Abstract
Cholesteryl ester transfer protein (CETP) mediates the transfer of cholesterol esters (CE) from atheroprotective high-density lipoproteins (HDL) to atherogenic low-density lipoproteins (LDL). CETP inhibition has been regarded as a promising strategy for increasing HDL levels and subsequently reducing the risk of cardiovascular diseases (CVD). Although the crystal structure of CETP is known, little is known regarding how CETP binds to HDL. Here, we investigated how various HDL-like particles interact with CETP by electron microscopy and molecular dynamics simulations. Results showed that CETP binds to HDL via hydrophobic interactions rather than protein-protein interactions. The HDL surface lipid curvature generates a hydrophobic environment, leading to CETP hydrophobic distal end interaction. This interaction is independent of other HDL components, such as apolipoproteins, cholesteryl esters and triglycerides. Thus, disrupting these hydrophobic interactions could be a new therapeutic strategy for attenuating the interaction of CETP with HDL.
Similar content being viewed by others
Introduction
An elevated level of low-density lipoprotein-cholesterol (LDL-C) and/or a low level of high-density lipoprotein cholesterol (HDL-C) in human plasma are major risk factors for cardiovascular disease (CVD). Cholesteryl ester transfer protein (CETP), with a molecular mass of 53 kDa (composed of 476 amino acids) before post-translational modification1, mediates the cholesteryl ester (CE) transfer from high-density lipoprotein (HDL) to low-density lipoprotein (LDL) and very low-density lipoprotein (VLDL) and in exchange for triglyceride (TG). CETP deficiency has been shown to be associated with elevated HDL cholesterol levels2,3. Loss-of-function mutations in the CETP gene were negative associated with premature atherosclerosis and have been implicated in longevity in some studies2. On the other hand, there is also evidence that CETP mutations are associated with an increased incidence of coronary heart diseases4. Despite these inconsistencies and because of the urgent public desire to expand treatment options beyond statins, the most financially successful drug to reduce LDL-C levels to date, CETP has been used as a promising drug target for designing inhibitors in order to treat heart disease5,6,7,8,9,10. Four large clinical trials of CETP inhibitors5,8,9,10 have been undertaken to date. The first two CETP inhibitors failed in phase III clinical trials due to increase mortality related to off-target effects and lack of efficacy. Two other CETP inhibitors9,10 are currently being investigated in large clinical outcome trials. As such, an improved understanding of CETP's molecular interactions could eventually provide beneficial and definitive descriptions of CETP function, thereby leading to new CETP-related drug design.
The structure of CETP, revealed by X-ray crystallography, resembles a banana shape with dimensions of roughly 3 × 3 × 13 nm and contains four structural components: an N-terminal β-barrel domain, a C-terminal β-barrel domain, a central β-sheet and a C-terminal extension (a distorted amphipathic helix, helix X, Glu465-Ser476 at the C terminus)11,12. Electron microscopy (EM) has shown that CETP forms a bridge between HDL and LDL, with its N-terminal β-barrel domain penetrating the HDL surface and its C-terminal β-barrel domain penetrating the LDL surface13. A molecular dynamics (MD) simulation has revealed that the distal portions of the N- and C-terminal β-barrel domains of CETP remain highly flexible in solution14. This flexibility may be necessary for conformational changes to occur at the distal ends, a necessary step for the formation of a tunnel through the entire molecule13,15.
Although CETP has been intensively studied, a detailed understanding of how CETP senses and binds to HDL remains unknown due to the heterogeneity and dynamics of HDL. HDLs vary in size, shape and composition16,17. Considering that most plasma CETPs are naturally bound to HDLs18, an investigation of how different components of HDL affect the interaction of spherical HDL with CETP is essential for a complete understanding of CETP function.
Here, we studied CETP interactions with various HDLs and liposome vesicles using optimized negative-staining (OpNS), cryo-electron tomography (Cryo-ET) and molecular dynamics simulations in order to understand how the different HDL components affect CETP binding.
Results
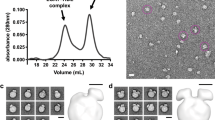
EM images of CETP bound to plasma HDL2
Based on previous reports, the spherical HDL2 in plasma varies in diameter (~9 to ~15 nm) and in density (from 1.063 to 1.125 g/ml)19,20. Earlier studies18,19,21 showed the average molecular weight of HDL2 is 360 kDa and that the particles contain three major surface components: apolipoprotein A-I and A-II (apoA-I and apoA-II) (total ~40.2% of MW) and phospholipids (~31.3% of MW), as well as core lipids: cholesterol esters (~17.6% of MW) and triglycerides (~4.2% of MW). A small amount of free cholesterol (~5.8% of MW) is distributed between the particle surface and the core21.
In the present study, CETP was incubated with isolated HDL2 at a molar ratio of 1:4 (HDL2: CETP), then prepared as described for optimized negative-staining (OpNS) EM method22,23 (a method that minimizes rouleaux formation) and examined by electron microscopy (EM)23,24,25. A large field EM micrograph and representative particles showed that CETP-HDL2 complexes had the appearance of rod shaped CETP penetrating spherical shaped HDL2 (Fig. 1a). No CETP was found to bridge two HDL2 particles or to adhere to the convex surface of HDL2 via its concave surface as hypothesized by crystallography11. The HDL2 bound to CETP had an average diameter of 12.7 ± 1.6 nm (Fig. 1e), similar to HDL2 alone (12.8 ± 1.3 nm) (Supplementary Fig. 1e). The bound CETP particles were 8.7 ± 1.7 nm in length and 3.2 ± 0.5 nm in width (Fig. 1f). Although the width of bound CETP was similar to that of CETP itself (~3.5 nm), the length of the bound CETP was much shorter (~8.7 vs. ~12.5 nm) (Supplementary Fig. 1e), suggesting that ~3–4 nm of the CETP molecule length had been inserted into the HDL2 surface (Supplementary Fig. 1e). The EM micrographs and statistical analyses showed that more than 50% of the HDL2 bound to CETP, in which, ~30.3% ± 2.7% (mean ± sd) of the HDL2 particles bound to one CETP molecule, while 15.7% ± 4.9% (mean ± sd) of the HDL2 bound to two CETP molecules and 4.7% ± 4.3% (mean ± sd) of the HDL2 bound to more than two CETP molecules. Among these bound HDL2, ~40% bound to two or more CETP molecules. In contrast, no CETP molecule bound to more than one HDL2 particle simultaneously.
