Abstract
The checkpoint kinase Rad53 is crucial to regulate DNA replication in the presence of replicative stress. Under conditions that interfere with the progression of replication forks, Rad53 prevents Exo1-dependent fork degradation. However, although EXO1 deletion avoids fork degradation in rad53 mutants, it does not suppress their sensitivity to the ribonucleotide reductase (RNR) inhibitor hydroxyurea (HU). In this case, the inability to restart stalled forks is likely to account for the lethality of rad53 mutant cells after replication blocks. Here we show that Rad53 regulates replication restart through the checkpoint-dependent transcriptional response and more specifically, through RNR induction. Thus, in addition to preventing fork degradation, Rad53 prevents cell death in the presence of HU by regulating RNR-expression and localization. When RNR is induced in the absence of Exo1 and RNR negative regulators, cell viability of rad53 mutants treated with HU is increased and the ability of replication forks to restart after replicative stress is restored.
Similar content being viewed by others
Introduction
Eukaryotic cells employ checkpoint pathways to coordinate DNA replication, DNA repair and cell cycle progression after DNA damage or replication stress1,2,3,4,5. Mec1 and Rad53, the homologues of mammalian ATR and Chk2, respectively, are the central protein kinases of the S phase checkpoint in the budding yeast Saccharomyces cerevisiae. Their activation triggers a global checkpoint response regulating different processes, including transcriptional induction of DNA damage response genes, inhibition of late replication origin firing, stabilization of stalled DNA replication forks and inhibition of cell cycle progression6,7. Of these, replication fork stabilization appears to be most important to prevent cell lethality after genotoxic stress8,9,10,11. Previous work indicated that the primary role of Rad53 in fork stabilization after DNA damage is to prevent Exo1-dependent replication fork breakdown11,12. Deletion of EXO1 suppresses replication fork catastrophe in rad53 mutants treated with the alkylating agent methyl methanosulfonate (MMS) and reduces the sensitivity of rad53 mutants to a wide variety of DNA damaging agents, including UV and ionising radiation11. Strikingly, however, deletion of EXO1 is completely ineffective in suppressing the sensitivity of rad53 mutants to the ribonucleotide reductase (RNR) inhibitor hydroxyurea (HU)11, despite the fact that EXO1 deletion suppresses the degradation of replication forks in HU12. This argues for the existence of an Exo1-independent mechanism, which is regulated by Rad53 and required for cell viability after HU arrest. This mechanism might involve direct effects on replisome components12,13,14 or some other function of Rad53.
Previous work has indicated that this additional mechanism is unlikely to be the well-documented role of checkpoints in inhibiting mitosis, since the microtubule inhibitor nocodazole, which prevents mitotic entry, does not markedly rescue the lethality of rad53Δ or rad53Δexo1Δ mutant strains after HU treatment11. Rad53 inhibits late origin firing via Sld3 and Dbf4 phosphorylation; however, this inhibition of late origin firing makes at best a small contribution to cell viability in the presence of HU15,16 and is therefore also unlikely to represent this other function. Rad53 is a critical regulator of transcriptional responses after DNA damage and replicative stress17,18. Although inhibiting protein synthesis does not seem to compromise viability or recovery after fork stalling in wild-type cells8,19, it is required to restore normal rates of DNA synthesis after replication blocks8, indicating that new protein synthesis and, perhaps, checkpoint-dependent transcriptional induction may play some role in replication resumption. Rad53 regulates transcription by activating the downstream Dun1 kinase, which phosphorylates and inactivates the transcriptional repressor Crt120. This leads to the induction of a large number of damage-inducible genes, including those encoding the RNR subunits Rnr2, Rnr3 and Rnr421,22. In S. cerevisiae, RNR is a tetrameric enzyme composed of two dimeric subunits, a large R1 and a small R2. The RNR large subunit contains the catalytic site and it is encoded by the RNR1 and RNR3 genes23,24. The small subunit is a heterodimer formed by the products of the genes RNR2 and RNR425,26. Recent reports indicate that RNR1 transcription is activated in response to genotoxic stress via Ixr1, in a Mec1– Rad53-dependent but Dun1-independent manner27. Rad53 and Dun1 also regulate RNR activity post-transcriptionally by phosphorylation of small protein regulators of RNR activity (Sml1)28,29 and localization (Dif1)30,31. It has been shown too that Rnr2 and Rnr4 redistribute from the nucleus to the cytoplasm after DNA damage or replicative stress32. Rnr2-Rnr4 nuclear localization is controlled by the inhibitor Dif1, whose phosphorylation and degradation in response to DNA damage promotes Rnr2-Rnr4 cytoplasmic localization30,31. Similarly, dynamic changes in RNR localization have also been reported in fission yeast33, although it is unclear whether nuclear translocation is an RNR-regulatory mechanism conserved in mammalian cells34,35,36,37.
In this study, we sought to test the possibility that the Exo1-independent mechanism described above involves RNR regulation. Our work indicates that Rad53 has a role in promoting fork restart by inducing the expression of RNR genes and by regulating RNR subcellular localization after HU-induced replicative stress.
