Abstract
Flavonoids are well known as a large class of polyphenolic compounds, which have a variety of physiological activities, including anti-influenza virus activity. The influenza A/WSN/33 infected A549 cells have been used to screen anti-influenza virus drugs from natural flavonoid compounds library. Unexpectedly, some flavonoid compounds significantly inhibited virus replication, while the others dramatically promoted virus replication. In this study, we attempted to understand these differences between flavonoid compounds in their antivirus mechanisms. Hesperidin and kaempferol were chosen as representatives of both sides, each of which exhibited the opposite effects on influenza virus replication. Our investigation revealed that the opposite effects produced by hesperidin and kaempferol on influenza virus were due to inducing the opposite cell-autonomous immune responses by selectively modulating MAP kinase pathways: hesperidin up-regulated P38 and JNK expression and activation, thus resulting in the enhanced cell-autonomous immunity; while kaempferol dramatically down-regulated p38 and JNK expression and activation, thereby suppressing cell-autonomous immunity. In addition, hesperidin restricted RNPs export from nucleus by down-regulating ERK activation, but kaempferol promoted RNPs export by up-regulating ERK activation. Our findings demonstrate that a new generation of anti-influenza virus drugs could be developed based on selective modulation of MAP kinase pathways to stimulate cell-autonomous immunity.
Similar content being viewed by others
Introduction
Influenza virus is a global human and animal respiratory pathogen, causing mild to severe illness in seasonal outbreaks and periodic world-wide pandemics1,2. Approximately 20% of the world's population was infected by influenza virus every year, resulting in considerable morbidity and mortality3. Although antiviral drug therapy is essential to reduce disease progression and virus transmission, the number and effectiveness of antiviral drugs are limited. One of the reasons is the most current antiviral drugs directly targeting specific viral proteins4,5. However, the high frequency of RNA virus genome mutation leads to current antiviral drugs that are vulnerable to the rapid emergence of viral resistance and frequent evasion of host immune system6,7,8. In addition, genome of influenza virus contains eight segmented RNA that encoding a total of eleven proteins (HA, NA, NP, NS1, NS2, PA, M1, M2, PB1-F2, PB1 and PB2), but only NA and M2 have been proved the effective targets for anti-influenza viral therapy9,13. Compared to previous approaches to target viral genome for developing antiviral drugs, recently two novel strategies have been proposed by manipulating cellular factors that are required for the viral life cycle or by stimulating the host antiviral responses10,11,12,13. Host antiviral responses refer to not only immune system activation, but also cellular self-defense. Cellular self-defense also termed as cell-autonomous immunity, is developed by all organisms to protect host against microbial pathogens during evolution14,15. Unlike immune cells' specialized antiviral abilities, cell-autonomous immunity exists in the most cell lineages, providing the first line of host defense16,17. Even before immune system is triggered and immune cells are recruited to the location of virus infection, epithelial cells, the initial site of influenza virus infection, have already mounted antiviral responses to resist virus invasion by preventing entry, uncoating, replication and release at each stage of the viral life cycle and to restrict virus spread through paracrine of cytokines and chemokines rousing the uninfected bystander cells. The effectors of antiviral responses also include constitutive host defense factors18,19,20,21. These mechanisms contribute to efficiently restraining viral replication and spread as well as decreasing tissue damage.
Mitogen-activated protein kinase (MAPK) cascades are well known as signal transducers in the conversion of various extracellular signals into cellular response, which regulate numerous genes expression involved in proliferation, differentiation and also cell death and immune response22,23,24. There are three major members which have been identified to date including extracellular-signal-regulated kinase (ERK), JUN N-terminal kinase (JNK) and p38 kinase, each of them is organized in one cascade. Upon influenza virus infection, all of three pathways are activated25. Recent works have been focused on revealing the functions of p38, JNK and ERK signaling pathways in influenza virus life cycle. Several studies showed that p38 and JNK but not ERK have been linked with the expression of cytokines and chemokines, while ERK affected nuclear export of viral ribonucleoprotein complexes (RNPs)26,27,28,29. JNK and p38 are activated in human bronchial epithelial cells upon influenza virus infection. These kinases regulate RANTES expression, which is a part of the innate antiviral response of the cell30,31.
If one agent such as the small molecular compound can stimulate host defense factors production in the intracellular pattern or in the pattern of exocytosis, this compound could have an antiviral potential. To test this hypothesis, we used TCID50 (50% tissue culture infective dose) method in the influenza A/WSN/33 infected A549 cells and MDCK cells to screen anti-influenza viral drugs from our established natural flavonoid compounds library. A549 cell and MDCK cell, derived from lung epithelia of cancer patient and kidney epithelia of dog respectively, are two host cells easily infected by influenza A/WSN/33. We have found that some flavonoid compounds possess an ability of anti-influenza virus, one of which is hesperidin. Hesperidin potentially inhibited influenza virus production in A549 cells and MDCK cells. In this study, we investigated the mechanisms underlying the anti-influenza virus activity of hesperidin. Our data demonstrated that hesperidin enhanced cell-autonomous immunity by modulating MAP kinase signaling pathways via up-regulating p38 and JNK activation while down-regulating ERK activation. Unexpectedly, we demonstrated that kaempferol, one of the other kind of flavonoids, inhibited cell-autonomous immunity also by modulating MAP kinase signaling pathways with the completely opposite mechanisms through down-regulating p38 and JNK activation and up-regulating ERK activation, thus in turn increased influenza virus replication and spread both in vitro and in vivo. Our findings suggest that cell-autonomous immunity plays a pivotal role to prevent virus replication and spread. Targeting the signaling pathways related to regulating cell-autonomous immunity may be a promising therapy for virus infection.
Results
Flavonoids exhibit the opposite effects on influenza A/WSN/33 virus replication in MDCK and A549 cells
To evaluate the effects of flavonoids on influenza A virus replication, we first tested whether these natural compounds can inhibit the production of infectious progeny virus in MDCK and A549 cells. The effects of ten representative flavonoids on influenza virus replication were examined. The results showed that some flavonoids had an ability to inhibit virus replication while the other flavonoids promoted virus replication (Fig. S1 and S2). We chose hesperidin and kaempferol as individual representatives of both sides with the opposite effects to be further studied. Virions from the hesperidin-treated MDCK cells were 148 fold less than that of the untreated MDCK cells infected by influenza virus (positive control cell, PC), while the kaemperol-treated MDCK cells produced 100 fold more virions than that of PC (Fig. 1A). The opposite effects of hesperidin and kaempferol on virus replication were further confirmed in A549 cells by western blot using antibodies for recognizing viral nucleoprotein (NP) and non-structure protein 1 (NS1), respectively. NS1 and NP proteins were highly expressed in the cells treated with kaempferol, while the hesperidin-treated cells showed much lower level of both proteins, compared to the untreated cells (Fig. 1C). The increased viral RNA level in the kaempferol-treated cells and the decreased viral RNA level in the hesperidin-treated cells were also observed, which were consistent with the levels of viral proteins (Fig. 1B). In addition, for cytopathic effect (CPE) progress 48 h post-infection, the cells treated with hesperidin showed a significant delay compared to PC, while kaempferol treatment promoted CPE formation (Fig. S3). Our results suggest that the differences between these flavonoids in their chemical structures may lead to an enormous disparity in regulating influenza virus replication.
