Abstract
Recent studies have demonstrated that immune cells play an important role in the pathogenesis of many neurological conditions. Immune cells constantly survey the brain microvasculature for irregularities in levels of factors that signal homeostasis. Immune responses are initiated when necessary, resulting in mobilisation of the microglial cells resident in the central nervous system (CNS) and/or of infiltrating peripheral cells. However, little is known about the kinetics of immune cells in healthy and diseased CNS, because it is difficult to perform long-term visualisation of cell motility in live tissue with minimal invasion. Here, we describe highly sensitive in vivo MRI techniques for sequential monitoring of cell migration in the CNS at the single-cell level. We show that MRI combined with intravenous administration of super-paramagnetic particles of iron oxide (SPIO) can be used to monitor the transmigration of peripheral phagocytes into healthy or LPS-treated mouse brains. We also demonstrate dynamic cell migration in live animal brains with time-lapse MRI videos. Time-lapse MRI was used to visualise and track cells with low motility in a control mouse brain. High-sensitivity MRI cell tracking using SPIO offers new insights into immune cell kinetics in the brain and the mechanisms of CNS homeostasis.
Similar content being viewed by others
Introduction
For more than a century, scientists have believed that the blood-brain barrier (BBB) is a sacred, impermeable wall that maintains central nervous system (CNS) homeostasis and protects the brain from harmful substances1. Recent studies have demonstrated, however, that certain immune cells play an important role in multiple neuroinflammatory diseases, such as multiple sclerosis2,3, which suggests that these cells can breach the BBB. In addition to neurological diseases, there is now growing evidence that neural-immune crosstalk may even occur in non-disease conditions of the healthy brain4. Immune responses are initiated when necessary, resulting in mobilisation of the microglial cells5 resident in the CNS and/or of infiltrating peripheral cells6. The contributions of these cells to homeostasis in normal and in damaged brains are poorly understood.
The development of non-invasive monitoring such as magnetic resonance imaging (MRI) holds promise for the study of the immune system in vivo3,7. MRI in particular offers a significant advantage in imaging deep sections with good spatial resolution, which overcomes a limitation of fluorescence imaging. To overcome inherent limitations in the signal to noise ratio (SNR) of conventional in vivo MRI, a stronger magnetic field can be applied. In addition, super-paramagnetic nanoparticles of iron oxide (SPIO), a contrast agent for MRI, improves MRI contrast to noise ratio (CNR) and detectability in the stronger magnetic fields by shortening the T2/T2* relaxation time, further advancing MRI application in cell imaging and tracking7,8,9. Combining high-magnetic-field strength with high-sensitivity radio-frequency (RF) coils and optimal contrast agents enables the visualisation of cell populations and molecular events in vivo in both animals and humans9,10. In the current study, we used an 11.7 T high-field MRI scanner in combination with a high sensitivity coil and SPIO to detect and monitor peripheral immune cell migration into healthy and lipopolysaccharide (LPS)-treated mouse brains at the single-cell level without surgical invasion.
We focused on the labelling and tracking of endogenous and highly phagocytic monocytes and macrophages. Monocytes and macrophages are important cells of the innate immune system and regulate tissue homeostasis, infection, inflammation and repair11,12. They are also highly motile and migrate to regions of inflammation within 12–48 h13. Some reports have shown that circulating monocytes and macrophages have a natural ability to traverse the intact or compromised BBB14.
To the best of our knowledge, this is the first study to demonstrate that high-magnetic-field MRI along with SPIO administration can be applied in non-invasive monitoring of single immune cell distribution and migration with time-lapse videos.
Results
To evaluate the potential of cell tracking by high-field MRI in the living mouse brain, we intravenously administered SPIO to normal control and LPS-treated mice. Circulating blood monocytes and tissue macrophages absorb intravenously administered SPIO, which allows them to be labelled in vivo15 (in vivo labelling). Blood vessels in the brain appeared as strongly hypointense regions immediately after SPIO injection (see Supplementary Fig. S1 online), which then gradually faded out over a period of 6–8 h after SPIO administration and had completely disappeared by 1 day after administration, due to the short half-life of SPIO in blood16. In control mice, several T2* hypointense spots appeared in the brain at 1 day after administration (Fig. 1b, 656.2 ± 544.9). These spots were detected repeatedly and were found to be significantly increased at 2 days after SPIO administration (Fig. 1c, 864.0 ± 484.3). The spots showed a non-specific distribution throughout the brain. Furthermore, most of them had disappeared by 7 days after SPIO administration (Fig. 1d, 380.8 ± 120.7). To confirm disruption of the BBB, we carried out gadolinium (Gd)-enhancement MRI at each time point. Gd enhancement was not observed in the control brain (data not shown).