Results
EXO1 deletion improves replication restart in rad53 mutants after replicative stress
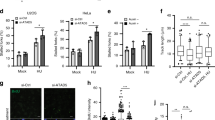
EXO1 deletion was able to suppress fork collapse but not the lethality of rad53Δ mutants treated with HU (Fig. 1a11). This led us to investigate whether Exo1 could influence replication restart after HU arrest. We first compared the replication pattern of RAD53+, rad53Δ and rad53Δexo1Δ cells during S phase in the presence of HU and after release from the block induced by this drug. Cells were synchronized in G1 and released into S-phase in the presence of 0.2 M HU (Fig. 1b). In the three strains, the cells entered normally into S-phase, as indicated by the estimation of the budding index in all cases (>95% budded cells after 120 min in HU, not shown), but the rate of replication was significantly reduced due to the depletion of dNTP levels caused by HU and the cells arrested with a DNA content close to 1C. Flow cytometry shows that, after HU removal, RAD53+ cells efficiently resumed and completed replication (Fig. 1b). In agreement with previous results38, rad53Δ cells showed a clear defect in replication resumption and thus in these cells the DNA content did not reach the 2C peak, even 120 min after HU removal. Although rad53Δexo1Δ cells were also defective in replication restart, the DNA peaks were clearly broader than in the rad53Δ mutant, suggesting that some replication might be occurring after HU-release in the absence of Exo1.
EXO1 affects replication fork restart in rad53Δ cells after HU arrest.
(a) Drop assays on YPD plus the indicated concentrations of HU were performed using 1:5 serial dilutions of exponentially growing cultures of RAD53+EXO1+, rad53ΔEXO1+ or rad53Δexo1Δ strains. SML1 was previously deleted in all of them to allow viability in the absence of RAD53. (b) The strains were synchronized in G1 with α−factor and released into YPD medium containing 0.2 M HU for 2 h. HU was then removed and cells were released into fresh YPD medium for 2 h. Samples were taken every 30 min and DNA content was measured by flow cytometry. (c) Replication fork progression was followed by density transfer, as described in the text. Cells were blocked in G1 with α-factor and released into medium containing HU for 90 min. HU was then removed and cultures were released into HU-free medium for 120 min. DNA content at the indicated time points is shown in Supplementary Fig. S1a. The sizes and positions of the restriction fragments 1 and 3 of the analysed replicon on chromosome VI are represented in the scheme. The relative amounts of radioactivity in the hybridized DNA are plotted against the gradient fraction number. The position of unreplicated (HH) and replicated (HL) DNA peaks is indicated at the top. Black arrows indicated fork movement from ARS607. The position of the initial HH peak is shown as a reference (gray area). (d) The graph shows the percentage of replicated DNA at positions ARS607 and fragment 3 in the RAD53+ strain (white bars), rad53Δ strain (gray bars) and rad53Δexo1Δ strain (black bars) at the end of the experiment shown in Figure 1C.
To test whether the increase in DNA-content observed in rad53Δexo1Δ cells after release from a HU-block reflected replication fork recovery, we used density transfer (Fig. 1c), a technique that allows the study of on-going DNA synthesis across a specific genomic region8,10,39. Using this approach, we examined the progression of replication forks along a previously characterized replicon on chromosome VI10, from the ARS607 origin (fragment 1) to a position located 20 kb away (fragment 3) (Fig. 1c). Cells were pre-grown in the presence of heavy isotopes (13C glucose, 15N ammonium sulphate) during seven generations, to generate Heavy-Heavy (HH) DNA. After G1 arrest with the α-factor mating pheromone, the cells were released into fresh medium containing HU in the presence of light isotopes (12C glucose, 14N ammonium sulphate) for 90 min and finally cells were released into fresh medium for 120 min (Fig. 1c and Supplementary Fig. S1). Semi-conservative DNA replication generates products of the hybrid, Heavy-Light (HL) density. HH-DNA (unreplicated) was separated from HL-DNA (replicated) by ultracentrifugation of DNA in CsCl gradients after restriction endonuclease digestion. The localization of restriction fragments in fractions from the gradient was performed by hybridization with specific probes.
In RAD53+ cells, fragment 1 was partially replicated in HU-arrested cells, as seen by the appearance of some HL DNA, while fragment 3 remained unreplicated (HH DNA) (Fig. 1c). This indicated that the ARS607 origin had fired and that replication forks stalled within fragment 1 as a consequence of the HU treatment. After release from HU arrest (120 min), both fragments 1 and 3 were fully replicated (HL DNA), indicating that the majority of replication forks resumed DNA synthesis after HU removal. In rad53Δ cells, considerably less replication of fragment 1 was detected during HU treatment (Fig. 1c, d), consistent with previous results obtained with rad53 and mec1 checkpoint mutants in the presence of HU40,41. Moreover, in contrast to wild type cells, rad53Δ cells showed little replication of fragment 3 (25%) and only partial replication of fragment 1 (68%) after release (120 min) from the HU arrest (Fig. 1c, d). This result indicates that most DNA replication forks were unable to restart in the rad53Δ strain after fork stalling. Remarkably, deletion of EXO1 in the rad53Δ background increased the amount of replication of fragment 3 after release from HU arrest from 25% to 41% (Fig. 1c, d), indicating more efficient replication restart in this strain than in the rad53Δ mutant. It is worth noting that the small proportion of replication forks that resume replication after release from the HU block in both rad53Δ and rad53Δexo1Δ mutants were previously undetected by other techniques such as two dimensional electrophoresis or alkaline agarose gels of BrdU-labelled DNA9,11,12, indicating the usefulness of density transfer in quantitative analysis of DNA replication. Taken together, these experiments confirm that Rad53 is critical for resumption of replication after HU arrest and show that the Exo1 pathway makes a small but detectable contribution to fork recovery. Nevertheless, although EXO1 deletion improves fork restart in cells lacking Rad53, it is not sufficient for these mutant cells to fully resume DNA replication after replicative stress and therefore other players must be involved in this process.
