Abstract
Measuring the binding kinetics of antibodies to intact membrane proteins by surface plasmon resonance has been challenging largely because of the inherent difficulties in capturing membrane proteins on chip surfaces while retaining their native conformation. Here we describe a method in which His-tagged CXCR5, a GPCR, was purified and captured on a Biacore chip surface via the affinity tag. The captured receptor protein was then stabilized on the chip surface by limited cross-linking. The resulting chip surface retained ligand binding activity and was used for monoclonal antibody kinetics assays by a standard Biacore kinetics assay method with a simple low pH regeneration step. We demonstrate the advantages of this whole receptor assay when compared to available peptide-based binding assays. We further extended the application of the capture-stabilize approach to virus-like particles and demonstrated its utility analyzing antibodies against CD52, a GPI-anchored protein, in its native membrane environment. The results are the first demonstration of chemically stabilized chip surfaces for membrane protein SPR assays.
Similar content being viewed by others
Introduction
G-protein-coupled receptors (GPCRs) belong to a large superfamily of multispanning membrane proteins that represent an important drug class1. Compared to small molecule drugs, the development of therapeutics monoclonal antibodies targeting GPCRs has been slow2. Although several promising therapeutic monoclonal antibodies targeting GPCRs are being developed for various indications, nothing has been approved for clinical use2,3. One reason that an anti-GPCR antibody cannot progress as fast as those mAbs targeting soluble protein targets is the difficulty in candidate analysis and selection. In particular, progress in the development of label-free ligand/antibody binding assays for GPCRs has so far been challenging largely because of the inherent difficulties in capturing membrane proteins on analytical surfaces while retaining their native conformation2,4,5. Surface plasmon resonance (SPR) biosensor-based assays, such as Biacore assays, have been used to study protein interactions in real time without labeling6. Biacore pioneered commercial SPR biosensors offering a unique technology for collecting high-quality, information-rich data from biomolecular binding events6,7,8. Since the release of the first instrument in 1990, researchers around the world have used Biacore optical biosensors to characterize binding events with samples ranging from proteins, nucleic acids and small molecules to complex mixtures, lipid vesicles, viruses, bacteria and eukaryotic cells8. However, the application of SPR assays to study membrane-associated systems such as GPCRs is still in its infancy. One approach, developed by Karlsson and others, is to immobilize a purified receptor onto the sensor surface and then reconstitute a membrane environment on the surface9,10,11. Myszka and co-workers extended this method by showing that it was possible to capture receptors out of crude preparations and directly study the binding of antibodies12,13,14. However, with these methods, the lipid bilayer reconstituted on the chip surface is unstable and cannot be regenerated and is therefore rarely used.
Another approach is to engineer membrane proteins specifically for immobilization. For example, in studies by Myszka and co-workers, GPCRs were engineered using point mutations, yielding improved thermostability and conformational homogeneity15. After immobilization onto Biacore sensor chip surfaces, the engineered GPCRs retained activity. However, this approach is time consuming and a stable engineered molecule may not always be achievable; multiple mutations may alter the native structure of the receptor. In addition, this method requires pre-activation of nickel-nitrilotriacetic acid (Ni-NTA) SPR surfaces by a non-specific amine coupling reagent prior to capturing the purified receptor. Therefore, any contaminating proteins can be immobilized on the activated NTA surface via this process as long as they contain a primary amine group and this yields low binding activities for the surface-immobilized target protein. Furthermore, after binding, the surface can only be regenerated with a low affinity binder, which makes it impossible for analyzing large high affinity molecules such as antibodies.