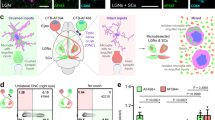
Cellular MRI of the brain with intravenous administration of super-paramagnetic particles (SPIO).
(a–h), Representative 300-μm single slices of mouse brain are shown at each time point: before (a, e), 1 day post (dp), (b, f), 2 dp (c, g) and 7 dp (d, h) SPIO administration. The upper row (a–d) shows brain MRIs of one control mouse; the lower row (e–h) shows brain MRIs of one lipopolysaccharide (LPS)-treated mouse. Scale bar, 1 mm. After administration of SPIO, T2*-weighted MRI reveals tiny, non-specific hypointense spots (b, c, f, g). Almost all spots disappear by 7 dp (d, h). (i), Quantification of spots (mean ± standard deviation [s.d.]) in the whole brain at each time point. Significantly greater spot counts are found in LPS-treated brains at 1 dp and 2 dp (n = 6) than in controls. Delayed significant increases are even found in normal control brains (n = 6). Data were analysed with a one-way ANOVA for significant inter-group differences and Bonferroni-corrected post-tests. Significant changes relative to baseline (0 dp) are indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.005. (j), (k), Ex vivo MRI scan after perfusion fixation shows hypointense spots post mortem in brains of the normal (j) and LPS-treated groups (k). (l), Histological sections show co-localisation of fluorescent dye-cross-linked SPIO and F4/80+ cells in the brain of an LPS-treated mouse. F4/80 is a macrophage marker. SPIO itself and SPIO-labelled cells are rarely found in control normal brains ((m), life sized). (n), SPIO-labelled cells are separate from CD31+-labelled blood vessels.
We then conducted the same examination in mouse brains with systemic inflammation induced by intraperitoneal LPS treatment. We observed extensive T2* hypointense spots throughout the brain at 1 and 2 days after SPIO administration (Figs. 1f and 1g, 3,591.6 ± 1,203.3 and 3,042.9 ± 1,937.6, respectively). BBB leakage was not detected with Gd enhancement (data not shown). In addition, most of the spots had disappeared by 7 days, as in the control (Fig. 1h, 736.2 ± 542.3).
To confirm the incorporation of labelled cells into the brain parenchyma, we carried out ex vivo MRI scans after perfusion fixation, which was intended to remove most circulating contrast agents and circulating labelled cells from the blood vessels17. We observed T2* hypointense spots in ex vivo MRI both in control (Fig. 1j) and LPS-treated mice (Fig. 1k). The number of hypointense spots in ex vivo MRI was much less than that in in vivo MRI (Supplementary Fig. S2 online). We speculate that this divergence was due to the washout mechanism having removed SPIO and SPIO-labelled cells remaining in the blood vessels during the perfusion fixation process used to make the ex vivo brain samples. In addition, the histology of the fixed brains showed co-localisation of SPIO and cells positive for F4/80, a murine macrophage marker (Fig. 1l,m and Supplementary Fig. S3 online). Most of the SPIO-labelled cells were separated from regions positive for CD31 (a micro vessel endothelial cell marker), interpreted as blood vessels (Fig. 1n).
Although some small molecules can pass through the BBB, the barrier limits the transport of unwanted molecules14. SPIO has a molecular weight of >20 kDa, which means that by itself it cannot cross the BBB into the brain parenchyma. However, the barrier could be circumvented if SPIO were absorbed by phagocytes, which can pass the BBB. Accordingly, we asked whether high-field MRI with in vivo SPIO labelling could detect the recruitment of peripheral cells into the brain parenchyma.