Hesperidin and kaempferol cause the opposite effects on influenza virus replication and antiviral state in the infected MDCK and A549 cells.
(A) MDCK cells were infected with influenza A/WSN/33 viruses at MOI of 0.001 for 3 h and then incubated with 100 μM of hesperidin or kaempferol for another 48 h, the cells were harvested and virus titer of each treatment was measured by TCID50 method. A549 cells were infected by Influenza A/WSN/33 virus for 3 h and then treated with hesperidin or kaempferol for 24 h, 0.05% DMSO as a control (PC). Cellular viral RNA (vRNA) level was examined by analyzing NP gene expression using Q-PCR (B), cellular influenza virus proteins NP and NS1 were detected by western blot (C), full-length blots are presented in Supplementary Figure 15. Anti-viral state associated genes expression in the infected A549 cells (D) and uninfected A549 cells (E). These genes, including interferon α (IFNα), interferon β (IFNβ) and interferon γ (IFNγ), monocyte chemotactic protein 1 (MCP1), regulated on activation, normal T cell expressed and secreted (RANTES) and interferon-inducible protein 10 (IP10) were determined by Q-PCR. Results were presented as means ± SEM of three independent experiments,*p < 0.05; **p < 0.01; ***p < 0.001, compared with untreated cell, mRNA expression of each target gene was normalized to GAPDH mRNA expression. H represented hesperidin; K represented kaempferol; 0.05% DMSO as a control (NC).
The opposite effects of kaempferol and hesperidin on viral replication may be due to exerting different influences on cell growth. To rule out the cytotoxicity induced by hesperidin and kaempferol treatments, we determined the cytotoxicity of kaempferol and hesperidin to A549 and MDCK cells. Up to the concentrations of 100 μM, hesperidin and kaempferol did not show any cytotoxicity to both cells, implying that the opposite effects of hesperidin and kaempferol on viral replication are not on account of disrupting cell growth, but probably due to interfering some definite cellular pathways, which are related to viral replication (Fig. S4).
Hesperidin and kaempferol show the opposite effects on antiviral state in A549 cells
To address whether the opposite effects of hesperidin and kaempferol on influenza virus replication were caused by producing different responses of epithelial cells to virus infection, we determined the antiviral state of the infected A549 cells treated with or without hesperidin and kaempferol. Q-PCR assay showed that antiviral state-associated genes expression in the infected A549 cells, including RANTES, IP10, MCP1, IFNα, IFNβ, IFNγ, etc., were significantly increased by hesperidin treatment but dramatically suppressed by kaempferol treatment, compared to the untreated cells (Fig. 1D and Fig. S5). We also detected antiviral state-associated genes expression in the uninfected A549 cells treated with hesperidin or kaempferol. We found that hesperidin treatment also enhanced antiviral state-associated genes expression in the uninfected A549 cells, but the levels of these genes expression were relatively lower than in the infected A549 cells (Fig. 1E and Fig. S6). In addition, kaempferol treatment still decreased antiviral state-associated genes expression in the uninfected A549 cells (Fig. 1E and Fig. S6). These results suggest that the opposite antiviral state-associated genes expression induced by hesperidin and kaempferol in A549 cells may be contributed to their opposite effects on influenza virus replication.
Hesperidin and kaempferol modulate transcriptional factors activation and antiviral genes expression through IFNs
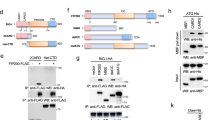
To further clarify whether IFN pathways participate in the hesperidin- and kaempferol-induced the opposite antiviral state, we detected the expression of Iκb, c-Jun, IRF3 and ATF-2 proteins using western blot, these proteins are required to initiate transcription of type I and type II IFN genes32,33,34. Our data showed that the levels of IκB, c-Jun, IRF3 and ATF-2 proteins in the nucleus were elevated by hesperidin-treatment while reduced by kaempferol-treatment compared to untreated control, indicating that both type I IFN and type II IFN signaling pathways are activated by hesperidin but suppressed by kaempferol (Fig. 2A). These results were consistent with the increased IFNα/β and IFNγ mRNA levels in the hesperidin-treated cells and the decreased IFNα/β and IFNγ mRNA levels in the kaempferol-treated cells (Fig. 1D). Activation of NFκB, c-Jun, IRF3 and ATF-2 are also responsible for other cytokines transcription such as RANTES30, suggesting that activation of these transcriptional factors are important for primary antiviral genes expression.
Interferon signaling pathways participate in hesperidin- and kaempferol-induced opposite antiviral state.
A549 cells were infected with Influenza A/WSN/33 virus for 3 h and then treated with hesperidin or kaempferol for 24 h, 0.05% DMSO as a control (PC). The proteins related to interferon signaling pathways were detected by western blot. The total and the nuclear proteins of activating transcription factor 2 (ATF-2), JUN gene coded protein (C-jun) and interferon regulatory factor 3 (IRF3) (A). Inhibitor of nuclear factor kappa-light-chain-enhancer of activated B cells (IκB), signal transducers and activators of transcription 1 (STAT1) and signal transducers and activators of transcription 3 (STAT3) proteins and their phosphorylation levels (B), GAPDH gene expression level is a control as in Fig. 1C. Full-length blots are presented in Supplementary Figure 15. Interferon-induced expression of antiviral genes, including: interferon-induced GTPase (MX-1), immunity-related GTPase family M protein (IRGM), interferon regulatory factor 7 (IRF7) and interferon activated gene 204 (IFI204) was analyzed by Q-PCR (c), data were presented as means ± SEM of three independent experiments, * p < 0.05; **p < 0.01;***p < 0.001, compared with untreated cells, mRNA expression of each target gene was normalized to GAPDH mRNA expression. H represented hesperidin; K represented kaempferol.