Negative-staining electron micrographs of HDL subclasses and liposome incubation with CETP by optimized negative-staining (OpNS).
The samples of CETP incubated with, (a) plasma HDL2, (b) plasma HDL3, (c) reconstituted spherical HDL (rHDL) and (d) 1-Palmitoyl-2-oleoylphosphatidylcholine (POPC) liposome vesicles were prepared by optimized negative-staining (OpNS). Survey view (left panel), representative conformations of particles (top right panel) and schematics of the particle structures and percentages of each conformation (bottom right panels). (e) Histogram of particle diameter (left panel, sampling step 0.5 nm, fitted with sixth-degree polynomial functions) and shape (right panel, sampling step 0.04, fitted with sixth-degree polynomial functions). The diameter is calculated based on the geometric mean of two measured diameters (in which one is the longest direction while the other is perpendicular to it); the shape is calculated based on the ratio of these two diameters. (f) The average diameters of CETP, lengths vs. widths. Particle window size is 37 nm in a, b and c. Particle window size is 59 nm in d. All scale bars, 90 nm.
TEM images of CETP bound to plasma HDL3
To investigate how three major HDL surface components: phospholipids, apoA-I and apoA-II affected the CETP binding, the above experiment was repeated with HDL3 that contain a different surface protein and lipid ratio than HDL2. Earlier studies showed21 the HDL3 vary in size, but have an average molecular weight of 175 kDa, a density of ~1.125 to ~1.210 g/ml and contain apoA-I and apoA-II (~55.5% of MW), phospholipids (~22.7% of MW), free cholesterol (~2.8% of MW), cholesterol esters (~14.7% of MW) and triglycerides (~3.4% of MW)21. Thus, HDL3 has a lower surface percentage of lipids, but a higher surface percentage of proteins than HDL2.
In this study, the HDL3 sample was incubated with CETP at a molar ratio of 4:1 (CETP:HDL3), prepared for OpNS and examined by EM. The EM micrographs showed that the CETP-HDL3 complexes were similar to CETP-HDL2 complexes, with the rod-shaped CETP adhering to the spherical shaped surface of HDL3 (Fig. 1b). As before, no CETP bridged two HDL3 particles or adhered to the convex surface of HDL3 via its concave surface. The diameter of HDL3 that was bound to CETP (Fig. 1e) was ~11.4 ± 1.7 nm, similar to that of HDL3 alone (~10.9 ± 1.0 nm) (Supplementary Fig. 1e). The rod shaped CETP in the complexes had a similar width to CETP itself (Supplementary Fig. 1e), but were shorter in length (~8.6 ± 1.2 nm, Fig. 1f). The micrographs and statistical analyses showed more than 40% of the HDL3 particles were bound to CETP (compared to ~50% for HDL2), in which 32.8% ± 7.0% (mean ± sd) of the HDL3 particles bound to one CETP molecule, while 8.2% ± 1.4% (mean ± sd) of the HDL3 particles bound to two and more CETP molecules. Among these bound HDL3, ~20.2% was also bound to two or more CETP molecules (Fig. 1b). Considering that HDL3 and HDL2 have the same surface components (apoA-I, apoA-II and phospholipids), the lower percentages (~41.1% vs. ~50.7%) of CETP bound to HDL that have a lower percentage (22.7% vs. 31.3%) of surface phospholipids suggest that the surface phospholipids/protein ratio may have contributed to some variation in CETP binding.
TEM images of CETP bound to spherical recombined HDL (rHDL)
To investigate which HDL surface component dominates CETP binding, one of three surface components, apoA-II, was excluded by repeating the above experiment using reconstituted HDL (rHDL)26. The rHDL contain apoA-I as the only protein (~43% in MW) and POPC as the only phospholipid (~32.9% in MW). Cholesteryl esters were the only core lipid components (~21.7% in MW)26 and a small amount free cholesterol (~2.4%) was distributed between the surface and the core.
In this study, the rHDL sample was incubated with CETP (molar ratio of 4:1 for CETP:rHDL) prepared for OpNS23 and examined by EM. The EM micrographs showed ~42.9% of rHDL particles were attached to CETP. No CETP was found to connect two rHDL particles or bind to the rHDL surface via its concave surface. The average diameter of rHDL particles in rHDL-CETP complexes was 10.2 ± 1.2, similar to rHDL (10.3 ± 0.7 nm) alone (Supplementary Fig. 1e). The CETP protrusion length of ~9.2 ± 1.3 nm was shorter than that of CETP alone, but slightly longer than the CETP protrusions on the surfaces of HDL2 and HDL3 (~9.2 nm vs. ~8.6 nm, Fig. 1f). The micrographs and statistical analyses showed ~34.6% ± 5.3% (mean ± sd) of the rHDL particles bound to one CETP molecule and ~7.6% ± 3.8% and ~0.7% ± 1.3% (mean ± sd) of the rHDL particles bound to two and more CETP molecules. Approximately 19.4% of the CETP-rHDL complexes were bound to two or more protruding CETP molecules. Notably, most multi-CETP binding complexes (~75%) showed the protruding CETP sharing the same hemisphere of rHDL (Fig. 1c). Moreover, the total number of CETP molecules bound to rHDL can exceed the total number of apoA-I molecules in each rHDL particle (Supplementary Fig. 2), which is consistent to the previous observation13. These observations showed that the apoA-II in HDL does not play a significant role in binding to CETP.