We used two methods to confirm that peripheral macrophages and monocytes are the main transporters of SPIO into the brain: intraperitoneal administration of clodronate liposomes, which deplete peripheral macrophages and monocytes18; and systemic administration of stealth magnetic particles, which are hardly absorbed by phagocytes due to the suppression of non-specific protein binding caused by their “brush-afforded” structure19. Administration of SPIO to LPS-treated mice caused a marked increase in T2* hypointense spots throughout the brain as described above (Fig. 2a). Meanwhile, clodronate intervention markedly reduced spots following SPIO administration (Fig. 2b). It has been shown that clodronate liposomes are efficiently taken up by circulating phagocytes and clodronate sodium is released during enzymatic breakdown of the liposomes within the cells, inducing apoptosis and selective depletion of phagocytes18. In addition, clodronate liposomes themselves do not cross the BBB; thus, they cannot deplete brain-resident phagocytic microglia when delivered systemically20. Therefore, the T2* hypointense spots seen in the brain following SPIO administration are likely peripheral phagocytes that transmigrate into the brain. Next, we substituted stealth magnetic particles for SPIO19. Though the phagocytes still existed in the body, we found the T2* hypointense spots in the brain to be highly restricted when we administered stealth magnetic particles in place of SPIO (Fig. 2c). This result indicates that the spots appear in the brain only because of phagocytosis of SPIO by macrophages and monocytes in the periphery, but not in the brain. In addition, ex vivo MRI showed that the T2* hypointense spots remained after washing of the vessel lumens. These results raised the possibility that SPIO-labelled peripheral phagocytes could transmigrate into normal and LPS-treated brains by passing through the BBB.
Confirmation studies: MRI after macrophage depletion and stealth magnetic particle administration.
(a), the brain of an LPS-treated mouse shows numerous T2* hypointense spots 2 days post (dp) SPIO administration. (b), In spite of SPIO administration to the LPS-treated mouse, markedly fewer hypointense spots are seen in the brain following intravenous administration of clodronate liposomes, a result of depletion of circulating macrophages and monocytes. (c), the number of hypointense spots is strongly reduced when using stealth magnetic particles (P-SPIO). (d), Quantification of spots (mean ± s.d.) in the whole brain of LPS − treated (n = 6), LPS + clodronate-treated (n = 4) and LPS + P-SPIO-treated groups (n = 3) at each time point. Data were analysed with a one-way ANOVA for significant inter-group differences and Bonferroni-corrected post-tests. Significant changes from baseline (0 dp) are indicated by *, P < 0.05; **, P < 0.01; and ***, P < 0.005.
To investigate whether the labelled cells retained SPIO after transmigration into the brain, peritoneal macrophages were harvested from green fluorescent protein (GFP)-expressing transgenic mice. After co-incubating GFP-positive macrophages with magnetic dextran nanoparticles cross-linked to a fluorescent dye (in vitro labelling), labelled cells were injected intravenously and tracked by both in vivo MRI and by post-mortem light microscopy. One and 2 days after transplantation of labelled cells into wild-type recipients, several T2* hypointense spots were found in the brain in a pattern similar to that observed with the in vivo labelling method described above (Fig. 3a,b). Following MRI scans, the brain was removed from the skull after perfusion fixation and the co-localisation of SPIO and GFP-positive cells was examined in histological sections (Fig. 3c). As expected, both in vitro and in vivo labelling approaches indicated cell transmigration into the brain through an intact BBB.
In vitro labelled cell transplantation.
(a),(b), the localization of injected SPIO-labelled peritoneal macrophages from EGFP-C57BL/6 mice is shown in in vivo MR images at the time points indicated: 1 day post (dp) (a) and 2 dp cell administration (b). (c), A histological section showing that GFP+ cells that infiltrate the brain contain SPIO particles. All images were obtained from the same mouse.