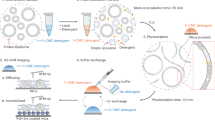
Recent developments in structural and functional analyses of GPCRs have allowed for more routine expression and purification of GPCR proteins in functional form without the need to modify their amino acid sequences16,17. Using unmodified receptor coding sequence, we added a 6xHis18,19,20 and HPC421,22 tandem tag to the C-terminus of the receptor to facilitate both purification and Biacore chip capturing processes. The tagged human CXCR5 receptor protein was expressed in insect Sf9 cells using the baculovirus expression system. The recombinant receptor protein was purified by a 2-step Ni-NTA/HPC4 affinity purification procedure and then captured on a Biacore NTA chip surface18,19. In order to regenerate the receptor surface for antibody binding kinetics assay, a limited cross-linking23 step was applied to stabilize the captured receptor. The resulting receptor surface retained ligand binding activity and was used for monoclonal antibody kinetics analysis by employing a standard Biacore kinetics assay method with a simple low pH regeneration step. We demonstrate here the advantages of this capture-stabilize approach in enabling a whole receptor-based assay. We further extended the application of the capture-stabilize approach to virus-like particles24,25 and demonstrate its utilities analyzing antibodies against CD52, a GPI-anchored protein, in its native membrane environment.
Results
Peptide-based binding kinetics assay
If an antibody binds to a linear portion of a GPCR molecule, for example, a contiguous sequence in the N-terminal extracellular domain, a synthetic peptide can be used for rapid binding evaluation. Two different assay formats can be used: the conventional format in which peptide flows over captured antibody at different concentrations or the “reverse format” which flows antibody over the immobilized peptide. Three anti-hCXCR5 mAb clones, 16D7, 79E7 and MAB190, were used to evaluate and compare these assay formats. As shown in Figure 1, in the conventional format, reproducible kinetics data (Table 1) were obtained. However, it did not provide sufficient resolution and sensitivity to differentiate the 3 mAb clones which is reflected in the Kon-Koff rate map shown in Figure 1a. In contrast, the reversed peptide-based assay format was able to distinguish mAb 16D7 with an almost 10-fold slower Koff-rate (Table 1), which was not revealed by the conventional format assay. The Kon-Koff rate map (Figure 1b) also indicated that this reversed peptide-based assay format displayed good reproducibility.
Biacore Kon-Koff rate map showing the differences between two peptide-based assay formats.
(a) The standard format in which the CXCR5 N-terminal peptide was flowed over captured antibody at different concentrations. Note that the differences among the three antibodies are within a single log range. (b) The reverse assay format in which antibodies were flowed over immobilized peptide at different concentrations. Note that 16D7 was differentiated from the two other antibodies.
However, since antibodies are dimeric molecules, it is possible that one antibody molecule could bind to two peptides on the chip surface thereby resulting in artificially high affinity through the avidity effect. Affinity (KD) data generated by the reverse format indeed were higher than the data generated by the conventional assay format. Therefore, the higher affinity data for the same antibody, at least in part, is attributed to the avidity effect. In order to evaluate the avidity effect in more detail, different levels (5, 20 and 100 RU, equivalent to 0.78, 3.1 and 15.6 fM/mm2, respectively) of biotinylated peptides were captured on a SA chip surface and kinetics assays were performed. As shown in Figure 2a, although the overall antibody binding RUs correlated to the peptide level captured, the shape of the sensorgrams for each antibody at different peptide densities were similar. As shown in Figure 2b, at three different peptide immobilization levels, MAB190 showed similar affinity results whereas clone 16D7 was dramatically different. The highest level of peptide (100 RU) did indeed alter the overall affinity (KD), mainly due to artificial Koff (9.85 × 10−8, Table 2), which was outside of the Biacore accuracy range (Koff ≥ 10−5S−1).
Peptide-based assays for anti-GPCR antibodies are convenient; however, they can only be used for analyzing antibodies recognizing linear epitopes. The conventional assay format lacks resolution as it can not distinguish between the three mAb clones we tested. Whereas the reversed assay format lacks accuracy due to the avidity effect. The development of a whole-receptor assay was able to address these issues.