IFNs, key components of cell-autonomous immunity response, induce the expression of hundreds of genes to eliminate pathogens and protect host in multiple cell types21. We then determined the expression and activation of STAT1 and STAT3, which are primary components of IFN signaling cascade and major activators of secondary antiviral genes transcription. STAT1 and STAT3 forming homodimers and heterodimers bind to DNA sequences known as gamma activated sites (GAS) in which are control elements of a wide variety of cytokine-inducible genes33. Highly phosphorylated STAT1 and STAT3 were observed in the hesperidin-treated cells but not in the kaempferol-treated cells (Fig. 2B). The expression of MX-1 and IRF7 was also markedly increased by hesperidin but suppressed by kaempferol (Fig. 2C). In addition, the expression of MX-1 and IRF7 induced by hesperidin or kaempferol was slightly affected in the uninfected A549 cells compared to the infected cells (Fig. S7). These data demonstrated that hesperidin effectively increased primary and secondary antiviral genes expression, therefore built an antiviral state of cell-autonomous immunity both in the infected cells and in the uninfected cells. In contrast, kaempferol impaired an antiviral state of cell-autonomous immunity both in the infected cells and in the uninfected cells.
Reason of hesperidin- and kaempferol-induced the opposite effects on influenza virus replication is selectively up-regulating or down-regulating MAP kinase signaling pathways
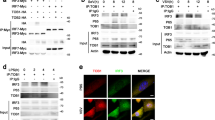
To identify whether the different antiviral states built by hesperidin and kaempferol are associated with MAP kinase signaling pathways (p38, JNK and ERK), we used specific inhibitors to determine how hesperidin and kaemperol regulate three MAP kinase groups. Addition of p38 inhibitor (SB203580) or JNK inhibitor (SP600125) to the infected A549 cells substantially promoted virus replication represented by virus titer as well as viral RNA level (Fig. 3A, B, C, D). These changes corresponded to the level of phosphorylated p38 and JNK proteins (Fig. S8). However, Addition of ERK inhibitor (U0126) to the infected A549 cells significantly reduced production of virions and prevented export of NP protein from the nucleus (Fig. S10). These results suggest that all of three MAP kinase signaling pathways are involved in regulating viral replication but they have different or even opposite effects on viral production. Then we ask whether hesperidin and kaempferol exert their effects on influenza virus replication through up- or down-regulating one or all of three MAP kinase signaling pathways. Indeed, three MAP kinase signaling pathways were activated during influenza virus infection (Fig. 3E, F and Fig. S9). Hesperidin treatment further improved activation of both p38 and JNK signaling pathways but attenuated ERK activation, whereas kaempferol treatment damped activation of p38 and JNK and reinforced ERK signaling pathway activation (Fig. 3E, F). In the infected cells, the increased export of RNP complexes from the nucleus and M1 protein translocation by kaempferol treatment and the decreased export of RNP complexes from the nucleus and M1 protein translocation by hesperidin treatment were also observed (Fig. 4A, B and Fig. S11). In the cells treated by U0126 alone and combination with kaempferol or hesperidin, we found that kaempferol offset the ability of U0126 (retaining RNPs in the nucleus) by up-regulating ERK activation, promoted the export of NP and M1 proteins from the nucleus and enhanced their expression levels (Fig. 4A, B, C and Fig. S10). In contrast, Hesperidin and U0126 combination was synergistic in inhibiting the export of NP and M1 proteins from the nucleus and decreased their expressions (Fig. 4A, B, C). To further confirm the effects of hesperidin and kaempferol on virus replication in the different regulation by ERK kinase cascade, we tested virions production in the treatment with hesperidin, kaempferol, U0126 alone or with the combination of U0126 with hesperidin or kaempferol (Fig. 4D). From the growth curves of influenza virus under indicated treatments, we found that U0126 or hesperidin alone had an ability to suppress influenza virus replication, while kaempferol offset the ability of U0126 to suppress influenza virus replication in the cells co-treated by U0126 and kaempferol. In contrast, the combination of U0126 and hesperidin was synergistic in inhibiting influenza virus replication. These results further demonstrated that hesperidin and kaempferol exert the opposite effects on influenza virus replication. Taken together, hesperidin and kaempferol selectively act on every MAP kinase signaling pathway by up- or down-regulating the one of which, thus in turn leading to the opposite effects of hesperidin and kaempferol on influenza virus replication.
JNK and p38 MAP kinase pathways are involved in hesperidin- and kaempferol-induced the opposite effects on influenza virus replication.
A549 cells were infected with influenza A/WSN/33 virus for 3 h and then treated with JNK inhibitor (SP600125) or p38 inhibitor (SB203580) for 24 h, 0.05% DMSO as a control (PC). Viral RNA levels in the SP600125-treated cells (A) and in the SB203580-treated cells (C) were measured by NP gene expression using Q-PCR, mRNA expression of NP gene was normalized to GAPDH mRNA expression. Virus titers in the SP600125-treated cells (B) and in the SB203580-treated cells (D) were measured by TCID50 method. The proteins (JNK, p38 and ERK) related to three MAP kinase signaling pathways along with their phosphorylation were detected by western blot (E) and analyzed using Image J software (F), GAPDH gene expression level is as a control. All of data were presented as means ± SEM of three independent experiments. H represented hesperidin; K represented kaempferol.
ERK signaling pathway participates in the hesperidin- and kaempferol- induced the opposite effects on influenza virus replication.
A549 cells were infected with influenza A/WSN/33 virus for 3 h and then treated by ERK inhibitor (U0126) or hesperidin or kaempferol, respectively, or by combination of U0126 with hesperidin or kaempferol for 24 h, 0.05% DMSO as a control (PC). (A) Cells were stained by indirect immunofluorescence using NP or M1 specific rabbit antibody and FITC-coupled anti-rabbit IgG to determine cellular distribution and expression of NP and M1 proteins (Green), DAPI staining shows cell nucleus (Blue). (B) Statistically analysis of expression of NP and M1 proteins in nucleus using confocal laser scanning microscope FV1000 (OLYMPUS). (C) Statistically analysis of expression of NP and M1 proteins using flow cytometry (BD FACSCalibur). Data presented as means ± SEM of five visual fields for confocal and three independent experiments for FACS. H represented hesperidin; K represented kaempferol. (D) Virus growth curves. A549 cells were infected with influenza virus at a multiplicity of infection of 0.001 for multi-cycle replication and then treated with 100 μM of hesperidin or kaempferol or 0.05% DMSO, or with 10 μM of U0126 alone or 10 μM of U0126 combined with 100 μM hesperidin or kaempferol. At the indicated times, supernatants were collected and then plaque titres were determined on MDCK cells. Each growth curve was represented by the average of three independent experiments.
p38 signaling pathway plays a main role to discriminate between hesperidin and kaempferol induced effects on cell-autonomous immunity
In view of hesperidin and kaempferol treatments induced more significant changes in expression and phosphorylation of p38 than in that of JNK and ERK, we proposed the hypothesis that p38 may mainly be in charge of cell-autonomous immunity mediated by hesperidin and kaempferol. To address this hypothesis, we detected activation of p38 protein in the infected A549 cells treated with hesperidin or kaempferol. Hesperidin treatment led to the increase of both p38 and phosphorylated p38 proteins, while kaempferol treatment reduced the levels of p38 protein and its phosphorylation (Fig. 3E, F).