To observe the dynamic movement of labelled cells in the brain, the brain of a live mouse was scanned continuously for 24 h for time-lapse acquisition (Fig. 4 and Supplementary Video 1 online). We injected SPIO into the tail vein and recorded multi-slice images from 16 to 40 h later. A representative 300-μm-thick single slice of the brain at different time points is shown in Figure 4. The blood half-life of SPIO is less than 0.5 h in rodents, due to rapid uptake by the reticuloendothelial system16. Therefore, the many migrating single spots found in time-lapse MRI recorded after 16 h post SPIO administration must have been motile immune cells that had absorbed SPIO. In addition, some SPIO-induced T2* hypointense spots, indicative of macrophage infiltration, remained stationary for several hours, then vanished from the images (e.g., Fig. 4 and Supplementary Video 1 online; blue circle). The frame interval of this time-lapse MRI was relatively long (20 min/frame), so that fast-moving cells flowing through the bloodstream could not be detected. Meanwhile, the hypointense spots in the brain on MRI that moved along the blood vessels were probably slower motile cells crawling on the endothelium (e.g., Fig. 4 and Supplementary Video 1 online; green arrowhead). The migration velocity of motile SPIO-labelled cells in the plane of the slice was estimated at less than 5 μm/min (e.g., Fig. 4 and Supplementary Video 1 online; yellow arrowhead). This result is in good agreement with a previous study that examined monocytes in the vascular lumen of normal mice and found the median velocity during cell crawling on endothelial cells to be 4 μm/min21.
Representative 300-μm-thick single MRI slice of the same mouse brain at different time points.
Time-lapse MR images of a single representative slice from 30 to 36 h post SPIO administration shows SPIO-labelled cells as T2* hypointense spots. Although many spots remain stationary (blue circle), a few motile cells migrate along the visible sites of blood vessels (green arrowhead). Another cell appears and migrates to another location (yellow arrowhead).
Although vascular endothelial cells bind to each other closely using numerous tight junctions, which limit the transport of materials between the blood and the brain parenchyma, some kinds of leukocytes can cross this barrier by diapedesis under physiological conditions14. It has been shown that motile SPIO-containing cells interact with endothelial cells before diapedesis, better known as tethering, rolling and stopping on the endothelial cells22. On the other hand, a previous report has demonstrated that phagocytosing microglia remain stationary or have low motility after nerve infiltration23,24. Our time-lapse MRI detected a similar phenomenon, as some spots remained stationary for several hours (Fig. 4, blue circle). These spots likely represented cells after infiltration into the brain and were the counterparts of those observed in in vivo and ex vivo experiments (Fig. 1). The observation of slowly motile cells might indicate interactions with vessel walls (Fig. 4, yellow and green arrowheads).
Discussion
Here, we demonstrate non-invasive detection of immune cell distribution with high-magnetic-field MRI using in vivo SPIO administration. In addition, time-lapse MRI videos allowed us to perform dynamic single-cell tracking in a live animal brain for hours and days. To our knowledge, this is the first report of single-cell monitoring by MRI of peripheral immune cells in healthy and LPS-treated mouse brains of live animals. The existence of the BBB is in part responsible for the early concept that the brain is an immune-privileged site, with restricted passage of immune cells into the brain for immune surveillance of the CNS25. However, the incidence of cells transmigrating into the brain is merely very low and is estimated to be 100-fold lower than that observed for the spleen or lung14,26. This sparseness makes it difficult to detect such cells in brains using histological sections. For example, following intravascular injection of 5 × 107 labelled, antigen-activated T cells into mice, an average of only 14 extravasated cells were found per 100 μm in coronal brain sections25. However, MRI can detect these cells much more easily due to the greater slice thickness (300 μm) and multi-slice scanning allows us to survey the whole brain. Our results using cellular MRI confirm the conclusions of the authors of the microscopic study25 and extend their findings to in vivo whole-brain tracking of monocytes and macrophages. We found that some hypointense spots temporarily seen in the brain slices vanished from the brain at 7 days after SPIO administration both in normal and in LPS-treated groups. We postulate that this phenomenon reflects migration from their previous position to other slices or tissues. We believe that this phenomenon does not reflect cell lysis. If this vanishing were due to cell lysis, hypointense spots should still exist throughout the brain because of uptake of the SPIO by other phagocytic cells. However, the spots disappeared from the slice of interest and very seldom reappeared in adjacent slices. Unfortunately, the slice thickness is relatively great in comparison with in-plane resolution at this moment, so it is difficult to certify a spot as being the same spot that migrated from another slice. The vanishing process remains an unresolved issue. In addition, we found that in stroke-model mice (Supplementary Fig. S4 online), spots remained in and around damaged brain regions for a relatively longer period. This means that SPIO-labelled cells remain for a relatively long time at one location when the brain has some abnormalities; we did not observe such persistence in the brains of both normal and LPS-treated mice. We therefore speculate that the LPS stimulation method we used did not induce abnormalities in the brain. If LPS stimulates the leukocytes, including macrophages and neutrophils and if those cells effectively take up SPIO and patrol the brain similarly to normal conditions, then we should find the spots easily in the brains of the LPS group. For the present, we can conclude that our in vivo phagocyte labelling technique has proven useful. However, a more comprehensive understanding of it is required.