Whole receptor-based binding kinetics assay
In order to develop GPCR whole receptor assays, a tandem 6xHis/HPC4 tag was added to the C-terminus of full-length human CXCR5 coding sequence and expressed in insect Sf9 cells using the baculovirus expression vector system. Following the methods described by Jaakola V.-P. et al.16, the human CXCR5 protein was purified as a mixture of monomer, dimer and trimer (Figure 3a, 3b). The purified receptor was directly captured on an NTA SPR sensor chip via Ni-mediated affinity capturing. In order to generate a stable chip surface, the commonly used amine coupling reagent NHS/EDC at 20 μM and 5 μM, respectively, was used to stabilize the captured receptor. The surface was then tested for both ligand binding (Figure 3c) and antibody binding (Figure 3d). High quality kinetics data were obtained following injection of ligand or antibody at different concentrations over the receptor surface.
Preparation of CXCR5 protein for whole receptor assay.
(a) SDS-PAGE analysis showing purified CXCR5 protein containing monomer as well as different degrees of oligomers. (B) Western blot analysis showing the reactivity of monomer, dimer and trimer by anti-CXCR5 antibody. (c) Representative Biacore kinetics sensorgram of CXCL13 ligand binding to CXCR5. (d) Representative Biacore kinetics sensorgram of anti-CXCR5 mAb 16D7 binding to CXCR5.
In order to assess the impact of the avidity effect on this capture and stabilize approach, chip surfaces with two different receptor densities (300 and 700 RUs) were prepared. As shown in Figure 4a, at both receptor densities, high quality kinetics sensorgrams were generated with good reproducibility. The Kon-Koff rate map (Figure 4b) further highlighted the reproducibility of triplicate data points. Three antibody samples as well as the ligand were well-differentiated by this assay format, which was not possible using the two other peptide-based assay formats. The kinetics data shown in Table 3 further demonstrated the reliability of the assay regardless of the difference in receptor density. Therefore, this limited chemical crosslinking step generated a stable sensor chip surface with full ligand binding activity. Using this method, the stabilized chip surface was regenerated with 50 mM HCl for up to 2000 repeated cycles without any noticeable loss of binding activity.
Capture-stabilized CD52 in virus-like particles
We further extended the application of the capture-stabilize approach to virus-like particles for analyzing other types of membrane protein targets in their native membrane environment. Human CD52, a heavily glycosylated glycosylphosphatidylinositol (GPI)-anchored protein composed of only 12 AA residues26,27, was used as an example. As shown in Figure 5, CD52 virus-like particles (VLPs) were prepared from 293FT cells. After Western blot analysis to verify the presence of CD52 in the VLP prep, VLPs containing CD52 were captured by the anti-CD52 antibody Alemtuzumab26 on a C1 chip surface. The captured VLPs were then stabilized by the same approach. Three different anti-CD52 monoclonal antibodies with slightly different binding epitopes (Chu, et al., 2014, unpublished data), including the capturing antibody Alemtuzumab, demonstrated good binding activity in kinetic assays. Similar to Ni-affinity capturing of the purified hCXCR5, the capture of CD52 VLPs by Alemtuzumab is target specific. The post capture cross-linking step stabilizes CD52 in the VLP surface and enables multiple kinetic binding assay cycles to be performed.
Capture-stabilize CD52 VLP on C1 chip for anti-CD52 antibody analysis.
(a). SDS-PAGE analysis of samples produced by HEK293FT cell transient expression. Lanes: 1. Sf9 cell negative control membrane prep, 2. Purified CD52 positive control protein produced in Sf9 cell, 3. HEK293FT negative control total cell lysates, 4. HEK293FT total cell lysates transiently expressing CD52, 5. A negative control VLP prep, 6. CD52 VLP prep from HEK293FT cells. (b). Western blotting with anti-CD52 antibody Alemtuzumab. The samples are the same as shown in panel A. (c). Biacore kinetics assay sensorgrams of anti-CD52 mAbs generated using CD52 VLPs captured and stabilized on a C1 chip surface.