To further explore the rule of p38 in influenza virus replication in hesperidin- and kaempferol-treated cells, we detected influenza virus production in A549 cells by treatment with SB203580 or by over-expression of p38 gene. The results showed that the progression of influenza virus was promoted when A549 cells treated with SB203580, however, SB203580 induced virus production was significantly reversed by addition of hesperidin (Fig. 5A, B). Antiviral genes expression was corresponded to the p38 protein expression in the cells treated by hesperidin, SB203580 and SB203580 and hesperidin combination, suggested that hesperidin restricted influenza virus replication and spread by enhancing cell-autonomous immunity partly through up-regulating p38 signaling pathway (Fig. 5E and Fig. S12). We constructed p38-full length expression plasmids, p-eGFP plasmid as control and transfected these plasmids into A549 cells. Over expression of p38 in the infected cells dramatically reduced kaempferol-induced viral RNA (vRNA) expression compared to control plasmid (Fig. S13). The enhanced autonomous immunity (highly expressing antiviral genes) in the kaempferol-treated infected cells was associated with high expression of the transfected p38 gene (Fig. S13). Additionally, we also detected the effects of co-treatment of SP600125 with hesperidin on virus production and found that this co-treatment reduced SP600125-induced virus production as well as vRNA expression, corresponding to the increased activation of JNK (Fig. S12 and Fig. 5C, D). However, the reduced virus production by co-treatment of SP600125 with hesperidin was much less than that by co-treatment of SB203580 with hesperidin, which probably related to the recovered activation of p38 by hesperidin was much more than that of JNK (Fig. S12). These results indicate that hesperidin exerting its effects on virus replication is mainly through p38 signaling pathway. Taken together, our results strongly demonstrate that p38 signaling pathway plays a definitive role in hesperidin- and kaempferol-induced cell-autonomous immunity to adopt differential responses to influenza virus infection.
p38 MAP kinase pathway has a critical role in hesperidin-induced cell-autonomous immune responses.
A549 cells were infected with influenza A/WSN/33 virus for 3 h and then treated by p38 inhibitor (SB203580), JNK inhibitor (SP600125) and hesperidin, respectively, or by combination of hesperidin with SB203580 or SP600125, 0.05% DMSO as a control (PC). Viral RNA levels in the SB203580-treated cells (A) and in the SP600125-treated cells (C) were measured by NP gene expression using Q-PCR, mRNA expression of NP gene was normalized to GAPDH mRNA expression. Virus titers in the SB203580-treated cells (B) and in the SP600125-treated cells (D) were measured by TCID50 method. (E) mRNA expression of genes associated with cell-autonomous immunity was determined by Q-PCR, including : IFNα, IFNβ and IFNγ, MCP1, RANTES, IP10, *p < 0.05; **p < 0.01; ***p < 0.001, compared with infected cells in presence of SB203580, mRNA expression of each target gene was normalized to GAPDH mRNA expression. All of data were presented as means ± SEM of three independent experiments. H represented hesperidin.
Hesperidin protects mice from influenza-induced lethality but kaempferol promotes mice death
To assess the effects of hesperidin and kaempferol on influenza virus replication in vivo, we established an influenza virus infected mouse model using intranasal administration of influenza virus at a concentration of 6.8 × 103 pfu/mouse. The infected mice received 100 mg/kg/day of hesperidin or kaempferol 48 h before infection until the mice died. The mice with hesperidin treatment were significantly protected from influenza-induced lethality, whereas all of the mice with kaempferol treatment died on day 10 after infection, earlier than the mice administered vehicle only (all mice died on day12; Fig. 6A). The shortest survival in the kaempferol-treated mice was due to the most severe weight loss compared to the control mice and the hesperidin-treated mice (Fig. 6B). Hesperidin treatment also led to a statistically significant reduction in influenza-induced lung pathology. The infected mice with kaempferol treatment showed extensive lung damage characterized by fragments, collapse and inflammatory cells infiltration such as neutrophils, monocytes and lymphocytes on day 6. In addition, the infected mice without any treatment were lesser extent damage in their lungs than that of the infected mice with kaempferol treatment. However, on day 3 after infection, each lung section of the infected mice with hesperidin treatment exhibited nearly normal lung architecture, even on day 6, few inflammatory cells infiltration could be observed in the lungs (Fig. 6E). The degrees of lung injury were paralleled by the high weight ratio of lungs to bodies in the kaempferol-treated mice versus the control mice versus the hesperidin-treated mice (Fig. 6C). Dissemination of influenza virus in the lungs of mice was detected by immunohistochemical staining using antibodies for recognizing NS1 and NP proteins. On day 3 after infection, the lung sections of hesperidin-treated mice showed virions restricted in superior segmental bronchus, whereas the kaempferol-treated mice and the control mice showed more virons staining in distal portions of bronchus in the lungs, indicating that influenza viruses reach deeper lung with more extensive spread in the kaempferol-treated mice and the untreated mice compared to the hesperidin-treated mice (Fig. 6F). On day 6 after infection, viral titers in serum showed that hesperidin treatment resulted in a statistically significant decrease, however kaempferol treatment led to approximate 100 fold increases than the untreated mice infected with influenza virus (Fig. 6D). Our data demonstrated that hesperidin treatment markedly reduced influenza virus replication and dissemination not only in the lungs but also in the blood, in contrast, kaempferol treatment dramatically promoted influenza virus replication and dissemination. We further in vivo confirmed the opposite effects of hesperidin and kaempferol on influenza virus infection.
Hesperidin and kaempferol have differential influences on the survival and lung pathology of mice challenged with a lethal dose of influenza virus.
(A) The survival of mice challenged by nasal administration of 6.8 × 103 pfu of influenza A/WSN/33 virus per mouse with pre-treatment of 100 mg/kg of hesperidin or kaempferol once daily for 2 days, 1% CMCNa as a vehicle, **p < 0.01, compared with 1% CMCNa treatment. Systemic and lung damages were evaluated by loss of body weight (B) and changes in lung/body weight ratio (C). (D) Viral RNA (NP gene) in serum on day 6 post-infection was determined by Q-PCR. (E) Lung pathology from infected mice with or without treatments was examined by H&E staining on days 3 and 6 post-infection, normal lungs as control. (F) Localization of virus within the lungs was determined by immunohistochemistry for influenza virus NP and NS1 proteins, indicated by red arrows. Original magnifications were ×100.