MRI with SPIO for cell tracking has several advantages compared with other imaging modalities such as PET, CT and conventional MRI: the combination of non-invasiveness and a durable contrast agent, which allows long-term data acquisition; non-use of radioactive isotopes, permitting long-term tracking; an ability to track cells in deep tissue; and an ability to quantify cell numbers. At the same time, however, because of the inherent limitations in the signal to noise ratio (SNR) of MRI, sensitivity is a key concern for single-cell detection in in vivo experiments. We successfully detected single cells in live animal brains by achieving images of high spatio-temporal resolution because of a high SNR. In addition to the high- magnetic-field, improvements in MR hardware, such as the custom-made RF coil used here, were a significant factor in this achievement.
Many questions about in vivo labelling and its application to biology remain unanswered. For example, the optimal choice of the particle and loading pathway is not established. We have focused on phagocytic cells such as macrophages, as they can easily load detectable amounts of particles. The hypointensities in our MR images would be affected not only by the number of labelled cells but also by the iron content of the cells. We conjecture that some cells were not detected due to insufficient SPIO uptake. The number of hypointense spots in our MR images does not always indicate the number of cells. On the other hand, the iron contents were affected by antigenic stimulation of the cells. The M1-M2 paradigm is commonly associated with properties of mature macrophages and classically macrophages (M1) are activated through LPS or IFN-γ stimulation27. Our result that the number of hypointense spots was increased in the LPS-challenge group raises the possibility that not only an increase in number of infiltrating cells but also an increase in macrophage phagocytic activity contributes to the detectability of hypointense spots, the latter factor causing an increase in the iron content of each cell.
Observing the movements of non-phagocytic cells remains a greater challenge. Recently, micrometre-sized particles of iron oxide (MPIO) have been used to label and observe non-phagocytic cells28. Further improvements in nanoparticle labelling or in the use of MPIO should be translatable into observations of other cell types using MRI. Even so, at high magnetic fields, a single-time-point MR image does not have the ability to discriminate between labelled cells inside and outside the BBB. Our time-lapse MRI movie could discriminate both several spots that remained stationary for several hours and spots with low motility. The detailed velocity and acceleration analysis of cell migration may help to discriminate between cells inside and outside the BBB. Furthermore, combining nanoparticles and high-field MRI permits the specific visualisation of immune cell migration in the brain, whereas other MR images such as T2-weighted (T2W) and diffusion weighted (DW) images show temporal and spatial changes in tissue contrast in neuroinflammatory and neurodegenerative diseases29,30. A single instrument can therefore potentially show both the damage due to the disease and the response of the macrophages to it. The T2W or DW techniques give us the functional information such as the level of cerebral damage30. After using DW image (DWI) to classify regions as curable or not, SPIO administration has the potential to detect differences in cell behaviour between curable areas and irreversibly damaged ones. Thus, future studies employing this non-invasive single-cell monitoring method may contribute to resolve how immune cells act in neuroinflammatory and neurodegenerative diseases.
Methods
Contrast media for MRI
Three kinds of iron oxide particles were used as contrast media: a commercially available superparamagnetic iron oxide particle (SPIO; Resovist®, I'rom Pharmaceutical Co, Ltd, Shinagawa-ku, Tokyo, Japan); red fluorescent-cross-linked SPIO (nano-screen MAG/R, Chemicell GmbH, Berlin, Germany); and stealth magnetic particles originally synthesised and grafted with a hydrophobic polymer brush of high grafting density, as previously described19. The mean particle diameters of the iron oxide particles were 50 nm, 100 nm and 100 nm, respectively and iron concentrations were 28 mg/mL, 25 mg/mL and 4.5 mg/mL, respectively. For gadolinium-enhanced magnetic resonance imaging (MRI), a stock solution was prepared by diluting commercially available gadoteridol (ProHance®, 279.3 mg/mL, Bracco Eisai Co, Ltd, Tokyo, Japan) 10-fold with saline.