Co-induction of large and small RNR subunits and EXO1 deletion contribute to suppress the lethality of rad53 mutants treated with HU
The inability of EXO1 deletion to suppress the sensitivity of rad53 cells to HU as well as to abolish the replication resumption defect of these checkpoint mutant cells after treatment with this drug (Fig. 1), suggests that Rad53 might allow cells to cope with this kind of replicative stress through an Exo1-independent mechanism. The results above pointed out that only some replication forks were able to restart after HU release in cells lacking Rad53 (Fig. 1c), even when fork degradation was prevented in the absence of Exo1, arguing that a critical factor for replication fork restart might be limiting. Rad53 has a crucial role in regulating RNR, which is the target of HU42 and therefore, we reasoned that this enzymatic complex could be a strong candidate for such a potential limiting factor. Thus, we investigated the contribution of checkpoint-dependent RNR expression to cell viability and DNA replication resumption after HU arrest.
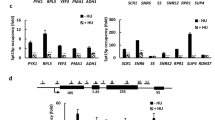
It is known that expression of the RNR1 gene from the inducible GAL1-10 promoter increases dNTP levels43. Therefore, we first induced the expression of RNR1 in rad53Δ mutant cells using a pGAL-RNR1 construct (Supplementary Fig. S2a) and tested the sensitivity of this strain to HU. Figure 2a shows that RNR1-overexpression did not rescue the lethality of rad53 mutant cells to HU. Similar results were obtained by expression of the other RNR large subunit, encoded by the RNR3 gene (Supplementary Fig. S2a, b), which is specifically induced after genotoxic stress23,24. These results were in agreement with a previous report showing that overexpression of RNR1 or RNR3 allows mec1Δ or rad53Δ cells to survive but it does not suppress their DNA damage sensitivities33. In addition, we also found that the individual overexpression of RNR2 or RNR4 did not rescue the viability of rad53Δ cells treated with HU either (Supplementary Fig. S2a, b). On the contrary, the simultaneous expression of RNR2 and RNR4, as well as the co-expression of large and small subunits of RNR (RNR3-RNR2-RNR4 or RNR1-RNR2-RNR4) significantly increased the resistance of rad53Δ cells to HU (Fig. 2b). This occurred despite the fact that overexpression of several RNR subunits has a toxic effect that causes greatly reduced growth of the rad53Δ strain (Fig. 2c, YPGal). Interestingly, all the strains that were able to suppress rad53 HU-sensitivity expressed both RNR2 and RNR4 (Fig. 2b and Supplementary Fig. S2c, d), strongly suggesting that expression of the RNR small subunit is essential for survival after HU-induced replicative stress in the absence of Rad53. Nevertheless, co-induction of RNR2-RNR4 plus RNR3 or RNR1 conferred higher resistance to HU, indicating that co-expression of large and small RNR subunits is highly important for the viability of rad53 mutant cells under the assayed conditions. These results are not just due to co-expression of a certain number of RNR subunits, irrespective of which of them are induced, as overexpression of RNR1-3-2 or RNR1-3-4 did not increase the resistance to HU of rad53Δ cells (Supplementary Fig. S2d). Furthermore, a rad53Δ strain carrying three inducible RNR1 copies placed at different positions in the genome (rad53ΔGAL-RNR1-1-1) was as sensitive to HU as a rad53Δ strain (Supplementary Fig. S2e). Therefore, we conclude that the regulation of specific-RNR subunits is critical for the viability of rad53Δ mutants exposed to HU.
EXO1 deletion and checkpoint-dependent RNR expression contribute to cell viability in the presence of replicative stress.
(a) 1:5 serial dilutions of exponential cultures of the indicated strains were tested for sensitivity to 10 mM HU, by using a drop assay on YPD plates (top) or YPGAL plates (bottom). (b) The indicated strains were tested for sensitivity to 10 mM and 20 mM HU as described in (a). (c) Drop assays on YPD or YPGal were performed with the indicated strains as described in (a). (d) Sensitivity to 10 mM, 15 mM or 20 mM HU was analyzed as described in (a). (e) Drop assays of the indicated strains as in (b).
As shown above, although EXO1 deletion was completely ineffective in suppressing the sensitivity of rad53 mutants to HU11, it made some contribution to fork restart after a HU block (Fig. 1c). Therefore, we next asked whether EXO1 deletion could contribute to the viability of rad53-deficient cells with induced RNR expression after HU treatment. As shown in Figure 2d, deletion of EXO1 increased the resistance to HU of the rad53ΔGAL-RNR2-4, rad53ΔGAL-RNR3-2-4 and rad53ΔGAL-RNR1-2-4 mutants when they are grown in the presence of galactose. These results strongly argue that both Exo1-prevention and RNR-induction mechanisms are important for the viability of rad53 mutants in the presence of HU-induced replicative stress.