Discussion
Cell-autonomous immunity confers the non-immune cells such as epithelial cells the capability to guard the host against immediate threat by restricting pathogens replication and spread and plays a vital role in antiviral response of the host cell14. Cell-autonomous effector mechanisms are essential for all multi-cellular organisms and appear conserved in different evolved species15. The pre-existing repertoire of constitutive host defense factors against virus (such as TRIM family) and the additional factors induced by cytokines and chemokines participate in different stages of pathogens' life cycle35,36. The main target of influenza virus is epithelial cells in the respiratory tract. These cells have an important role in host innate and adaptive immune responses to the virus infection. Influenza virus can be recognized by many PRRs (pattern recognition receptors), including RLRs (RIG-I-like receptors), TLRs (Toll-like receptors) and NLRs (NOD-like receptors) and then trigger cellular signaling pathways activation.
Previous studies have demonstrated several pathways regulating cell-autonomous immunity. The specific inhibitor of p38 protein inhibited cell-autonomous immunity by decreased cytokines and chemokines expression. By contrast, over expression of p38 protein enhanced cell-autonomous immunity (Fig. 3 and Fig. 5)37,38,39. Previous reports combined with our findings suggested MAP kinase pathways involved in regulating cell-autonomous immunity. Furthermore, activation of p38 and JNK is required for interferon expression40,41,42. In addition, inhibitors of ERK restricted RNPs export from the nucleus resulting in limited replication of influenza virus (Fig. 4)29,43. Taken together, we propose that exploiting cell-autonomous immunity by selectively modulating MAP kinase pathways could be a new strategy for developing anti-influenza virus drugs.
Flavonoids-based anti-influenza virus drugs screening revealed that flavonoid compounds like hesperidin and kaempferol showed the different antiviral activities by altering cell-autonomous immunity state mostly though IFNs. Type I and type II IFNs are well known as their antiviral effects for various viral infections32,44,45. Previous reports have demonstrated that p38 protein has a critical role in antiviral responses26,46. The phosphorylated p38 proteins translocate in nucleus to activate several transcription factors such as NFκB, AP1 and Jun and are also essential for STAT1 phosphorylation, which is directly involved in INFγ transcription47,48,49,50,51. NFκB/IκB complex is activated by phosphorylation of IκB allowing the release of NFκB52. With activated IRF3, they translocate in nucleus, forming complex with ATF-2 to assemble on IFNβ promoter, under the help of accessory factor HMG-I/Y to form enhanceosome, which recruit RNA polymerase II to the promoter and start the transcription of IFNβ32,34. C-Jun and ATF-2 are proximal regulation elements for IFNγ transcription, their activation is essential to IFNγ production32.
The differences between hesperidin- and kaempferol-induced cell-autonomous immunity are not in the degree but in the quality: cell-autonomous immunity enhanced by hesperidin but suppressed by kaempferol. Our further investigation revealed that the completely opposite cell-autonomous immune responses induced by hesperidin and kaempferol are due to their differences in the modulating MAP kinase pathways (Fig. S14). One of the most important aspects is that hesperidin up-regulates expression and activation of P38 and JNK, resulting in the enhanced cell-autonomous immunity. In contrast, kaempferol dramatically down-regulates expression and activation of p38 and JNK, thereby suppressing cell-autonomous immunity. These two opposite cell-autonomous immune responses were established via cytokines and chemokines expression levels, especially by IFN-β and IFN-γ as primary antiviral response and then much stronger and broader secondary antiviral responses were activated via STAT pathway. On the other aspect, hesperidin restricted RNPs export from the nucleus by down-regulating ERK activation while kaempferol promoted RNPs export by up-regulating ERK activation. Furthermore, we compared chemical structures of hesperidin and kaempferol to other flavonoids in our screening pool, which exhibited the opposite effects of anti-influenza virus and found that the opposite effects of these two kinds of flavonoids on influenza virus replication were probably due to their structural differences. Flavonoids possessing an ability of anti-influenza virus replication have a double hydrogen bond between C2 and C3 and a replaced group at C2 or a double bond between C2 and C3 and a replaced group at C3. In contrast, the flavonoids promoting influenza virus replication all have a double bond between C2 and C3 and a replaced group at C2, suggesting that the form of chemical bond between C2 and C3 and a replaced group at C2 or C3 in one flavonoid compound may be essential for its anti-influenza virus efficacy. The relationship between chemical structures of flavonoid compounds and their anti-viral activities should be investigated deeply in the future.
Interestingly, using H1N1 virus strain, we infected A549 cells and found kaempferol significantly promoted the virus replication. It seems to be a contradiction between our observation in the H1N1 infected A549 cell and Sithisarn's observation that kaempferol inhibited influenza A nucleoprotein production in A549 cell infected with the highly pathogenic avian influenza H5N1 virus strain A/Thailand/Kan-1/0453. However, they did not perform virus yield detection in the H5N1 infected A549 cells with kaempferol treatment and further investigation of possible mechanism. Based on the Sithisarn's observation, we can't infer that kaempferol has a potential to inhibit replication and spread of either H1N1 or H5N1 influenza virus both in vitro and in vivo. Previous data in transcriptomic study revealed that there appears to be no qualitative difference between H5N1 and H1N1 viruses in the signaling pathways triggered in infected cells54. The stronger proinflammatory cytokine cascades induced in H5N1 infection than in H1N1 infection is likely due to its potency in activating the innate immune sensors rather than differences in the type of innate immune sensors or signaling pathways activated55. In this study, we didn't conduct any research toward H5N1 strain, thus further study should be carried out to verify whether kaempferol can inhibit H5N1 infection by reducing virus production.
Flavonoids, widely exist in plant kingdom, including our daily diets such as vegetables and fruits, are also important components of many traditional herb medicines56,57. The better understanding of the functions of flavonoids, the better taking advantages of flavonoids in our daily lives and health care industry58,59. For example, we can choose fruits and vegetables rich in hesperidin as our main food source during influenza seasons to promote our autonomous immunity to defend influenza virus infection. Additionally, many traditional antiviral drugs are made from plant extracts containing some antiviral components, but mechanisms of these molecules underlying their antiviral effects remain unclear. Our study may provide a promising strategy to find out a new generation of anti-influenza viruses drugs based on selectively modulating MAP kinase pathways to stimulate cell-autonomous immunity.