EM images of CETP bound to liposomes
To investigate which one of the remaining two HDL surface components, apoA-I or phospholipids, dominates the CETP binding, a lipid vesicle without apoA-I was further used to repeat the above experiments. The lipid vesicle (a POPC-only liposome) was incubated with CETP and the sample was prepared by OpNS and examined by EM. The EM micrographs and representative liposomes showed the liposomes had a distribution of ~7.7 to ~116.2 nm in diameter (Fig. 1d), which is within the diameter range of all types of lipoproteins (from small HDL to large VLDL). Zoomed in views of a micrograph (Supplementary Fig. 3) and representative particle images (Fig. 1d, top right panel) showed spherical liposomes with surface CETP protrusions with an average length of ~8.5 ± 0.6 nm (Fig. 1f). The liposome particles retained their shape and structure after CETP insertion. Incubation with CETP did not cause particle disruption. Additionally, CETP did not form a bridge between two liposome particles, or bind to a liposome so that its concave surface was adjacent to the convex surface of the liposome. The micrographs and statistical analyses showed ~19.1% ± 3.2% of the liposome particles bound to 1–2 CETP molecules, ~6.0% ± 2.7% of the liposome particles bound to 3–4 CETP molecules and ~5.1% ± 3.6% and 2.7% ± 1.7% bound to 5–6, or 7 and more CETP molecules respectively. In total, ~32% of the liposomes contained at least one surface CETP protrusion. Among these CETP bound liposomes, ~66% were bound to two or more CETP molecules (Fig. 1d). Notably, smaller liposomes had more CETP molecules bound (Fig. 2). For instance, 5–7 CETP molecules were bound to liposomes ~15 nm in diameter, while liposomes over ~40 nm were rarely bound to CETP (Fig. 2b and Supplementary Fig.4). This relationship was analyzed based on a linear fitting the number of bound CETP against the liposome diameters (Fig. 2a). This result suggests that the smaller liposomes had a higher binding affinity to CETP. Moreover, it confirmed that apoA-I in HDL do not dominate the CETP binding.
Distributions of the number of bound CETP on liposome against liposome diameter.
Panel (a) shows a linear plot of mean of liposome diameters versus the number of its binding CETP molecules. The mean diameter was calculated within each CETP binding number group. The data were approximated by linear regression (R = −0.98, p = 2.06E-05). Panel (b) shows a plot of the mean number of binding CETP molecules versus their binding liposome diameter (black line). As some CETP molecules that are located behind/in-front of the liposomes may not be counted, a geometric model (shown in supplementary Fig. 4) was used to adjust the binding number. The adjusted/calibrated number of bound CETP molecules versus the liposome diameter was also plotted (shown in blue line). The data was fitted with a sixth-degree polynomial function (histograms have a sampling step of 2.17 nm on liposome diameter) using a total of 3,282 liposome/liposome-CETP complexes. Each liposome diameter was calculated as the geometric mean (the square root of the product) of two perpendicular diameters.
Three dimensional structure of a complex of CETP bound to liposome by individual-particle cryo-electron tomography
To confirm that the conformation of liposome-CETP complex was not due to negative-stain artifact, the sample was also flash-frozen in vitreous buffer without any negative-staining. The frozen sample was then examined by cryo-electron microscopy (cryo-EM) at −170°C and under a low-dose imaging condition (Fig. 3a). The micrographs and selected particles showed the CETP liposome surfaces adhering with rod-shape CETP (arrows in Fig. 3b), similar to that from the OpNS (Fig. 1d). To reveal the detailed structure in three dimensions (3D), the sample was also imaged by cryo-electron tomography (cryo-ET) through a series of tilting angles (angle range of −57° to 60° in steps of 1.5°). A 3D structure of a representative complex was reconstructed by individual-particle electron tomography (IPET)27. IPET was designed for 3D reconstruction of an asymmetric individual particle rather than averaging different particles like in conventional single-particle reconstruction. The naturally varying particle diameter of liposomes made it impossible to achieve any 3D reconstruction by conventional cryo-EM single-particle reconstruction methods.
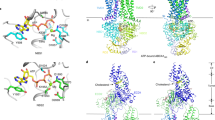
Electron cryo-micrograph (cryo-EM) and 3D reconstruction of a CETP-liposome complex.
(a) Cryo-EM images of the sample of POPC liposomes incubated with CETP. (b) Four representative liposome-CETP complexes. (c) Ab-inito 3D reconstruction of an individual liposome-CETP complex by individual-particle electron tomography (IPET). The complex was imaged by electron cryo-tomography (cryo-ET, tilting angles ranging from −57° to 60° in steps of 1.5°). Seven representative tilting views (contrast reversed) of a targeted liposome-CETP complex (CTF corrected) were compared to the projections of the intermediate 3D reconstructions (column 2 to 3) and final reconstruction (column 4) at their corresponding angles. The final 3D reconstructions (before and after missing wedge correction) viewing from the corresponding angles were also displayed in last two columns. (d) The final 3D reconstruction of a targeted liposome-CETP complex (resolution of 3.5 nm–7.2 nm based on the Fourier shell correlation, FSC, curve declined to a value of 0.5 and 0.143). (e) Ridge-body docking a CETP crystal structure (PDB ID: 2OBD) into an obvious protrusion of the 3D reconstruction. (f) Schematic of a CETP-liposome complex. The CETP protrusions in all above images were indicated by arrows. Scale Bars: a–c, 50 nm; d, 10 nm.