Discussion
With this “capture-stabilize” approach, purified GPCR molecules are first captured directly on the Ni-NTA chip surface via a C-terminal 6xHis-tag. Alternatively, an anti-6xHis antibody can be first immobilized on a CM5 chip and then similar levels of the receptor protein are captured indirectly on the chip surface. If necessary, more receptor can be captured to meet specific requirements, such as small molecule compound binding assays. In both cases, receptor molecules are captured specifically via the 6xHis-tag. The subsequent limited cross-linking step further stabilizes the captured receptor molecules. Therefore, it is essential that the starting material, purified GPCR protein, is tested for activity by ligand binding. The purification methods used here, adapted from Jaakola V. et al. for determining the crystal structure of the human A2A adenosine receptor in complex with a high-affinity subtype-selective antagonist16, yield active pure GPCRs without the need to engineer stabilized receptors which require a substantial amount of effort15.
During the stabilization step, some of the receptor binding sites may have been blocked by the cross-linking reaction. A 10-fold dilution of the standard amine-coupling reagent from GE (200 μΜ NHS/50 μM EDC) gave the best results. Over cross-linking with 200 μΜ NHS/50 μM EDC resulted in significant loss of binding activity (data not shown). We also tested five other cross-linking reagents for receptor stabilization on both NTA and CM5 chip surfaces. In each case, similar results were obtained (data not shown). Therefore, we chose the 20 μΜ NHS/5 μM EDC as our standard stabilization reagent. The receptor surface stabilized by this simple cross-linking step yielded sufficient binding capacity for analyzing both antibody and ligand binding. More importantly, the limited cross-linking step kept the hCXCR5 whole receptor surface stable. The surface was not only resistant to the surfactant (0.05% Tween-20) present in the running buffer, but also allows for low pH (pH 1.5) regeneration steps. These features enabled the binding assay to be performed with the same conditions used for soluble proteins, which resulted in high quality kinetics sensorgrams and excellent reproducibility (CV<25%). This assay has been applied successfully to eight different GPCR targets (data not shown) and is expected to be applicable to most, if not all, GPCR molecules.
We further extended the application of the capture-stabilize approach to virus-like particles and demonstrated its utilities analyzing antibodies against CD52. Alemtuzumab (Campath-1H), specific to human CD52, is currently approved as a treatment for B-cell chronic lymphocytic leukaemia. Although Alemtuzumab is capable of binding to naked synthetic CD52 peptide, other anti-CD52 mAb clones require glycan as part of their binding target and the synthetic peptide method with these mAbs is not a viable option. The virus-like particles were captured by Alemtuzumab and further stabilized by limited cross-linking and this stabilized surface worked well in binding kinetics assays for comparing all types of antibodies, including the capturing antibody. VLPs produced by recombinant expression systems in Saccharomyces cerevisiae have been reported at 30–50 nM in size28. We also determined that the CD52 VLPs produced in HEK293FT cells were similar in size (30–50 nM, data not shown). It is presumed that only a portion of the CD52 molecules displayed on VLP surface is bound by the capturing antibody; those unoccupied CD52 molecules are therefore available for binding by the analyte antibodies. Similarly, we expect that this approach can be applied to other types of membrane targets, such as multi-transmembrane domain protein ion channels.
In conclusion, we have demonstrated the first successful use of a simple chemical cross-linking step to stabilize captured membrane proteins on Biacore chip surfaces. The resultant membrane protein surfaces remains stable allowing multiple rounds of consistent binding analyses with low pH regeneration steps. The capture-stabilize approach should provide a very useful tool for analyzing antibodies against a variety of membrane protein targets.
Methods
Conventional peptide-based Biacore assay
A mouse antibody capturing kit from GE was used for Biacore CM5 chip surface preparation. Following standard protocols provided by the manufacturer, the rabbit anti-mouse polyclonal antibody (GE) was first immobilized on the chip surface via amine coupling to the free carboxyl groups on the CM5 chip surface using standard NHS/EDC procedures. Mouse monoclonal antibodies against hCXCR5 clones 16D7, 79E7 and MAB190 (R&D Systems) were then captured by the anti-mouse antibody. Next, the hCXCR5 N-terminal peptide (MNYPLTLEMD LENLEDLFWE LDRLDNYNDT SLVENHLCPA TEGPLMASFK AVFVP) was injected over the captured anti-hCXCR5 antibodies at various concentrations to generate kinetics sensorgrams.
