Biol Chem 284, 21338–46 (2009).
Neault, M., Mallette, F. A., Vogel, G., Michaud-Levesque, J. & Richard, S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res 40, 9513–21 (2012).
Moumen, A., Masterson, P., O’Connor, M. J. & Jackson, S. P. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123, 1065–78 (2005).
Krietsch, J. et al. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res 40, 10287–301 (2012).
Polo, Sophie E. et al. Regulation of DNA-End Resection by hnRNPU-like Proteins Promotes DNA Double-Strand Break Signaling and Repair. Mol Cell 45, 505–516 (2012).
Adamson, B., Smogorzewska, A., Sigoillot, F. D., King, R. W. & Elledge, S. J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 19, 318–28 (2012).
Chowdhury, D., Choi, Y. E. & Brault, M. E. Charity begins at home: non-coding RNA functions in DNA repair. Nat Rev Mol Cell Biol 14, 181–9 (2013).
Francia, S. et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488, 231–5 (2012).
Pryde, F. et al. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J Cell Sci 118, 2043–55 (2005).
Gabler, S. et al. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 72, 7960–71 (1998).
Kzhyshkowska, J. et al. Heterogeneous nuclear ribonucleoprotein E1B-AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem J 358, 305–14 (2001).
Kzhyshkowska, J., Kremmer, E., Hofmann, M., Wolf, H. & Dobner, T. Protein arginine methylation during lytic adenovirus infection. Biochem J 383, 259–65 (2004).
Côté, J., Boisvert, F. O.-M., Boulanger, M.-C., Bedford, M. T. & Richard, S. Sam68 RNA Binding Protein Is an In Vivo Substrate for Protein Arginine N-Methyltransferase 1. Molecular Biology of the Cell 14, 274–287 (2003).
Boisvert, F. M. et al. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J Cell Biol 159, 957–69 (2002).
Laffitte, M. C. et al. Formation of linear amplicons with inverted duplications in Leishmania requires the MRE11 nuclease. PLoS Genet 10, e1004805 (2014).
Boisvert, F. M., Chénard, C. A. & Richard, S. Protein Interfaces in Signaling Regulated by Arginine Methylation. Sci. STKE 271, re2 (2005).
Glisovic, T., Bachorik, J. L., Yong, J. & Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582, 1977–86 (2008).
Romig, H., Fackelmayer, F. O., Renz, A., Ramsperger, U. & Richter, A. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J 11, 3431–40 (1992).
Britton, S. et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res 42, 9047–62 (2014).
Herrmann, F., Bossert, M., Schwander, A., Akgün, E. & Fackelmayer, F. O. Arginine methylation of scaffold attachment factor A by heterogeneous nuclear ribonucleoprotein particle-associated PRMT1. J Biol Chem 279, 48774–9 (2004).
Acknowledgements
We thank Éric Bonneil at Université de Montréal for performing the mass spectrometry analysis. This work was supported by grant MOP-93811 from the Canadian Institutes of Health Canada (CIHR).
Author information
Authors and Affiliations
Contributions
G.G., Z.Y and Y.C. performed experiments analyzed data and wrote the manuscript. G.G. and Z.Y. generated Figure 1, 2, 3 and Table 1 and Y.C. generated Figure 4. J.Y.M interpreted and analyzed the data for Figure 4 and edited the manuscript. S.R. conceived the study, analyzed data and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Gurunathan, G., Yu, Z., Coulombe, Y. et al. Arginine methylation of hnRNPUL1 regulates interaction with NBS1 and recruitment to sites of DNA damage. Sci Rep 5, 10475 (2015). https://doi.org/10.1038/srep10475
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep10475
This article is cited by
-
PRMT1 and PRMT5: on the road of homologous recombination and non-homologous end joining
Genome Instability & Disease (2022)
-
Altered gene expression and PTSD symptom dimensions in World Trade Center responders
Molecular Psychiatry (2022)
-
Synergistic effects of type I PRMT and PARP inhibitors against non-small cell lung cancer cells
Clinical Epigenetics (2021)
-
PBX1: a key character of the hallmarks of cancer
Journal of Molecular Medicine (2021)
-
PRMT1-dependent regulation of RNA metabolism and DNA damage response sustains pancreatic ductal adenocarcinoma
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.