Animal experiments
All experimental protocols were approved by the Research Ethics Committee of Osaka University. All experimental procedures involving animals and their care were carried out in accordance with the Osaka University Guidelines for Animal Experimentation and the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All animal experiments were performed in 8- to 12-week-old (22.8 ± 2.6 g) male mice. Thirty-four wild-type C57BL/6 mice and four EGFP-C57BL/6 mice were obtained from Japan SLC (Hamamatsu, Japan). All mice were housed in a controlled vivarium environment (24°C; 12:12 h light-dark cycle) and fed a standard pellet diet and water adlibitum. Mice with abnormal brain morphology such as ventricular enlargement were excluded at the first MRI scanning (n = 2). The LPS-treated group comprised eighteen mice intraperitoneally (i.p.) administered lipopolysaccharide from Escherichiacoli O26 (LPS; 0.5 μg per gram of body weight; dissolved in saline with 10% ethyl alcohol, Wako Pure Chemical Industry, Tokyo, Japan), three days before SPIO administration. The control group comprised 14 mice administered saline (10 mL per kg of body weight). In addition, the mice were randomly assigned as follows:
-
1
In vivo labelling study (n = 8 from each group)
-
2
Gd-enhancement study (n = 3 from each group)
-
3
Macrophage depletion study (n = 4 from LPS group)
-
4
Stealth particle study (n = 3 from LPS group)
-
5
Time-lapse study (n = 3 from control group)
In vivo labelling study
Phagocytes were labelled in situ by direct intravenous injection to generate SPIO-labelled cells. Resovist (5 mL per kg of body weight) was administered through the tail vein to both LPS-treated and control groups (n = 6 in each group). To confirm the existence of SPIO phagocytosis by fluorescence microscopy, we added two subjects to each group (resulting in n = 8) and the same procedure was done with fluorescent dye-cross-linked SPIO instead of Resovist. The brains of live mice were scanned sequentially at 1, 2 and 7 days post SPIO injection to observe SPIO-labelled cells in the brain.
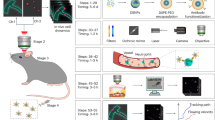
In vivo MRI protocols
All mice for the in vivo labelling study were anaesthetised with a mixture of air and 2.8% isoflurane (Abbott Laboratories, Abbott Park, IL, USA) and then placed in an MRI-compatible animal cradle. The isoflurane concentration was maintained at 1.5 ± 0.4%, adjusted to maintain the animal at 60 ± 10 breaths/min throughout the MRI sessions. Respiratory signals and body temperature were monitored using a physiological monitoring system (SA Instruments, Inc., Stony Brook, NY, USA). Body temperature was maintained by circulating water through warming pads. MRI was conducted using an 11.7 T vertical-bore Bruker Avance II imaging system (Bruker Biospin, Ettlingen, Germany) and a custom-made 15-mm diameter transmit/receive volume radio frequency (RF) coil (m2m imaging, Cleveland, OH, USA). MRI was performed 1, 2 and 7 days after SPIO administration. An axial T2* weighted fast, low-angle shot (FLASH) image (T2*WI; repetition time [TR] = 500 ms, echo time [TE] = 4 ms, flip angle [FA] = 30°, number of averages [NA] = 8, field of view [FOV] = 15 mm × 15 mm, matrix size = 256 × 256, slice thickness = 300 μm and acquisition time = 18 min) was acquired around the brain.