A total of 79 tilting images of a representative liposome-CETP complex were windowed from CTF-corrected (by TOMOCTF28) cryo-ET micrographs (Fig. 3c, left panel). Although the tilt images (but CTF corrected) were noisy, a protrusion of CETP was marginally visible (arrows in Fig. 3c, left panel). To confirm that the protrusion was from a bound CETP instead of noise, three images from consecutively tilted angles (28.5°, 30° and 31.5°) were averaged together to enhance the signal contrast and to reduce the noise level. The averaged image showed the protrusion was more clearly visible (arrow in Supplementary Fig. 5a) than in any individual tilt image, suggesting that the protrusion is indeed from CETP instead of noise. During IPET reconstruction, the tilt images were gradually aligned precisely to their “global center” via an iteration process27. The noise in the 3D reconstructions and projections was gradually eliminated as the signal was enhanced (Fig. 3c from second to last column). The protrusion remained visible in each intermediate 3D reconstruction and projection (arrows indicated in Fig. 3c from second to fifth column) as well as the final 3D reconstruction (Fig. 3d). To further confirm whether the protrusion in the final 3D reconstruction was from real signal instead of noise, five central slices (arrows indicated in Supplementary Fig. 5b) of the final 3D reconstruction were directly averaged together. This averaged image and its contour map showed the protrusion remained significant (Supplementary Figs. 5c and d). Fourier shell correlation (FSC) analysis showed the resolution is from ~35 Å to 72 Å based on Supplementary Fig. 6. By rigid-body docking the crystal structure of CETP (PDB entrance: 2OBD11) into the protrusion, a near perfect match to the protrusion diameter in width, but ~5 nm shorter in length, suggests a CETP penetrating in the liposome surface (Fig. 3e and f). This experiment confirmed the conformation where CETP penetrates the liposome surface is not due to a negative-staining artifact.
MD simulation study of the relationship between liposome size and surface hydrophobicity
To understand why CETP interacts with the lipid surface of HDL/liposome without apoA-I or apoA-II (Fig. 1) and why smaller liposomes had a higher binding affinity to CETP (Fig. 2), we hypothesized that a higher surface curvature in a smaller HDL/liposome29 will generate a higher degree of surface hydrophobicity (Supplementary Fig. 7b) and increase the affinity of the interaction of the relatively hydrophobic distal end of the CETP N-terminal domain with the liposome surface13 (Supplementary Fig. 7a).
To test this hypothesis, we employed MD simulation to study the liposome surface hydrophobicity against the liposome size by generating a series of liposome vesicles with diameters of ~12 nm, ~20 nm, ~27 nm, ~35 nm and ~42 nm (Fig. 4 and supporting Table 1). After energy minimization (Supplementary Fig. 8), the analyses on the surface hydrophobicity of each lipid vesicle showed that the percentages of surface hydrophobic area (measured by the solvent accessible surface area, SASA14) were ~12.9%, ~9.2%, ~8.0%, ~7.4% and ~7.3% respectively (Fig. 4b and d), suggesting that smaller liposomes had a higher hydrophobicity than the larger liposomes (Fig. 4e).
Hydrophobicity analysis of POPC liposome by molecular dynamics (MD) simulation.
(a) A cross-section view of initial model and the processes to achieve a 12.2 nm liposome by MD simulations. The initial model (cross-section view, left panel), a phospholipids double-layer sphere with an open pore, was used. The pore allows the outer- and inner-shell phospholipids exchange to maintain the balance between the outer and inner shells during MD simulation. By energy minimization, the pore was gradually closed while the liposome shrank (cross section views, right two panels). (b) After MD simulation for 90 ns, the pore was completely closed and an energy minimized liposome, 12.2 nm in diameter, was achieved. (c) Plot of the liposome surface hydrophobic area percentage against the MD simulation time. The percentages were computed by taking their hydrophobic surface area divided by the whole liposome surface area. The three triangles shown in the diagram indicated the three intermediate states of the above liposome during energy minimization. (d) Four larger size liposomes with diameters of 19.6 nm, 26.0 nm, 34.4 nm and 42 nm were also achieved by the energy minimization. (e) Plot of the percentages of surface hydrophobic area against the liposome diameters. The data was approximated by linear regression (R = 0.99, p = 7.99E-04). Cyan for the hydrophilic components of POPC molecule and yellow for the hydrophobic lipid tails. Scale Bars: a, 3 nm; b, d and e, 10 nm.
Discussion
Our experiments showed that, i) the similar morphology in CETP binding between plasma HDL and rHDL implied that the absence of ApoA-II and TG in rHDL did not affect CETP binding to HDL (Fig. 1); ii) the total number of CETP molecules bound to rHDL can exceed the total number of apoA-I molecules in each rHDL particle; iii) non-apoA-I containing liposomes showed similar CETP protrusions to apoA-I containing HDL (Fig. 1d); iv) that there is a significant correlation between liposome size and number of bound CETP molecules; and v) a significant correlation between liposome size and the percentage of surface hydrophobic area. Those results suggest that apoA-I and apoA-II may not be involved in CETP binding; however, the surface phospholipids and surface curvature likely dominate the interaction of CETP with the surface of HDL.
A larger surface lipid area provides a higher opportunity for CETP binding. We noticed that HDL2, which has has a larger diameter than HDL3 (~13 vs. ~11), also has a higher percentage of bound CETP particles than HDL3 (~50% vs. ~42%). This may due to the HDL2 surface lipids occupying a larger area than the surface lipids of HDL3 (~31.3% vs. ~22.7%). This is consistent with liposomes which are larger than HDL2 or HDL3 having ~6 bound CETP molecules (Fig. 1d and 2b), compared to the average of 1–2 CETP molecules that are bound to HDL2 and HDL3 (Fig. 1a and b). This suggests that the binding affinity of CETP is regulated by surface lipid area, as well as surface curvature.
Our results favor the tunnel mechanism rather than the shuttle mechanism for the transfer of CE from HDL to LDL via CETP. Both of these mechanisms were hypothesized to be involved in the CETP-mediated transfer of CE between HDL and LDL two decades ago30,31. In the shuttle mechanism, CETP interacts with HDL and acquires CEs, after which the “CE-enriched” CETP is released from the HDL for subsequent deposition into LDL. The “CE-poor” CETP is then released from the LDL surface for subsequent interaction with HDL in another cycle of CE transfer32. In the tunnel mechanism, CETP binds to HDL and LDL simultaneously to form a ternary complex. CETP then undergoes a conformational change to form a hydrophobic tunnel whereby CEs transfer from HDL to LDL.