In addition to regulating transcription, Rad53 also promotes removal of two negative regulators of RNR, Sml1 and Dif1. We routinely delete SML1 to suppress the lethality of RAD53 deletion. We next asked whether deletion of DIF1, which facilitates the localization of Rnr2-4 to the cytoplasm, would contribute to cell survival in the presence of HU. Figure 2e shows that the rad53ΔGAL-RNR3-2-4exo1Δdif1Δ mutant is indeed more resistant to HU than the rad53ΔGAL-3-2-4exo1Δ mutant, arguing that Rad53-dependent regulation of RNR cellular localization through Dif1 contributes to cell survival in the presence of HU.
RNR expression promotes replication resumption after fork stalling in rad53 mutants in the absence of EXO1
As the resistance to HU is expected to correlate with the ability of cells to resume and complete chromosome replication after the replicative stress induced by this drug, we next examined whether RNR expression and EXO1 deletion contribute to replication restart in rad53Δ mutant cells after HU arrest. The simple growth assay in the previous experiments cannot distinguish between gene expression requirements during or after HU treatment, because cells are exposed to HU chronically. We therefore used the GAL-inducible system to analyse replication resumption when RNR genes are expressed exclusively after HU removal during a single S-phase (Fig. 3).
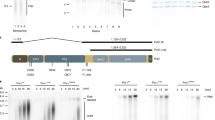
RNR expression allows replication resumption in rad53Δexo1Δ mutants after fork stalling.
(a) Cultures of RAD53+, rad53ΔGAL-RNR1 and rad53Δexo1ΔGAL-RNR1 strains were grown in YPRAF medium, arrested in G1 with α-factor and released from the G1 block into RAF-containing medium containing 0.2 M HU. After 3 h in HU, cultures were released into fresh medium in the presence of Glc (left panel) or Gal (right panel). DNA content was measured by flow cytometry at the indicated times. The expression of Rnr1 was followed by immunoblot analysis (bottom panel) as described in Methods. A Ponceau-S-stained membrane coincident with Rnr1 migration was used as a loading control. Full-length blots are presented in Supplementary figure S4. (b) Analysis of S phase progression of rad53ΔEXO1+GAL-RNR3-2-4 and rad53Δexo1ΔGAL-RNR3-2-4 strains as described in (a). The expression of Rnr3 was followed by immunoblot analysis (bottom panel) as described in (a). (c) Analysis of S phase progression of rad53ΔEXO1+GAL-RNR1-2-4 and rad53Δexo1ΔGAL-RNR1-2-4 strains as described in (a). (d) rad53Δexo1ΔGAL-RNR3-2-4 cells were grown in YPRAF medium and arrested in G1 with α-factor. The culture was divided in two and incubated in either YPD (white column) or YPRAF (black column) in the presence of 0.2 M HU for 3 h. The YPD-culture was then released from HU into fresh medium with Glc, while the YPRAF-culture was released in the presence of Gal. Cells were tested for viability after α−factor arrest, after 3 h in the presence of 0.2 M HU and 2 h after release from HU. (e) Analysis of S phase progression of rad53ΔEXO1+dif1ΔGAL-RNR3-2-4 and rad53Δexo1Δdif1ΔGAL-RNR3-2-4 strains as described in (a).
We analysed replication resumption by measuring DNA content by flow cytometry in rad53ΔEXO1+ or rad53Δexo1Δ strains overexpressing RNR1, RNR3-2-4 or RNR1-2-4 (Fig. 3a, b, c). Cells were grown in raffinose (RAF)-containing medium, synchronized in G1 phase with α-factor and released into fresh medium supplemented with 0.2 M HU. After 3 h, the cells were released from the HU arrest into fresh medium containing either glucose (repressive, GAL1,10 promoter OFF) or galactose (activating, GAL1,10 promoter ON). Figure 3a shows that whereas RAD53+ cells completed replication after HU-release, rad53Δ cells did not reach the 2C DNA content peak when RNR1 was induced, regardless of the presence or the absence of Exo1, indicating that these cells could not finish S-phase. By contrast, although induction of RNR3-2-4 did not allow by itself the completion of S-phase (Fig. 3b, left), it led to efficient replication resumption when it was combined with EXO1 deletion, allowing cells to reach a 2C DNA content by 180 min after HU removal (Fig. 3b, right). Similar results were obtained when EXO1 was deleted in rad53ΔGAL-RNR1-2-4 or rad53ΔGAL-RNR2-4 strains (Fig. 3c, Supplementary Fig. S3). Consistent with these data, a significant percentage of rad53Δexo1Δ cells (35%) survived after an acute HU-treatment when RNR3-2-4 genes were expressed after HU-removal (Fig. 3d). This is a notable increase in cell viability, taking into account that RNR overexpression severely affects growth of the rad53ΔRNR3-2-4 strain (Fig. 2c). Importantly, the viability remained low when cells were plated immediately after HU-treatment and it only recovered when cells were plated 2 h after HU-release in the presence of galactose (Fig. 3d). All these results indicate that both the expression of RNR genes and the deletion of EXO1 are necessary for the resumption of DNA synthesis after HU arrest in rad53Δ cells.