Gadolinium (Gd)-enhanced MRI protocols
To check for leakage of the blood-brain barrier (BBB), Gd-enhanced MRI was performed (n = 3 from each group) as previously described31,32. A lateral tail vein of the mouse was used as the route for continuous infusion of Gd. After filling a 150-cm polyethylene tube (PE10, 0.61 mm OD; Becton Dickinson, NJ, USA) with gadoteridol solution, a specially designed non-magnetic copper needle (0.36 mm in outer diameter) was placed into the tube end and a Gd-filled syringe was inserted into the another end. The needle was cannulated into a lateral tail vein of the anaesthetised mouse. After cannulation, the mouse was placed in an MRI-compatible animal cradle and an axial T1-weighted FLASH sequence (T1WI; TR/TE = 90/1.5 ms, FA = 80°, NA = 8, FOV = 15 mm × 15 mm, matrix size = 256 × 256, slice thickness = 300 μm) was performed immediately after a single intravenous injection of 0.25 mmol/kg of gadoteridol. The cannulation and MRI were performed at the same time points as in the in vivo MRI protocol described above.
Macrophage depletion study
Clodronate liposome suspensions were purchased from Katayama Chemical, Osaka, Japan. Peripheral macrophages and monocytes were depleted by treating mice with a single dose of 200 μL intraperitoneal clodronate liposomes 24 h after LPS treatment (n = 4). Depletion was confirmed by flow cytometric analysis of monocytes and macrophages in the blood and spleen. Resovist (5 mL per kg of body weight) was administered via the tail vein 48 h after liposome administration and in-vivo MRI scans were performed as described above.
Stealth magnetic particle study
A solution of stealth magnetic particles (10 mL per kg of body weight) was administered via the tail vein in place of Resovist, 3 days after LPS treatment (n = 3) and in-vivo MRI scans were performed as described above.
Spot quantitative and statistical analysis
We counted the number of T2* hypointense spots using an automatic analysis software program for spot counting that employed standard image processing techniques. The program was developed with Microsoft © Visual C++ 2008 (VC08E) and OpenCV on a Windows 7 SP1 (32 bit) machine. First, we applied a standard watershed algorithm to greyscale images to segment the brain into regions. Second, we applied a typical four-neighbour connected-component labelling algorithm to the brain regions after creating binary images by image thresholding for segmentation of the spots from the T2*W images. All T2* hypointense spots in these brains were successfully labelled, because their intensities were extremely low. Finally, we counted the number of the spots from the labels. Operators who subsequently handled the data were blinded to the group assignment. To determine differences in the number of T2* hypointense spots, analysis of variance (ANOVA) on ranks and a Bonferroni's all-pairwise multiple comparison test (KaleidaGraph 4.1, Hulinks, Japan) were used. In all comparisons, P < 0.05 was considered statistically significant.
Ex vivo MRI protocols
After performing in vivo imaging at 2 days post SPIO administration, blood was washed out by intracardiac perfusion with heparinised phosphate-buffered saline (PBS) after anaesthesia and then fixation with 2% paraformaldehyde (PFA) in PBS was performed (n = 2 for both control and LPS-treated groups). The brain was harvested and fixed overnight in 2% PFA. After fixation, tissues were impregnated in distilled water overnight and then suspended in a fluorine-based solvent (Fluorinert FC-43, Sumitomo 3M Ltd., Tokyo, Japan), a per fluorinated fluid, which provides a homogenous medium while yielding no proton signal. For ex vivo study, FLASH images were acquired using the 11.7 T AVANCE II system. A 10-mm diameter volume coil was used for radio frequency transmission and reception. The MRI experimental parameters were TR/TE = 600/4 ms, FA = 30°, NA = 16, FOV = 9.6 mm × 9.6 mm and matrix size = 256 × 256.
Immunohistological study
After performing ex vivo MRI, the brain was embedded in paraffin wax (n = 2 for both control and LPS-treated groups). Tissue sections were cut with a microtome at a thickness of 5 μm. Sections were dewaxed in xylene and rehydrated by a series of ethanol-water washes. Antigen retrieval was performed by incubating the sections in 10 mM citrate retrieval buffer solution at 120°C for 10 min. To block endogenous peroxidase activity, sections were incubated in peroxidase blocking solution (3% H2O2 in methanol) for 20 min and rinsed with distilled water, followed by a 5-min rinse in PBS. Sections were blocked in a solution of 10% foetal bovine serum (FBS) with 1% bovine serum albumin (BSA; Sigma Chemical, Saint Louis, MO, USA) for 1 h at room temperature. The sections were then incubated with primary antibodies to F4/80 (1:500, Alexa Fluor 488-conjugated, #123119, Biolegend, San Diego, CA, USA), a murine macrophage marker and CD31 (1:500, Alexa Fluor 647-conjugated, #102415, Biolegend, San Diego, CA, USA) diluted in 1% BSA-TBST (0.1% Tween 20 in Tris-buffered saline, pH 7.4) for 1 h at room temperature and then overnight in the dark at 4°C. The slides were then washed three times in TBST for 5 min each and mounted with ProLong® Gold Antifade Reagent with DAPI (Invitrogen). Images were captured from stained 5-μm paraffin-embedded sections using a Keyence BZ-8100 all-in-one fluorescence microscope.