Methods
Ethics Statement
All animal experiments were approved by the Animal Ethics and Experimentation Committee of the Institute of Biophysics with license number of SYXK (SPF) 2008-104 and performed according to the regulations for the administration of affairs concerning experimental animals approved by the state council of the People's Republic of China.
Cells and viruses
Madin-Darby canine kidney (MDCK) and human lung carcinoma (A549) cells (American Type Culture Collection, Manassas, VA) were grown in DMEM containing 10% fetal bovine serum (GIBCO) without antibiotics in a 5% CO2 incubator. Influenza A/WSN/33 was provided by Dr. Gegore Fu Gao, (CAS, Beijing). The virus was passaged and amplified in MDCK cells, in DMEM with 1 μg/ml TPCK-trypsin and 0.2% BSA, virions were tittered and stored in aliquots at −80°C for further studies.
Chemical reagents
Chrysin, daidzein, diosmetin, genistein, hesperidin, icraiin, kaempferol, naringenin, naringin dihydrochalcone and neosperidin dichdrochalcone were purchased from Shanxi Huike Botanical Development Co. Ltd in China and their purity is larger than 98%. Hesperidin and kaempferol used for in vivo animal studies were suspended in 1% carboxymethyl cellulose sodium (CMCNa), freshly prepared for daily use. All Compounds stocks were dissolved in dimethyl sulfoxide (DMSO) at a concentration of 200 mM for in vitro studies and diluted for further use.
Virus infection and compounds treatment
A549 cells were seeded at 24-well plate one day before challenged with influenza A/WSN/33 at MOI 0.001 for 3 h at 37°C, removed virus and washed once with PBS, then incubated with medium in the presence of 100 μM hesperidin or kaempferol or DMSO for 24 h. Mock infection was performed with DMEM medium. Viral gene (NP gene) expression, cytokines and chemokines expression were determined by Western Blot and Q-PCR.
Viral titration method
The 50% tissue culture infectious dose (TCID50) was determined in MDCK and A549 cells. MDCK and A549 cells were seeded in 96-well plate 24 h before infection, then cells were infected with 1:10 diluted virus solution in presence of 100 μM indicated compounds at 37°C, infectivity was determined by endpoint dilution titration on MDCK cells and detection of the cytopathogenic effects at 48 h post infection. TCID50 values were calculated by the method of Reed and Muench60.
Immunoblotting assay
Cells were lysed and cleared by centrifugation, protein concentration was determined by Bradford system (Therom) and protein was separated on SDS-poly-acrylamide gels, then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore) and stained with indicated primary antibodies. Species-specific secondary antibodies which conjugated to horseradish peroxidase (Cell signaling) were incubated with the membranes. Membranes were developed using enhanced chemiluminescence reaction (Transgene).
Flow cytometric staining assay
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% triton ×100. Cells were then incubated with blocking buffer (1%BSA/PBS) and stained with NP (SANTA CRUZ) or M1 (Abcam) antibody for 1 h on ice and washed twice with ice-cold PBS. Cells were incubated with species-specific secondary antibodies which conjugated to FITC (Sigma) for 30 min on ice. Cells were washed twice and resuspended in 200 μl PBS. Fluorescence of Antibody-stained cells was measured and analyzed using a FACSCalibur (BD Biosciences).
Immunostaining assay
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% triton ×100.Cells were then incubated with blocking buffer (1%BSA/PBS) and stained with NP (SANTA CRUZ) or M1 (Abcam) antibody for 1 h at 37°C and washed twice with PBS. Cells were incubated with species-specific secondary antibodies which conjugated to FITC (Sigma) for 30 min at 37°C. Cells were washed twice and incubated with 2 μg/ml DAPI (Invitrotgen) for 5 min at 37°C. Cells were then imaged at 60× oil microscope objective using confocal laser scanning microscope FV1000 (OLYMPUS).
RNA Quantification
Total RNA was isolated for quantitative real-time PCR (Q-PCR) using a standard guanidium isothiocyanate method. Viral RNA (vRNA) was isolated using starspin viral RNA kit (Genstar). RNA was quantitated and reversed transcripted by means of TransScript one-step gDNA Removal and cDNA Synthesis SuperMix (TransGen) with oligo-dT primers. Q-PCR was done using the step one plus Real-time PCR system (ABI). Q-PCR was conducted in a final volume of 20 μl containing: 1× pre-mix, gene specific primers and cDNA as template. Amplification conditions were: 95°C (10 min), 40 cycles of 95°C (30 s), 61°C (30 s), 72°C (30 s).
Growth curves
A549 cells were infected with influenza virus at a multiplicity of infection of 0.001 for multi-cycle replication and then treated with 100 μM of hesperidin or kaempferol or 0.05% DMSO, or with 10 μM of U0126 alone or 10 μM of U0126 combined with 100 μM of hesperidin or kaempferol. At the indicated times, supernatants were collected,and then plaque titres were determined on MDCK cells. Each growth curve was represented by the average of three independent experiments.
Plasmid construction
The full-length p38α expression plasmid was created by PCR amplification of p38α cDNA (NCBI munber: NM_001315.2). Fragments were cloned into the SpeI/BamHI sites of pSin-EF2-Oct4-puro vector.
Preparation of nuclear proteins and whole cell extracts
Nuclear fractions of A549 cells were prepared using Nuclear-Cytosol Extraction kit (Applygen). In brief, A549 cells were washed in PBS and lysed with cold buffer A on ice for 10 min, add 30 μl buffer B on ice for 1 min. The nuclear pellet was harvest and washed once with 100 μl buffer A by centrifugation at 12,000 × g for 5 min at 4°C. The nuclear pellet was resuspended in 75 μl of ice-cold buffer C and incubated on ice for 30 min with vortex 10 s every 5 min. The nuclear protein was extracted by centrifugation at 12,000 × g for 5 min at 4°C. Whole cell extracts were prepared by lysing A549 cells with ice-cold cell lysis buffer (Applygen), protease inhibitor (Sigma) and phosphatase inhibitor (Sigma) on ice for 20 min. The whole cell lysates were harvest by centrifugation at 13,000 × g for 20 min at 4°C. The protein content was determined with pierce BCA protein assay kit (Thermo) using BSA as a standard.
Viability assay
A549 or MDCK cells were seeded in 96-well plate and grown for 24 h. Seeded cells were treated with increasing concentrations of hesperidin or kaempferol (0, 5, 10, 25, 50, 100, 200 and 400 μM in medium containing 0.05% DMSO) for 48 h. Negative control wells contained 0.05% DMSO only. The medium was then replaced by 100 μl MTT-PBS solution and incubated for 4 h at 37°C. The supernatant was removed carefully and the tetrazolium crystals were dissolved by adding 100 μl of DMSO to each well. The plate were shaken for 10 min and analyzed in an enzyme-linked immunosorbent assay (ELISA) reader at 590 nm excitation.