Our results showed that CETP bound to only one HDL particle at a time and did not form a bridge between two HDL2 HDL3 or rHDL particles. This suggests that the binding of CETP to HDL is directional. When the fact that i) we were unable to demonstrate CETP bridging two HDL particles (Fig. 1), ii)small HDL/liposome particles with a higher surface curvature and hydrophobicity are more likely to bind to more CETP molecules (Fig. 1 and 2), iii) the N-terminal β-barrel domain of CETP binds to HDL while the C-terminal β-barrel domain binds to LDL13 and iv) the distal end of the N-terminal β-barrel domain of CETP is relatively hydrophobic11, are considered in light of the current results which show that CETP binds to only one liposome particle at a time and does not bridge two liposomes (Fig. 1, 2 and 3), it follows that the distal end of the CETP N-terminal β-barrel domain is most likely inserted into the liposome particle surface via a protein-lipid interaction. This interaction is different for LDL particles, where the distal end of the C-terminal β-barrel domain of CETP inserts to the LDL surface13, possibly via a protein-protein interaction. The conclusion that there is a reduced interaction of the distal end of the N-terminal domain of CETP with LDL surface lipids may be due to the fact that LDL particles have a flattened, ellipsoidal shape with planar opposing faces that, as shown in our earlier cryo-EM single-particle reconstruction33, are largely covered by apoB-100 (~500 kDa). The reduced lipid surface area combined with the reduced surface curvature of the particles is likely to have contributed to the low binding affinity of the hydrophobic distal end of the CETP N-terminal β-barrel domain to LDL.
CETP directionally bound to HDL is consistent with many earlier reports, including i) a CETP-mediated net transfer of CE into VLDL34, ii) CETP bridging HDL and LDL instead of bridging two HDL or LDL particles13, iii) the distal portion of the N-terminal β-barrel domain of CETP being more flexible in solution than is indicated by its crystal structure14; and iv) MD simulations showing that the penetration of the N-terminal β-barrel domain of CETP into the HDL surface generates an opening that allows CE to access the CETP tunnel15. However, our results are inconsistent with those of other investigators who have shown that CETP binds to the edge of discoidal HDL35 via an interaction with apoA-I36 and atomistic and coarse-grained simulations of CETP–HDL interaction which have shown that the N-terminus helix-X domain of CETP penetrates deeply into the HDL particle core37. Nonetheless, our results favor a tunnel, not a shuttle mechanism whereby CETP no longer binds to HDL when it has acquired an appropriate number of CEs from HDL.
While we have illuminated some of the mechanism behind CETP, several questions remain: i) how the CETP hydrophobic N-terminal β-barrel domain of CETP penetrates the HDL surface and with such high specificity (e.g. not using the C-term β-barrel domain? Normally, as hydrophobic interactions are not specific, it is unclear how much this affinity relate to the surface curvature interactions in concert with the N-term distal end to open up); ii) how CE molecules can be transferred through a ~10 nm channel; iii) how TGs can be transferred back to HDL from LDL; iv) how CE and TG exchange between LDL and VLDL; v) how CETP homo-exchanged the radiolabeled lipid transfer among HDL particles. Our results highlight the CETP N-terminal β-barrel domain hydrophobic distal end as a potential drug target, which may lead to a next-generation drug to treat CVDs.
Methods
Protein and liposome isolation
Recombinant human CETP (~53 kDa with no post-translational modifications) was expressed and purified from the Chinese hamster ovary cell line DG441, as previously described11. The CETP concentration was determined by absorbance at 280 nm. Native plasma HDL2 and HDL3 in phosphate buffered saline (PBS) were isolated from fresh, pooled samples of human plasma by ultracentrifugation as reported38. Pooled samples from multiple donors were used to make sure that the sample was representative and that the results could not be attributed to something that specifically related to a single individual. Spherical, reconstituted HDL (rHDL) in PBS were prepared39,40. Liposome vesicle samples were produced by Encapsula NanoSciences (Brentwood, TN). The sample contained 1 mg/ml 1-Palmitoyl-2-oleoylphosphatidylcholine (POPC, from Avanti Polar lipids) with a peak vesicle size of ~50 nm in a buffer containing 20 mM Tris-Cl, 154 mM NaCl, pH 7.4.
Specimen preparation for negative-staining EM
Specimens were prepared for EM using the optimized negative-staining (OpNS) protocol23,25, which minimizes the rouleaux artifact that is observed with lipoproteins23,24,25. In brief, CETP (0.28 mg/ml) was incubated with HDL2, HDL3, rHDL and liposomes, respectively at 37°C for 15 minutes at a molar ratio of ~4:1 (CETP: lipid macromolecular particle). CETP-lipid macromolecular complexes were diluted to 1.5 μg/ml with Dulbecco's phosphate buffered saline (DPBS). A 4 μl aliquot was placed on a thin-carbon-coated copper grid (300 mesh TEM grid, Cu-300CN, Pacific Grid-Tech, San Francisco, CA) that had been glow-discharged. After one minute, excess solution was blotted with filter paper, followed by washing and negative staining with 1% (w/v) uranyl formate (UF)23,25. After air drying, the grids were further dried for an hour at 40°C.
Electron microscopy data acquisition and image pre-processing
The OpNS micrographs were acquired under defocus between ~0.6 um to ~2.2 um on a Gatan UltraScan 4 K × 4 K CCD equipped on a Zeiss Libra 120 Plus transmission electron microscope (Carl Zeiss NTS GmbH, Oberkochen, Germany). The TEM was operated under a high-tension of 120 kV, energy filtering of 20 eV and magnification range of 31.5 K to 80 K, in which each pixel of the micrographs corresponded to 3.68 to 1.48 Å respectively. A total of 15–72 micrographs were imaged from each sample. The contrast transfer function (CTF) of each micrograph was determined and then corrected by the phase-flip option using ctfit (EMAN software)41. ~300–~3000 particles from each sample were selected and windowed by the boxer software in the EMAN software package41 and submitted for Gaussian low-pass filtering before size measurement.
Cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) data acquisition
Cryo-EM specimens were prepared on lacey carbon film coated copper grid (Cu-200LC, Pacific Grid-Tech, San Francisco, CA). Cryo-EM data of liposome binding to CETP specimens were acquired under a less than ~2.3 μm defocus with a high-sensitivity 4 K × 4 K pixel Gatan Ultrascan CCD camera at 50 K magnification by the same Zeiss Libra 120 TEM (each pixel of the micrograph corresponded to 2.4 Å in the specimens). Total dose for un-tilt 2D micrographs is about 10 to 30 e−/Å2. For cryo-ET data acquisition, the specimens mounted on a Gatan 915 high-tilt cryo-EM holder were tilted at angles ranging from −57° to 60° in steps of 1.5°. The total dose of electron illumination was up to ~120 e−/Å2 or slightly higher. The tilt series of tomographic data was controlled and imaged by manual operation and by Gatan tomography software (Zeiss Libra 120 TEM) that was preinstalled in the microscope.
Correction of contrast transfer function (CTF) for cryo-ET data
Tilting series of micrographs were initially aligned together with the IMOD software package42. The defocus near the tilt-axis area of each tilt micrograph was examined by fitting CTF parameters with its power spectrum by ctffind3 in the FREALIGN software package43 and then examined by ctfit (EMAN software package)41. The CTF was then corrected by TOMOCTF28. The tilt series of each CETP-liposome image in windows of 200 × 200 pixels was tracked and selected by IPET software.
Individual-particle electron tomography (IPET) 3D reconstruction
Ab-initio 3D reconstruction of an individual liposome-CETP complex was conducted by the IPET method27. In IPET, a small image containing only a targeted liposome-CETP complex was windowed from each tilted whole-micrograph (CTF corrected). An ab-initio initial model was generated by directly back-projecting these small-images into a 3D map. The map was then refined via three rounds of refinement loops (including more than a hundred iterations) by the focused electron tomography reconstruction (FETR) algorithm27. In FETR, an automatically-generated dynamic Gaussian low-pass filter and an automatically generated soft-mask were applied to both the references and tilted images to achieve the final 3D reconstruction. Since the specimen holder has a limitation to tilt to ±90° angle, a wedge shaped data was missing in final 3D reconstruction, which resulted in a certain level of artifact, especially along the Z direction. The missing wedge data was estimated via our newly developed interactive algorithm (related manuscript in preparation). As an implementation of this algorithm, we computed the missing wedge data and contributed to the final reconstruction. The crystal structure (PDB entry 2OBD11) was fitted into the final IPET 3D reconstruction by using a rigid-body fitting option in UCSF Chimera44.
Fourier shell correlation (FSC) analysis
To analyze tomographic 3D reconstruction resolution, the center-refined raw ET images were split into two groups based on having an odd- or even-numbered index in the order of tilt angles27. Each group was used to generate a 3D reconstruction image; the two 3D reconstructions from both groups were then used to compute the FSC curve over their corresponding spatial frequency shells in Fourier space (using the “RF 3” command in SPIDER45) (Supplementary Fig. 5). The frequency at which the FSC curve declined to a value of 0.5 and 0.143 (golden standard46) was used to represent the resolution of the final reconstruction.
Statistical analyses of CETP binding to HDL particles and liposomes
Particle size was determined by measuring the diameter in two orthogonal directions, as described before25. In brief, the geometric mean of the perpendicular diameters was used to represent the particle diameter. The aspect ratio of the long and perpendicular diameters was used to represent particle shape. Histograms of the particle diameters were generated with 2.17 nm sampling steps. Each histogram was fitted with a 9th degree polynomial function in R for data analysis. Since the CETP bound to the liposome may have been blocked from certain viewing directions, we computed the probability based a geometric model (Supplementary Fig. 3). The probability ( ) is a function of the liposome diameter (
) is a function of the liposome diameter ( ) and CETP protrusion length (l), in equation of
) and CETP protrusion length (l), in equation of  . The final histogram was adjusted by taking the measured histogram divided by this variable probability.
. The final histogram was adjusted by taking the measured histogram divided by this variable probability.
Liposome structures by molecular dynamics (MD) simulation and hydrophobicity analysis
Based on the size range of liposomes in our experiment, five initial liposome models with diameters of 12 nm, 20 nm, 27 nm, 35 nm and 42 nm were simulated. The initial models to simulate the liposomes were generated as below, the lipid bilayer vesicles (~65 Å2/lipids47 and the average thickness ~36.8 Å48) with an open pore (with 13% of surface area)49 were surrounded by a solution containing water molecules and NaCl at 0.1 M. These pores on the vesicles would allow the lipid flipping and transferring between inner and outer membranes during energy minimization and MD simulations50,51.
As the liposomes contained a large number of molecules, we used the Residue-based Coarse Graining (RBCG) MD method to simplify our initial models in order to enable a long time-scale simulation52. In RBCG, about every 10 atoms were grouped together and assigned as a bead model based on their chemical functionality. One POPC molecule was assigned 13 beads (one bead for choline, one for phosphate, two beads for glycerol and nine for two fatty acid chains) based on Marrink's CG lipid model53. The interaction potentials between beads and the bead masses were used as previously reported54.