As shown in Fig. 2e, deleting DIF1 contributes to increase HU-resistance in the rad53Δexo1ΔGAL-RNR3-2-4 strain. Therefore we tested whether the absence of Dif1 could also contribute to replication resumption after release from a HU-block. Figure 3e shows that rad53Δexo1ΔGAL-RNR3-2-4 cells appeared to complete replication earlier in the absence of Dif1 and thus the rad53Δexo1ΔGAL-RNR3-2-4 dif1Δ mutant strain showed a 2C DNA content 2 h after release from HU arrest in medium with galactose. This result indicates that Dif1 inhibition and therefore the correct RNR subcellular localization, is also important for replication resumption after fork stalling.
Taken together, these experiments show that Rad53-dependent regulation of the expression and subcellular localization of RNR, in conjunction with Rad53-dependent inhibition of Exo1, are critical for the resumption of replication after HU arrest.
RNR expression restores replication fork restart in rad53Δexo1Δ cells after replicative stress
Flow cytometry analysis indicated that rad53Δexo1Δ mutants were able to resume replication and reached a 2C DNA content when RNR2-4, RNR3-2-4 or RNR1-2-4 were expressed after fork stalling induced by HU (Fig. 3). However, replication resumption could be due to the restart of DNA replication forks blocked during HU treatment or, instead, global replication could be rescued by new origin firing. To address this issue, we used density transfer and examined replication fork resumption within a single replicon on chromosome VI. In this experiment, we followed replication from ARS607 (fragment 1) to positions 3, 4 and 6 located at 20 kb, 40 kb or 65 kb away respectively (Fig. 4). Importantly, we used a strain that lacks ARS608 and ARS609 origins and therefore, the examined replicon can only be replicated from ARS607 and from a subtelomeric origin associated with the X-element at the end of the chromosome, which is only activated in the absence of Rad53-dependent inhibition of late origin firing10,11.
New DNA synthesis in rad53Δexo1ΔGAL-RNR2-3-4 mutants after replication blocks is due to fork restart.
(a) The experiment was performed as in Fig. 3a, with slight modifications described in (b). DNA content of rad53Δexo1ΔGAL-RNR2-3-4 cells was determined by flow cytometry at the indicated time points. (b) Replication fork progression was analysed in the rad53Δexo1ΔGAL-RNR2-3-4 strain by density transfer. Cells were initially grown in 0.1% Glc in medium with heavy isotopes and then blocked in G1 with α−factor. After G1 arrest, cells were released into medium containing HU and light isotopes, under conditions that prevented GAL1,10-dependent expression. Cells were held in HU for 3 h and then released from the HU arrest into fresh medium with either Glc or Gal. The position of the four ClaI/SalI restriction fragments from chromosome VI that were analysed is shown at the top, coincident with ARS607 and 20, 40 and 65 kb away from the origin. (c) Percentage of replicated DNA in the rad53Δexo1ΔGAL-RNR2-3-4 strain after HU-release in the presence of GLC or GAL (from Fig. 3b). The amount of replicated DNA at each fragment was plotted against distance from the ARS607 sequence. White bars represent RNR-repressing conditions (GLC) and black bars RNR-expressing conditions (GAL).
rad53Δexo1ΔGAL-RNR3-2-4 cells were initially grown in the presence of heavy isotopes, synchronized in G1 with α-factor and then released into fresh medium with light isotopes in the presence of 0.2 M HU. After 3 h in HU, these cells were released into fresh medium in the presence of glucose or galactose for 2 h. Flow cytometry (Fig. 4a) shows that the cells reached a 2C DNA content when released into galactose-containing medium after release from the HU-block, in agreement with data in Fig. 3b. Density transfer experiments (Fig. 4b) showed that when the cells were released from the G1-block in the presence of HU, there was some replication (HL peak) in fragments 1 and 6, indicating that the ARS607 origin fired normally and that the subtelomeric origin located at the end of the chromosome was also activated due to the absence of Rad53. As mentioned above, the severe phenotype of rad53 mutants exposed to high concentrations of HU led to very little replication of fragments 1 and 6 (Fig. 4b). When HU-release took place under conditions of RNR repression (GLC), fork restart was highly defective, as judged by the high amounts of unreplicated (HH) DNA that remained 2 h after HU-release in all restriction fragments. Remarkably, galactose induction of RNR3-2-4 (GAL) after HU arrest significantly increased the amount of replication of fragments 1, 3, 4 and 6 in the rad53Δexo1Δ background. Importantly, the extent of replication of the origin-distal fragments 3 and 4, which can only be passively replicated, increased up to 68 and 67% (Fig. 4c), indicating that a high fraction of stalled-replication forks indeed recovered from the HU-block when RNR was expressed. These data indicate that replication resumption after RNR induction in a rad53Δexo1Δ background is due to fork restart.