Reversed peptide-based Biacore assay
C-terminal biotinylated peptide (MNYPLTLEMD LENLEDLFWE LDRLDNYNDT SLVENHLCPA TEGPLMASFK AVFVPC-Biotin) was captured by streptavidin on the Biacore SA chip surface at specified density. Unused biotin-binding sites were blocked by free biotin. The same set of anti-hCXCR5 monoclonal antibodies used in the conventional assay was injected at various concentrations to generate Biacore kinetics sensorgrams.
Expression of hCXCR5
Human CXCR5 receptor was expressed in Sf9 cells using the FastBac expression system (Invitrogen). Sf9 cells were grown in suspension in flasks with serum-free medium. Cells were infected with recombinant virus at a density of 6 × 106 cells/ml; virus was added at the multiplicity of infection (MOI) of 10. An equal volume of fresh medium was added immediately afterward. Cells were harvested by centrifugation 72 hours post-infection.
Purification of hCXCR5
Following Jaakola V.-P.'s protocols16, The harvested Sf9 cells were initially disrupted by homogenization in a hypotonic buffer containing 10 mM HEPES (pH 7.5), 20 mM KCl and 10 mM MgCl2. Extensive washing of the isolated raw membranes was performed by repeated centrifugation (typically six to nine times) in a high-osmotic buffer containing 1.0 M NaCl, 10 mM HEPES (pH 7.5), 10 mM MgCl2, 20 mM KCl and protease inhibitor cocktail (Roche), followed by Dounce homogenization to resuspend the membranes in fresh wash buffer thereby separating soluble and membrane-associated proteins from integral transmembrane proteins. Highly purified membranes were solubilized by incubation in the presence of 0.5% (w/v) n-dodecyl-D-maltopyranoside (DDM) (Sigma) and 0.01% (w/v) cholesteryl hemisuccinate (CHS) (Sigma) for two to three hours at 4°C. After solubilization, the unsolubilized material was removed by centrifugation at 150,000 × g for 45 minutes. The supernatant was separated, supplemented with 25 mM buffered imidazole and incubated with Ni-NTA resin (Qiagen) overnight at 4°C; typically, 1.5 ml of resin per one liter of original culture volume was used. After binding, the resin was washed with ten column volumes of 25 mM HEPES (pH 7.5), 800 mM NaCl, 10% (v/v) glycerol, 55 mM imidazole, 0.05% (w/v) DDM and 0.001% (w/v) CHS. The receptor was eluted with 25 mM HEPES (pH 7.5), 800 mM NaCl, 10% (v/v) glycerol, 0.05% (w/v) DDM, 0.001% (w/v) CHS and 200 mM imidazole. The eluted receptor was directly added to anti-HPC4 mAb coupled affinity resin in the presence of 1 mM CaCl2 overnight, washed 12.5 mM HEPES (pH 7.5), 400 mM NaCl, 5% (v/v) glycerol, 0.025% (w/v) DDM, 0.001% (w/v) CHS, 1 mM CaCls and eluted with 12.5 mM HEPES (pH 7.5), 400 mM NaCl, 5% (v/v) glycerol, 0.025% (w/v) DDM, 0.001% (w/v) CHS, 1 mM EDTA. The eluted receptor was buffer exchanged to 12.5 mM HEPES (pH 7.5), 400 mM NaCl, 5% (v/v) glycerol, 0.025% (w/v) DDM, 0.001% (w/v) CHS and stored at −80°C.
Capturing hCXCR5 on Biacore sensor chip surface
For direct NTA chip capture, following 1 min injection of 0.5 mM NiCl2, purified hCXCR5 receptor at concentration of 50 ng/ml diluted in HBS-N buffer was injected with a flow rate of 5 μl/min to achieve capture level between 300-700 resonance units (RU), depending on experiment purpose.
Stabilization of hCXCR5
Ni-affinity captured hCXCR5 receptor protein was stabilized with limited cross-linking by flow over the mixtures of 20 μM EDC and 5 μM NHS for 7 min. Subsequently, excess cross-linkers were quenched with 1M ethanolamine for 10 min. The surface stabilized receptor chip was primed with running buffer before subsequent assay steps.