Energy minimization was conducted by Nanoscale Molecular Dynamics (NAMD)55. The criteria to determine whether the liposomes were stabilized were as follows: i) the surface pores were completely closed; ii) liposomes should have a spherical shape determined by Radial Distribution Function (RDF), which is computed with respect to the liposome's center of mass (supplementary Fig. 8); and iii) stabilized surface hydrophobicity. The area of the hydrophobic surface on each stabilized liposome was measured by the Solvent Accessible Surface Area (SASA) function in Visual Molecular Dynamics (VMD)56. The percentages of surface hydrophobic area on each liposome surface were computed by taking their hydrophobic surface area and dividing by the whole liposome surface area. The average of the last 300 frames (12 ns) was used to calculate the hydrophobicity.
References
Drayna, D. et al. Cloning and sequencing of human cholesteryl ester transfer protein cDNA. Nature 327, 632–4 (1987).
Inazu, A. et al. Increased high-density lipoprotein levels caused by a common cholesteryl-ester transfer protein gene mutation. N Engl J Med 323, 1234–8 (1990).
Brown, M. L. et al. Molecular basis of lipid transfer protein deficiency in a family with increased high-density lipoproteins. Nature 342, 448–51 (1989).
Zhong, S. et al. Increased coronary heart disease in Japanese-American men with mutation in the cholesteryl ester transfer protein gene despite increased HDL levels. J Clin Invest 97, 2917–23 (1996).
Barter, P. J. et al. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med 357, 2109–22 (2007).
Funder, J. W. The off-target effects of torcetrapib. Endocrinology 150, 2024–6 (2009).
Stein, E. A. et al. Safety and tolerability of dalcetrapib. Am J Cardiol 104, 82–91 (2009).
Derks, M., Anzures-Cabrera, J., Turnbull, L. & Phelan, M. Safety, tolerability and pharmacokinetics of dalcetrapib following single and multiple ascending doses in healthy subjects: a randomized, double-blind, placebo-controlled, phase I study. Clin Drug Investig 31, 325–35 (2011).
Gotto, A. M., Jr et al. Evaluation of lipids, drug concentration and safety parameters following cessation of treatment with the cholesteryl ester transfer protein inhibitor anacetrapib in patients with or at high risk for coronary heart disease. Am J Cardiol 113, 76–83 (2014).
Friedrich, S. et al. The pharmacokinetics and pharmacokinetic/pharmacodynamic relationships of evacetrapib administered as monotherapy or in combination with statins. CPT Pharmacometrics Syst Pharmacol 3, e94 (2014).
Qiu, X. et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat Struct Mol Biol 14, 106–13 (2007).
Liu, S. et al. Crystal structures of cholesteryl ester transfer protein in complex with inhibitors. J Biol Chem 287, 37321–9 (2012).
Zhang, L. et al. Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nat Chem Biol 8, 342–9 (2012).
Lei, D. et al. Structural features of cholesteryl ester transfer protein: a molecular dynamics simulation study. Proteins 81, 415–25 (2013).
Cilpa-Karhu, G., Jauhiainen, M. & Riekkola, M. L. Atomistic molecular dynamics simulation reveals the mechanism by which CETP penetrates into HDL enabling lipid transfer from HDL to CETP. J Lipid Res (2014).
van Antwerpen, R., La Belle, M., Navratilova, E. & Krauss, R. M. Structural heterogeneity of apoB-containing serum lipoproteins visualized using cryo-electron microscopy. J Lipid Res 40, 1827–36 (1999).
van Antwerpen, R. et al. Cryo-electron microscopy of low density lipoprotein and reconstituted discoidal high density lipoprotein: imaging of the apolipoprotein moiety. J Lipid Res 38, 659–69 (1997).
Pattnaik, N. M. & Zilversmit, D. B. Interaction of cholesteryl ester exchange protein with human plasma lipoproteins and phospholipid vesicles. J Biol Chem 254, 2782–6 (1979).
Jeyarajah, E. J., Cromwell, W. C. & Otvos, J. D. Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy. Clin Lab Med 26, 847–70 (2006).
Otvos, J. D., Jeyarajah, E. J. & Cromwell, W. C. Measurement issues related to lipoprotein heterogeneity. Am J Cardiol 90, 22i–29i (2002).
Lund-Katz, S., Liu, L., Thuahnai, S. T. & Phillips, M. C. High density lipoprotein structure. Front Biosci 8, d1044–54 (2003).
Orlova, E. V. et al. Three-dimensional structure of low density lipoproteins by electron cryomicroscopy. Proc Natl Acad Sci U S A 96, 8420–5 (1999).
Zhang, L. et al. An optimized negative-staining protocol of electron microscopy for apoE4 POPC lipoprotein. J Lipid Res 51, 1228–36 (2010).
Zhang, L., Tong, H., Garewal, M. & Ren, G. Optimized negative-staining electron microscopy for lipoprotein studies. Biochim Biophys Acta 1830, 2150–9 (2013).
Zhang, L. et al. Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. J Lipid Res 52, 175–84 (2011).
Zhang, B. et al. Effects of reconstituted HDL on charge-based LDL subfractions as characterized by capillary isotachophoresis. J Lipid Res 48, 1175–89 (2007).
Zhang, L. & Ren, G. IPET and FETR: experimental approach for studying molecular structure dynamics by cryo-electron tomography of a single-molecule structure. PLoS One 7, e30249 (2012).
Fernandez, J. J., Li, S. & Crowther, R. A. CTF determination and correction in electron cryotomography. Ultramicroscopy 106, 587–96 (2006).
Jensen, M. B. et al. Membrane curvature sensing by amphipathic helices: a single liposome study using alpha-synuclein and annexin B12. J Biol Chem 286, 42603–14 (2011).
Barter, P. J. & Jones, M. E. Kinetic studies of the transfer of esterified cholesterol between human plasma low and high density lipoproteins. J Lipid Res 21, 238–49 (1980).
Ihm, J., Quinn, D. M., Busch, S. J., Chataing, B. & Harmony, J. A. Kinetics of plasma protein-catalyzed exchange of phosphatidylcholine and cholesteryl ester between plasma lipoproteins. J Lipid Res 23, 1328–41 (1982).