Animals and treatments
Four to six-week-old female BALB/c mice were purchased from the Vital River Laboratory Technology Co.Ltd (Beijing, China). The welfare of all animals housed and studied were under the relevant guidelines and Institutional Animal Care and Use Committee-approved protocols. Total 72 mice were divided into 4 groups (uninfected, hesperidin-treated, kaempferol-treated and untreated). Each group was re-divided into three sub-groups (day 3, day 6 p.i. and lifespan), 6 mice per each sub-group. Mice were anesthetized intraperitoneally by pentobarbital sodium solution (6 mg/ml) and inoculated intranasally with 6.8 × 103 pfu of influenza A/WSN/33 viruses diluted in 50 μl PBS or the uninfected group with 50 μl PBS only. Hesperidin or kaempferol was suspended in 1% CMCNa solution (10 mg/ml) and treated mice 48 h before infection and every 24 h thereafter by intragastric administration (100 mg/kg). Blood was drawn from orbit before mice sacrificed for tissue analysis and the separated serum was stored at −80°C for viral RNA analysis. Mice were sacrificed on day 3 or day 6 post infection. The removed lungs were immediately weighted and subsequently fixed by 4% Paraformaldehyde (PFA) for immunohistochemistry (IHC).
Pathology of infected mice
After the lung tissue were embedded in low-melting-point paraffin wax, 5 μm sections were cut and stained with haematoxylin and eosin (HE) for general histopathology assessment by microscopically examined. The staining of viral proteins (NP or NS1), were using standard IHC protocol. In brief, sections were blocked in 10% sheep serum for 1 h at room temperate (RT) after deparaffinization and antigen unmasking. Viral proteins NP and NS1 were stained with their specific primary antibody (SANTA CRUS and Abcam) overnight at 4°C. Removed antibody solution and washed the sections with wash buffer (0.1 M PBS, pH7.2) three times for 5 min each. The sections were incubated with secondary antibody (ORIGENE) for 30 min at 37°C. After repeat the wash step, sections were developed with DAB solution and stained with haematoxylin solution, then were dehydrated and mounted in cover slip.
Statistical analysis
The results were presented with mean ± SEM of the indicated experiment. Statistical analysis was performed by student t test, values of P < 0.05 were considered statistically significant, *P < 0.05; **P < 0.01; ***P < 0.001. Survival analyses were analyzed using Kaplan-Meier method and a Log-rank (Mantel-Cox) test.
References
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M. & Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 56, 152–179 (1992).
Cox, N. J. & Subbarao, K. Influenza. Lancet 354, 1277–1282 (1999).
Moscona, A. Neuraminidase inhibitors for influenza. N. Engl. J. Med. 353, 1363–1373 (2005).
Gubareva, L. V., Kaiser, L. & Hayden, F. G. Influenza virus neuraminidase inhibitors. Lancet 355, 827–835 (2000).
Gubareva, L. V. & Hayden, F. G. The anti-influenza activity of M2 and neuraminidase inhibotors [Kawaoka Y. (Ed.)] [169–172] (Caister Academic, England, 2006).
Fleming, D. M. Managing influenza: amantadine, rimantadine and beyond. Int. J. Clin. Pract. 55, 189–195 (2001).
Hayden, F. Developing new antiviral agents for influenza treatment: what does the future hold? Clin. Infect. Dis. 48, S3–S13 (2009).
Moscona, A. Global transmission of oseltamivir-resistant influenza. N. Engl. J. Med. 360, 953–956 (2009).
Lamb, R. A. & Krug, R. M. [Orthomyxoviridae: the viruses and their replication]. Fields virology [Fields B. N., Knipe D. M., & Howley P. M. (eds.)] [1487–1531] (Lippincott-Raven, Philadelphia, 2001).
Muller, K. H. et al. Emerging cellular targets for influenza antiviral agents. Trends Pharmacol. Sci. 33, 89–99 (2012).
Konig, R. et al. Human host factors required for influenza virus replication. Nature 463, 813–817 (2010).
Ludwig, S. Disruption of virus-host cell interactions and cell signaling pathways as an anti-viral approach against influenza virus infections. Biol. Che. 392, 837–847 (2011).
Lee, S. M. Y. & Yen, H. L. Targeting the host or the virus: Current and novel concepts for antiviral approaches against influenza virus infection. Antiviral Res. 96, 391–404 (2012).
Randow, F., MacMicking, J. D. & James, L. C. Cellular self-defense: How cell-autonomous immunity protects against pathogens. Science 340, 701–706 (2013).
Ronald, P. C. & Beutler, B. Plant and animal sensors of conserved microbial signatures. Science 330, 1061–1064 (2010).
Levine, B., Mizushima, N. & Virgin, H. W. Autophagy in immunity and inflammation. Nature 469, 323–335 (2011).
Neil, S. J. D., Zang, T. & Bieniasz, P. D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008).
Yan, N. & Chen, Z. J. J. Intrinsic antiviral immunity. Nat. Immunol. 13, 214–222 (2012).
MacMicking, J. D. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 12, 367–382 (2012).
Holm, C. K. et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat. Immunol. 13, 737–743 (2012).
Sadler, A. J. & Williams, B. R. G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568 (2008).
Hazzalin, C. A. & Mahadevan, L. C. MAPK-regulated transcription: A continuously variable gene switch? Nat. Rev. Mol. Cell Biol. 3, 30–40 (2002).
Dong, C., Davis, R. J. & Flavell, R. A. MAP kinases in the immune response. Annu. Rev. Immunol. 20, 55–72 (2002).
Wada, T. & Penninger, J. M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 23, 2838–2849 (2004).
Ludwig, S. Influenza viruses and MAP kinase cascades- Novel targets for an antiviral intervention? Signal Transduction 7, 81–88 (2007).
Mikkelsen, S. S. et al. RIG-I-mediated activation of p38 MAPK is essential for viral induction of interferon and activation of dendritic cells dependence on TRAF2 and TAK1. J. Biol. Chem. 284, 10774–10782 (2009).
Hui, K. P. Y. et al. Induction of proinflammatory cytokines in primary human macrophages by influenza A virus (H5N1) is selectively regulated by IFN regulatory factor 3 and p38 MAPK. J. Immunol. 182, 1088–1098 (2009).
Waetzig, V. et al. c-Jun N-terminal kinases (JNKs) mediate pro-inflammatory actions of microglia. Glia 50, 235–246 (2005).