Regeneration of captured–stabilized hCXCR5 surfaces
At the end of each binding cycle, the GPCR surfaces were regenerated with injection of 50 mM HCl for 1 min at the flow rate of 50 ul/min.
Kinetic characterization of hCXCR5 ligand and antibodies
CXCL13 and 3 anti-hCXCR5 mAbs were each tested in triplicate in 2-fold dilution series for binding to hCXCR5 in HBS-EP+ running buffer. All samples were injected at a flow rate of 50 μl/min.
Capture-stabilize CD52 virus–like particles (VLP)
Full-length CD52 coding sequence (ATGAAGCGCTTCCTCTTCCTCCTACTCACCA TCAGCCTCCTGGTTATGGTACAGATACAAACTGGACTCTCAGGACAAAACGACACCAGCCAAACCAGCAGCCCCTCAGCATCCAGCAGCATGAGCGGAGGCATTTTCCTTTTCTTCGTGGCCAATGCCATAATCCACCTCTTCTGCTTCAGTTGA) was cloned into pEF_DEST51 vector (Invitrogen). Following the user guide for MembraneProTM Functional Protein Expression System from Life Technologies, CD52 VLP prep was obtained. After activation of the C1 chip surface by standard NHS/EDC procedure, the anti-CD52 antibody Alemtuzumab was immobilized and CD52 VLPs were then captured by the anti-CD52 antibody at 1300 RU level. The captured VLPs were further stabilized by limited crosslinking using 20 μM NHS/5 μM EDC for 7 min at flow rate of 5 μl/min and excess crosslinkers were quenched by injection of 1 M ethanolamine for 10 min. Kinetics assays with three anti-CD52 mAbs at concentrations from 0–80 nM were then performed.
Data processing and analysis
All biosensor data processing and analysis was performed using Biacore T100 Evaluation Software (GE, version 2.0.1). For kinetic analyses, data were locally fit to a 1:1 interaction model. CV (coefficient of variation) was calculated as the standard deviation (STDEV) divided by the mean.
References
Overington, J. P., Al-Lazikani, B. & Hopkins, A. L. How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 (2006).
Hutchings, C. J., Koglin, M. & Marshall, F. H. Therapeutic antibodies directed at G protein-coupled receptors. MAbs 2, 594–606 (2010).
Herr, D. R. Potential use of G protein-coupled receptor-blocking monoclonal antibodies as therapeutic agents for cancers. Int. Rev. Cell Mol. Biol. 297, 45–81 (2012).
Eisenstein, M. GPCRs: insane in the membrane. Nat. Meth. 6, 929–932 (2009).
Matthew, A. Cooper, Optical biosensors in drug discovery. Nat. Rev. Drug Discov. 1, 515–528 (2002).
Hahnefeld, C., Drewianka, S. & Herberg, F. W. Determination of Kinetic Data Using Surface Plasmon Resonance Biosensors. Meth. in Mol. Medic. 94, 299–320 (2004).
Phillips, K. S. & Cheng, Q. Recent advances in surface plasmon resonance based techniques for bioanalysis. Anal. Bioanal. Chem. 387, 1831–1840 (2007).
Jason-Moller, J., Murphy, M. & Bruno, J. Overview of Biacore Systems and Their Applications. Currt. Protoc. in Prot. Sci. 19.13.1–19.13.14– (2006).
Karlsson, O. P. & Löfås, S. Flow-mediated on-surface reconstitution of G-protein coupled receptors for application in surface resonance biosensors. Anal. Biochem. 300, 132–138 (2002).
Peter, J. et al. Direct analysis of a GPCR-agonist interaction by surface plasmon resonance. Eur. Bioph. J. 35, 709–712 (2006).
Sena, S. et al. A. Functional studies with membrane-bound and detergent-solubilized α2-adrenergic receptors expressed in Sf9 cells. Bioch. et Bioph. Act. (BBA) – Biomem. 1712, 62–70 (2005).