Discussion
In order to cope with replicative stress, cells need the efficient coordination of mechanisms that protect DNA replication forks and allow replication resumption after fork stalling, so that chromosome replication can be completed and genome stability is preserved. In this work, we show that the Rad53 checkpoint kinase promotes replication fork restart after HU treatment by inducing the expression of RNR and counteracting the RNR regulator Dif1. In addition, we show that replication fork restart after exposure to HU also requires Rad53-dependent inhibition of Exo1, which, as shown before, is necessary to avoid fork breakdown11,12. These results expand the view of how Rad53 controls several aspects of DNA replication under conditions of replicative stress and, taken with previous data, allows proposing the following model for the action of this checkpoint protein (depicted in Fig. 5): first, Rad53 inhibits the activation of additional replication origins by phosphorylating Sld3 and Dbf415,16; second, Rad53 prevents Exo1 from exerting a deleterious effect on replication forks11,12; and third, as shown here, Rad53 promotes replication restart by inducing the synthesis and activation of RNR, which is especially important after HU treatment.
Model for regulation of DNA replication by Rad53.
This model summarizes the different processes of DNA replication in which Rad53 regulation is involved. The replication forks activate the sensor kinase Mec1, which then phosphorylates the downstream kinase Rad53. Rad53 works through phosphorylation of Sld3 and Dbf4 to inhibit late origin firing. Rad53 works through inhibition of Exo1 to stabilize replication forks and through Ixr1 and Dun1-dependent induction and activation of RNR to promote fork restart.
It was previously shown that RNR1 or RNR3 overexpression suppresses rad53Δ lethality but not the extreme sensitivity of this checkpoint mutant to genotoxic stress38, which questioned the importance of increasing the expression of genes encoding RNR for cell survival after replicative stress. However, we show here that co-expression of specific RNR subunits increases the viability of rad53Δ mutants after replication blocks. In addition, we show that EXO1 deletion clearly contributes to the survival of rad53Δ cells in the presence of HU when RNR expression is induced, arguing that Exo1-regulation has a previously unnoticed role in the survival of rad53 mutants after replicative stress. Therefore, two Rad53 functions, preservation of fork integrity and up-regulation of RNR, are important for cell survival upon HU-induced replicative stress. Furthermore, we show that these two functions are necessary to promote fork restart after replication blockage. This essential role in fork restart explains why these Rad53 activities are required for cell viability and uncovers how the Rad53 checkpoint protein facilitates replication resumption from stalled forks.
It is currently unclear why new synthesis of RNR subunits after HU treatment is important for fork restart. One possibility is that the RNR-complex becomes irreversibly inactivated after prolonged HU treatment. In this respect, it is known that HU blocks DNA synthesis by reducing the RNR free radical contained in the small subunit44 and our results indicate that new expression of Rnr2-Rnr4 is essential for cell survival after replicative stress in the absence of Rad53. The fact that additional co-expression of RNR3 or RNR1 further increased the recovery after HU treatment could suggest that the expression of small and large RNR-subunits is required to form an enzymatically active complex that is able to supply appropriate dNTP levels. A second possibility to explain the requirement of RNR expression for replication resumption after fork stalling is that the increased dNTP concentration associated with the DNA damage response45 may be critical for some aspect of DNA polymerase function in fork restart.
Strikingly, while our results indicate that fork recovery in rad53 mutants is largely dependent on the new synthesis of RNR, it is known that new protein synthesis is dispensable for replication resumption after HU arrest in wild-type cells8,19. A possible explanation for these data is that Rad53-dependent RNR regulation might involve an active mechanism to re-activate RNR after HU treatment. In this scenario, wild-type cells would not be able to synthesize RNR in the absence of new protein synthesis, but would be capable of re-activating the remaining RNR pool. This could be also an explanation for the low rate of DNA synthesis after replication blocks that is observed when new protein synthesis is inhibited8. By contrast, all of these RNR-regulatory mechanisms would be impaired in rad53 cells, thus explaining why new RNR synthesis is strictly required for fork resumption in this checkpoint mutant.
Cells have evolved several strategies to regulate RNR activity, such as control of gene expression, inhibitor destruction and subcellular localization20,30,31. Additionally, protein-protein interactions between RNR and several cell cycle regulators could also modulate RNR activity35,46. Interestingly, a novel mechanism for the regulation of the activity of the mammalian RNR-subunit p53R2 via ATM-mediated phosphorylation has been reported47. Future examination of the possible interactions between the RNR-complex and the checkpoint machinery is required to explore this potential level of regulation in budding yeast.
DNA damage checkpoints act as a barrier to the development of cancer48,49 and, therefore, down-regulation of checkpoints may contribute to carcinogenesis. Besides, intense cell proliferation requires high rates of DNA synthesis and an elevated RNR activity has been associated with tumorogenesis50,51. Checkpoint-defective cancer cells may be especially sensitive to the chemotherapies that damage DNA, deplete nucleotide pools or otherwise interfere with replication fork progression. The loss of checkpoint targets like Exo1 and Dif1 may contribute to emerging drug resistance of such cancer cells.