Pleschka, S. et al. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3, 301–305 (2001).
Kujime, K., Hashimoto, S., Gin, Y., Shimizu, K. & Horie, T. p38 mitogen-activated protein kinase and c-Jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J. Immunol. 164, 3222–3228 (2000).
Ludwig, S. et al. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J. Biol. Chem. 276, 21990–21998(2001).
Randall, R. E. & Goodbourn, S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89, 1–47 (2008).
Levy, D. E. & Garcia-Sastre, A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12, 143–156 (2001).
Doyle, S. E. et al. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17, 251–263 (2002).
James, L. C., Keeble, A. H., Khan, Z., Rhodes, D. A. & Trowsdale, J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. 104, 6200–6205 (2007).
McEwan, W. A. et al. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat. Immunol. 14, 327–336 (2013).
Matsuzawa, T. et al. IFN-gamma Elicits Macrophage Autophagy via the p38 MAPK Signaling Pathway. J. Immunol. 189, 813–818 (2012).
Bolz, D. D., Tenor, J. L. & Aballay, A. A conserved PMK-1/p38 MAPK is required in caenorhabditis elegans tissue-specific immune response to yersinia pestis infection. J. Biol. Chem. 285, 10832–10840 (2010).
Kawli, T., He, F. L. & Tan, M. W. It takes nerves to fight infections: insights on neuro-immune interactions from C. elegans. Dis. Model Mech. 3, 721–731 (2010).
Takauji, R. et al. CpG-DNA-induced IFN-alpha production involves p38 MAPK-dependent STAT1 phosphorylation in human plasmacytoid dendritic cell precursors. J. Leukoc. Biol. 72, 1011–1019 (2002).
Tripathi, A. & Sodhi, A. Prolactin-induced production of cytokines in macrophages in vitro involves JAK/STAT and JNK/MAPK pathways. Int. Immunol. 20, 327–336 (2008).
Fejer, G. et al. Key role of splenic myeloid DCs in the IFN-alpha/beta response to adenoviruses in vivo. PLoS Pathog. 4, e1000208 (2008).
Tamrakar, A. K. et al. NOD2 activation induces muscle cell-autonomous innate immune responses and insulin resistance. Endocrinology 151, 5624–5637 (2010).
Grandvaux, N., tenOever, B. R., Servant, M. J. & Hiscott, J. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infec. Dis. 15, 259–267 (2002).
Costa-Pereira, A. P. et al. The antiviral response to gamma interferon. J. Virol. 76, 9060–9068 (2002).
Malcolm, K. C. & Worthen, G. S. Lipopolysaccharide stimulates p38-dependent induction of antiviral genes in neutrophils independently of paracrine factors. J. Biol. Chem. 278, 15693–15701 (2003).
Goh, K. C., Haque, S. J. & Williams, B. R. G. p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J. 18, 5601–5608 (1999).
Ramsauer, K. et al. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc. Natl. Acad. Sci. 99, 12859–12864 (2002).
Saccani, S., Pantano, S. & Natoli, G. p38-dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat. Immunol. 3, 69–75 (2002).
Humar, M. et al. The mitogen-activated protein kinase p38 regulates activator protein 1 by direct phosphorylation of c-Jun. Int. J. Biochem. Cell Biol. 39, 2278–2288 (2007).
Vignola, M. J., Kashatus, D. F., Taylor, G. A., Counter, C. M. & Valdivia, R. H. cPLA(2) regulates the expression of type I interferons and intracellular immunity to chlamydia trachomatis. J. Biol. Chem. 285, 21625–21635 (2010).
Gaur, P., Munjal, A. & Lal, S. K. Influenza virus and cell signaling pathways. Med. Sci. Monit. 17, 148–154 (2011).
Sithisarn, P., Michaelis, M., Schubert-Zsilavecz, M. & Cinatl, J. Differential antiviral and anti-inflammatory mechanisms of the flavonoids biochanin A and baicalein in H5N1 influenza A virus-infected cells. Antiviral Res. 97, 41–48 (2013).
Lee, S. M. Y. et al. Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 Influenza viruses in primary human macrophages. PloS One 4, e.0008072 (2009).
Hui, K. P. Y. et al. H5N1 influenza virus-induced mediators upregulate RIG-I in uninfected cells by paracrine effects contributing to amplified cytokine cascades. J. Infect. Dis. 204, 1866–1878 (2011).
Nikolova, M. & Asenov, A. Surface flavonoid aglycones in newly studied plant species. Nat. Prod. Res. 20, 103–106 (2006).
Havsteen, B. H. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 96, 67–202 (2002).
Albers, R. et al. Markers to measure immunomodulation in human nutrition intervention studies. Brit. J. Nutr. 94, 452–481 (2005).
Fulop, T. et al. Dysregulation of T-cell function in the elderly-scientific basis and clinical implications. Drugs Aging 22, 589–603 (2005).
Reed, L. I. & Muench, H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27, 493–497 (1938).
Acknowledgements
This study was supported by a grant from Tianjin Science and Technology commission (13ZCDSY03800). The authors thank Dr. George Fu Gao (Institute of Microbiology, Chinese Academy of Sciences) for generous supply of influenza A/WSN/33 viruses.
Author information
Authors and Affiliations
Contributions
All experiments were designed by W.D. and W.L. The experiments were carried out by W.D., X.L.W. and F.Y.Z. participated in animal experiment. W.D., X.L.W., J.F.H., H.F., C.L.Z. and W.L. analyzed and discussed data. W.D. and W.L. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
supplemental info
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Dong, W., Wei, X., Zhang, F. et al. A dual character of flavonoids in influenza A virus replication and spread through modulating cell-autonomous immunity by MAPK signaling pathways. Sci Rep 4, 7237 (2014). https://doi.org/10.1038/srep07237
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07237
This article is cited by
-
Specialized metabolites from plants as a source of new multi-target antiviral drugs: a systematic review
Phytochemistry Reviews (2023)
-
Network pharmacology-based predictions of active components and pharmacological mechanisms of Artemisia annua L. for the treatment of the novel Corona virus disease 2019 (COVID-19)
BMC Complementary Medicine and Therapies (2022)
-
Flavonoids: A complementary approach to conventional therapy of COVID-19?
Phytochemistry Reviews (2021)
-
Recognition of Natural Products as Potential Inhibitors of COVID-19 Main Protease (Mpro): In-Silico Evidences
Natural Products and Bioprospecting (2020)
-
The upshot of Polyphenolic compounds on immunity amid COVID-19 pandemic and other emerging communicable diseases: An appraisal
Natural Products and Bioprospecting (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.