Navratilova, I., Sodroski, J. & Myszka, D. G. Solubilization, stabilization and purification of chemokine receptors using biosensor technology. Anal. Biochem. 339, 271–281 (2005).
Rich, R. L. & Myszka, D. G. Higher-throughput, label-free, real-time molecular interaction analysis. Anal. Biochem. 361, 1–6 (2007).
Rich, R. L. & Myszka, D. G. Survey of the year 2006 commercial optical biosensor literature. J. Mol. Recognit. 20, 300–366 (2007).
Rich, A. L., Errey, J., Marshall, F. & Myszka, D. G. Biacore analysis with stabilized G-protein-coupled receptors. Anal. Biochem. 409, 267–272 (2011).
Jaakola, V.-P. et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science 322, 1211–1217 (2008).
Grisshammer, R. Purification of recombinant G-protein-coupled receptors. Meth. in Enzymol. 463, 631–645 (2009).
Wear, M. A. & Walkinshaw, M. D. Determination of the rate constants for the FK506 binding protein/rapamycin interaction using surface plasmon resonance: an alternative sensor surface for Ni2+-nitrilotriacetic acid immobilization of His-tagged proteins. Anal. Biochem. 371, 250–252 (2007).
Yoshitani, N. et al. NTA-mediated protein capturing strategy in screening experiments for small organic molecules by surface plasmon resonance. Proteomics 7, 494–499 (2007).
Mersich, C. & Jungbauer, A. Generic method for quantification of FLAG-tagged fusion proteins by a real time biosensor. J. Biochem. Biophys. Meth. 70, 555–563 (2007).
Rezaie, A. R. & Esmon, C. T. The function of calcium in protein C activation by thrombin and the thrombin-thrombomodulin complex can be distinguished by mutational analysis of protein C derivatives. J. Biol. Chem. 267, 26104–26109 (1992).
Stearns, D. J. et al. The interaction of a Ca2+-dependent monoclonal antibody with the protein C activation peptide region. Evidence for obligatory Ca2+ binding to both antigen and antibody. J. Biol. Chem. 263, 826–832 (1988).
Hermanson, G. T. Bioconjugate Techniques (Academic Press, San Diego, 1996).
Vennema, H. et al. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15, 2020–2028 (1996).
Elizabeth, V., Grgacic, L. & Anderson, D. A. Virus-like particles: Passport to immune recognition. Meth. 40, 60–65 (2006).
Nguyen, T.-H. et al. Alemtuzumab induction of intracellular signaling and apoptosis in malignant B lymphocytes. Leukemia & Lymphoma, 53, 699–709 (2012).
Boutin, P. et al. Effects of anti-CD52 antibody treatment on immune cell depletion and progression of disease in murine systemic lupus erythematosus. J. Immunol. 188, 119.13 (2012).
Mach, H. et al. Disassembly and reassembly of yeast-derived recombinant human papillomavirus virus-like particles (HPV VLPs). J. Pharm. Sci. 95, 2195–2206 (2006).
Acknowledgements
We thank Alla Pritsker for preparing anti-CXCR5 monoclonal antibodies. We also thank Dr. Leila Sevigny for proof reading of the manuscript and helpful discussion.
Author information
Authors and Affiliations
Contributions
R.C. conducted all the experiments. R.C. and D.R. performed data analysis and wrote the manuscript with contributions from W.B. W.B. supervised the project.
Ethics declarations
Competing interests
R.C. and D.R. are authors on a patent application filed by the Genzyme Corporation on this intellectual property right, which is currently progressing through the international patenting process.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Chu, R., Reczek, D. & Brondyk, W. Capture-stabilize approach for membrane protein SPR assays. Sci Rep 4, 7360 (2014). https://doi.org/10.1038/srep07360
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07360
This article is cited by
-
Dissecting the conformation of glycans and their interactions with proteins
Journal of Biomedical Science (2020)
-
Biophysics in drug discovery: impact, challenges and opportunities
Nature Reviews Drug Discovery (2016)
-
Real-time and label-free analysis of binding thermodynamics of carbohydrate-protein interactions on unfixed cancer cell surfaces using a QCM biosensor
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.