Methods
Strains, Plasmids and Growth Conditions
All the yeast strains are derived from W303-1a and constructed by standard techniques. They are described in Supplementary Table S1. The ORFs encoding the different RNR subunits were amplified by PCR and cloned into pRS-GAL1,10 vectors. The resulting GAL1,10-RNR cassettes were integrated in the genome of a sml1Δ strain at the positions indicated in Supplementary Table S1. The resulting strains were then crossed with rad53Δ or rad53Δexo1Δ strains of the opposite mating type to generate diploids that were tetrad dissected after sporulation to obtain the strains used in the study.
Strains were grown regularly in YP medium plus glucose (2%) (YPD) at 30°C. For the induction experiments using the GAL1,10 promoter, raffinose (2%) was used to facilitate the induction of the relevant gene upon addition of galactose (2%). Cells were synchronized in G1 phase with the mating pheromone α-factor at 10 μg/ml. In cell cycle experiments, hydroxyurea (HU) was used at 0.2 M. In raffinose-medium, cell cycle progression is slower and thereby, cells were held in HU for 3 h to ensure S-phase onset.
Drop assays
Cultures growing exponentially were normalised and 1:5 serial dilutions were spotted onto YPD, YPGAL, YPD + HU or YPGAL + HU plates, depending on the experiment.
DNA replication analysis assays
Samples for flow-cytometry analysis were fixed with 70% ethanol and processed as described52. Density transfer assays were performed and analysed essentially as described53. All the strains used for these experiments lack the ARS608 and ARS60911. DNA was digested with ClaI and SalI before gradient centrifugation in CsCl. Probes corresponding to the analysed fragments were as described11. The extent of replication was calculated as: % replication = 100[0.5 HL/(HH + 0.5 HL)].
Protein electrophoresis and immunoblotting
Protein extracts were prepared as described previously54. For western blot analysis, proteins were separated by SDS-page and transferred to a nitrocellulose membrane (GE-healthcare). Membranes were cut in two halves and the upper or lower parts were used to detect RNR or PGK respectively. Detection of Rnr1 was performed using the rabbit polyclonal antibody AS09 576 (Agrisera; dilution 1:5000); Rnr3 was detected using the rabbit polyclonal antibody AS09 574 (Agrisera; dilution 1:1000); PGK was detected using the mouse polyclonal antibody A-6457 (Sigma; dilution 1:10000); Horseradish-peroxidase-coupled anti-rabbit or anti-mouse were used as the secondary antibodies. Immunoreactive bands were visualized by enhanced chemiluminescence (ECL, GE Healthcare) according to the manufacturer's instructions. Protein electrophoresis was carried out using 7.5% and 12% SDS-PAGE gels. The proteins were stained with Coomassie Blue after the electrophoresis.
References
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol Cell 40, 179–204 (2010).
Zeman, M. K. & Cimprich, K. A. Causes and consequences of replication stress. Nat Cell Biol 16, 2–9 (2014).
Segurado, M. & Tercero, J. A. The S-phase checkpoint: targeting the replication fork. Biol Cell 101, 617–627 (2009).
Labib, K. & De Piccoli, G. Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Philos Trans R Soc Lond B Biol Sci 366, 3554–3561 (2011).
Jossen, R. & Bermejo, R. The DNA damage checkpoint response to replication stress: A Game of Forks. Front Genet 4, 26 (2013).
Branzei, D. & Foiani, M. The checkpoint response to replication stress. DNA Repair (Amst) 8, 1038–1046 (2009).
Zegerman, P. & Diffley, J. F. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 8, 1077–1088 (2009).
Tercero, J. A., Longhese, M. P. & Diffley, J. F. A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11, 1323–1336 (2003).
Lopes, M. et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412, 557–561 (2001).
Tercero, J. A. & Diffley, J. F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412, 553–557 (2001).
Segurado, M. & Diffley, J. F. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22, 1816–1827 (2008).
Cotta-Ramusino, C. et al. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17, 153–159 (2005).
Cobb, J. A., Bjergbaek, L., Shimada, K., Frei, C. & Gasser, S. M. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22, 4325–4336 (2003).
Lucca, C. et al. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23, 1206–1213 (2004).
Zegerman, P. & Diffley, J. F. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467, 474–478 (2010).
Lopez-Mosqueda, J. et al. Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467, 479–483 (2010).
Allen, J. B., Zhou, Z., Siede, W., Friedberg, E. C. & Elledge, S. J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev 8, 2401–2415 (1994).
Weinert, T. A., Kiser, G. L. & Hartwell, L. H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev 8, 652–665 (1994).
Pellicioli, A. et al. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J 18, 6561–6572 (1999).
Huang, M., Zhou, Z. & Elledge, S. J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94, 595–605 (1998).
Zhou, Z. & Elledge, S. J. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75, 1119–1127 (1993).
Zaim, J., Speina, E. & Kierzek, A. M. Identification of new genes regulated by the Crt1 transcription factor, an effector of the DNA damage checkpoint pathway in Saccharomyces cerevisiae. J Biol Chem 280, 28–37 (2005).
Elledge, S. J. & Davis, R. W. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev 4, 740–751 (1990).
Domkin, V., Thelander, L. & Chabes, A. Yeast DNA damage-inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross-talking Rnr1/Rnr3 complex. J Biol Chem 277, 18574–18578 (2002).
Huang, M. & Elledge, S. J. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol Cell Biol 17, 6105–6113 (1997).