Tall, A. R. Plasma cholesteryl ester transfer protein. J Lipid Res 34, 1255–74 (1993).
Ren, G. et al. Model of human low-density lipoprotein and bound receptor based on cryoEM. Proc Natl Acad Sci U S A 107, 1059–64 (2010).
Morton, R. E. & Zilversmit, D. B. Inter-relationship of lipids transferred by the lipid-transfer protein isolated from human lipoprotein-deficient plasma. J Biol Chem 258, 11751–7 (1983).
Bruce, C. et al. Molecular determinants of plasma cholesteryl ester transfer protein binding to high density lipoproteins. J Biol Chem 270, 11532–42 (1995).
Dergunov, A. D., Shabrova, E. V. & Dobretsov, G. E. Cholesteryl ester diffusion, location and self-association constraints determine CETP activity with discoidal HDL: Excimer probe study. Arch Biochem Biophys 564, 211–8 (2014).
Koivuniemi, A., Vuorela, T., Kovanen, P. T., Vattulainen, I. & Hyvonen, M. T. Lipid exchange mechanism of the cholesteryl ester transfer protein clarified by atomistic and coarse-grained simulations. PLoS Comput Biol 8, e1002299 (2012).
Han, S. et al. Comparison of lipoprotein separation and lipid analysis methodologies for human and cynomolgus monkey plasma samples. J Cardiovasc Transl Res 5, 75–83 (2012).
Rye, K. A., Hime, N. J. & Barter, P. J. Evidence that cholesteryl ester transfer protein-mediated reductions in reconstituted high density lipoprotein size involve particle fusion. J Biol Chem 272, 3953–60 (1997).
Rye, K. A. & Barter, P. J. The influence of apolipoproteins on the structure and function of spheroidal, reconstituted high density lipoproteins. J Biol Chem 269, 10298–303 (1994).
Ludtke, S. J., Baldwin, P. R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128, 82–97 (1999).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–6 (1996).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142, 334–47 (2003).
Pettersen, E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12 (2004).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116, 190–9 (1996).
Scheres, S. H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat Methods 9, 853–4 (2012).
Kucerka, N., Nieh, M. P. & Katsaras, J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim Biophys Acta 1808, 2761–71 (2011).
Tahara, Y. & Fujiyoshi, Y. A new method to measure bilayer thickness: cryo-electron microscopy of frozen hydrated liposomes and image simulation. Micron 25, 141–9 (1994).
Risselada, H. J. & Marrink, S. J. Curvature effects on lipid packing and dynamics in liposomes revealed by coarse grained molecular dynamics simulations. Phys Chem Chem Phys 11, 2056–67 (2009).
de Vries, A. H., Mark, A. E. & Marrink, S. J. Molecular dynamics simulation of the spontaneous formation of a small DPPC vesicle in water in atomistic detail. J Am Chem Soc 126, 4488–9 (2004).
Marrink, S. J. & Mark, A. E. Molecular dynamics simulation of the formation, structure and dynamics of small phospholipid vesicles. J Am Chem Soc 125, 15233–42 (2003).
Marrink, S. J., de Vries, A. H. & Mark, A. E. Coarse grained model for semiquantitative lipid simulations. Journal of Physical Chemistry B 108, 750–760 (2004).
Marrink, S. J., Risselada, J. & Mark, A. E. Simulation of gel phase formation and melting in lipid bilayers using a coarse grained model. Chem Phys Lipids 135, 223–44 (2005).
Shih, A. Y., Freddolino, P. L., Arkhipov, A. & Schulten, K. Assembly of lipoprotein particles revealed by coarse-grained molecular dynamics simulations. J Struct Biol 157, 579–92 (2007).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–802 (2005).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J Mol Graph 14, 33–8, 27–8 (1996).
Acknowledgements
We thank Mr. Dongsheng Lei and Xing Zhang for discussion in MD simulation. This work was supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health (no. R01HL115153) and Merck fund (No: LKRD105435). Work at the Molecular Foundry was supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
This project was initiated and designed by R.C., L.Z., K.A.R., X.Q., D.G.J. and G.R.; and refined by M.Z. and M.A.C.; X.Q., D.G.J., M.P. and K.A.R. isolated the CETP and HDL samples; M.Z., L.Z. and R.C. conducted the experiments and acquired the OpNS data; H.T. and L.Z. acquired the cryo-EM data; M.Z. processed the NS data, computed the statistics and performed the molecular dynamics simulation;L.Z., M.Z. and F.W. processed the cryo-ET data; A.R. analyzed the projections and averages of 3D reconstruction; M.Z., L.Z., R.C., M.A.C., M.J.R. and G.R. interpreted and manipulated the data; M.Z., R.C., M.A.C. and G.R. drafted the initial manuscript, which was revised by L.Z., K.A.R., A.R., M.P., M.J.R., X.Q. and D.G.J.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supporting info
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, M., Charles, R., Tong, H. et al. HDL surface lipids mediate CETP binding as revealed by electron microscopy and molecular dynamics simulation. Sci Rep 5, 8741 (2015). https://doi.org/10.1038/srep08741
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep08741
This article is cited by
-
Structure-based mechanism and inhibition of cholesteryl ester transfer protein
Current Atherosclerosis Reports (2023)
-
Human cholesteryl ester transport protein transgene promotes macrophage reverse cholesterol transport in C57BL/6 mice and phospholipid transfer protein gene knockout mice
Journal of Physiology and Biochemistry (2021)
-
Study of Structural stability and formation mechanisms in DSPC and DPSM liposomes: A coarse-grained molecular dynamics simulation
Scientific Reports (2020)
-
LoTToR: An Algorithm for Missing-Wedge Correction of the Low-Tilt Tomographic 3D Reconstruction of a Single-Molecule Structure
Scientific Reports (2020)
-
Single-Molecule 3D Images of “Hole-Hole” IgG1 Homodimers by Individual-Particle Electron Tomography
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.