Chabes, A. et al. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc Natl Acad Sci U S A 97, 2474–2479 (2000).
Tsaponina, O., Barsoum, E., Astrom, S. U. & Chabes, A. Ixr1 is required for the expression of the ribonucleotide reductase Rnr1 and maintenance of dNTP pools. PLoS Genet 7, e1002061 (2011).
Zhao, X., Chabes, A., Domkin, V., Thelander, L. & Rothstein, R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J 20, 3544–3553 (2001).
Zhao, X. & Rothstein, R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci U S A 99, 3746–3751 (2002).
Wu, X. & Huang, M. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol 28, 7156–7167 (2008).
Lee, Y. D., Wang, J., Stubbe, J. & Elledge, S. J. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell 32, 70–80 (2008).
Yao, R. et al. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci U S A 100, 6628–6633 (2003).
Liu, C. et al. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 17, 1130–1140 (2003).
Tanaka, H. et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404, 42–49 (2000).
Xue, L. et al. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res 63, 980–986 (2003).
Engstrom, Y. & Rozell, B. Immunocytochemical evidence for the cytoplasmic localization and differential expression during the cell cycle of the M1 and M2 subunits of mammalian ribonucleotide reductase. EMBO J 7, 1615–1620 (1988).
Pontarin, G. et al. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc Natl Acad Sci U S A 105, 17801–17806 (2008).
Desany, B. A., Alcasabas, A. A., Bachant, J. B. & Elledge, S. J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev 12, 2956–2970 (1998).
Tercero, J. A. Density transfer as a method to analyze the progression of DNA replication forks. Methods Mol Biol 521, 203–213 (2009).
Cobb, J. A. et al. Replisome instability, fork collapse and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19, 3055–3069 (2005).
Feng, W., Bachant, J., Collingwood, D., Raghuraman, M. K. & Brewer, B. J. Centromere replication timing determines different forms of genomic instability in Saccharomyces cerevisiae checkpoint mutants during replication stress. Genetics 183, 1249–1260 (2009).
Harder, J. & Follmann, H. Identification of a free radical and oxygen dependence of ribonucleotide reductase in yeast. Free Radic Res Commun 10, 281–286 (1990).
Chabes, A. & Stillman, B. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104, 1183–1188 (2007).
Nyholm, S., Thelander, L. & Graslund, A. Reduction and loss of the iron center in the reaction of the small subunit of mouse ribonucleotide reductase with hydroxyurea. Biochemistry 32, 11569–11574 (1993).
Chabes, A. et al. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112, 391–401 (2003).
Piao, C. et al. MEK2 regulates ribonucleotide reductase activity through functional interaction with ribonucleotide reductase small subunit p53R2. Cell Cycle 11, 3237–3249 (2012).
Chang, L. et al. ATM-mediated serine 72 phosphorylation stabilizes ribonucleotide reductase small subunit p53R2 protein against MDM2 to DNA damage. Proc Natl Acad Sci U S A 105, 18519–18524 (2008).
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005).
Gorgoulis, V. G. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005).
Davidson, J. D. et al. An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res 64, 3761–3766 (2004).
Kuo, M. L., Hwang, H. S., Sosnay, P. R., Kunugi, K. A. & Kinsella, T. J. Overexpression of the R2 subunit of ribonucleotide reductase in human nasopharyngeal cancer cells reduces radiosensitivity. Cancer J 9, 277–285 (2003).
Labib, K., Diffley, J. F. & Kearsey, S. E. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol 1, 415–422 (1999).
Tercero, J. A., Labib, K. & Diffley, J. F. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J 19, 2082–2093 (2000).
Foiani, M., Marini, F., Gamba, D., Lucchini, G. & Plevani, P. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol 14, 923–933 (1994).
Acknowledgements
We thank Josep Clotet and Olga Calvo for providing antibodies and Andrei Chabes for providing strains. This work was supported by the Spanish Ministry of Economy and Competitiveness (MINECO) [BFU2013-43766-P and Consolider Ingenio CSD2007-00015 to J.A.T.; BFU2010-20034 and BFU2013-45182-P to M.S.] and Cancer Researck UK [to J.D.]. E.C. is the recipient of a pre-doctoral fellowship from MINECO. M.S. was supported by a JAE-DOC contract from CSIC and by a Ramón y Cajal contract from MINECO (RYC-2009-04407).
Author information
Authors and Affiliations
Contributions
M.S. and E.C.M. designed and carried out the experiments. J.D., J.A.T. and M.S. designed the study. J.A.T. and M.S. wrote the manuscript. All authors analysed and discussed the results and approved the final version of the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Morafraile, E., Diffley, J., Tercero, J. et al. Checkpoint-dependent RNR induction promotes fork restart after replicative stress. Sci Rep 5, 7886 (2015). https://doi.org/10.1038/srep07886
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07886
This article is cited by
-
Yeast Stn1 promotes MCM to circumvent Rad53 control of the S phase checkpoint
Current Genetics (2022)
-
RNases H1 and H2: guardians of the stability of the nuclear genome when supply of dNTPs is limiting for DNA synthesis
Current Genetics (2020)
-
S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion
Cellular and Molecular Life Sciences (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.