In vitro labelling study
Green fluorescent protein (GFP)-positive peritoneal macrophages elicited by intraperitoneal injection of 4% thioglycollate were isolated from EGFP-C57BL/6 mice. GFP+ macrophages were cultured as a monolayer in 10-cm RepCell culture plates (CellSeed Inc. Tokyo, Japan) in complete RPMI 1640 medium containing 10% FBS, 100 mg/mL streptomycin, 100 U/mL penicillin and 2 mM glutamine at 37°C, as described elsewhere33. At 90% confluence, macrophages were incubated with fluorescent dye-cross-linked SPIO particles (10 μg Fe/mL) in culture medium for 3 h. After incubation, macrophages were washed three times with PBS to remove excess labelling agent. Macrophages were then incubated with 5 mL of trypsin-EDTA at room temperature for 5 min and centrifuged (1,000 rpm for 10 min). The cells were then washed twice with PBS. The cells were counted and the number of dead cells was confirmed to be less than 10% using the trypan blue exclusion assay. A total of 1 × 106 SPIO-labelled macrophages were prepared in a 1.0-mL micro centrifuge tube and then a 0.1-ml suspension of SPIO-labelled cells was injected into the tail vein of recipient wild-type C57BL/6 mice for 10 s (n = 4). Invivo MRI scans at 1 day and 2 days post-cell-transfer were performed as described above. After in vivo MRI, histological sections were prepared as described above and the distribution of GFP+ cells and SPIO was observed with an Olympus FSX100 all-in-one fluorescence microscope.
Time-lapse MRI protocols
MRI was conducted using an 11.7 T vertical-bore Bruker Avance II imaging system and a custom-made 15-mm diameter transmit/receive volume RF coil. Three mice from the control group were subcutaneously injected with 5 mL of saline to avoid dehydration and atropine sulphate (0.05 mg per kg via subcutaneous injection) was administrated before anaesthesia. Mice were anaesthetised with a mixture of air and 2.8% isoflurane and then placed in an MRI-compatible animal cradle. The isoflurane concentration was maintained at 1.5 ± 0.4% to produce a breathing rate of 60 ± 10 breaths/min throughout the MRI sessions. Respiratory signals and body temperature were monitored with a physiological monitoring system. Body temperature was maintained by circulating water through warming pads. Sequential MRI was started 16 h after SPIO administration. Axial T2*-weighted FLASH images (T2*WI; TR/TE = 500/4 ms, FA = 30°, NA = 8, FOV = 15 mm × 15 mm, matrix size = 256 × 256, slice thickness = 300 μm and acquisition time = 18 min) were continuously acquired for 24 h at 2-min intervals (3 frames/h).
Imaging processing for time-lapse MRI
The post-processing method realigned the acquired time-series MR volumes and adjusted the intensity to make it homogeneous across volumes. The image alignment method used is based on a rigid image registration technique. The base volume is the geometric centre of the time-series data and the other volume data, called floating volumes, are aligned to the base volume. Using the steepest descent algorithm, the method finds the optimum 3D translation vector, defined as producing the maximum mutual information between the base volume and the translated floating volume.
References
Obermeier, B., Daneman, R. & Ransohoff, R. M. Development, maintenance and disruption of the blood-brain barrier. Nat Med 19, 1584–1596 (2013).
Arima, Y. et al. Regional neural activation defines a gateway for autoreactive T cells to cross the blood-brain barrier. Cell 148, 447–457 (2012).
Mori, Y. et al. Early pathological alterations of lower lumbar cords detected by ultrahigh-field MRI in a mouse multiple sclerosis model. Int Immunol 26, 93–101 (2014).
McAllister, A. K. & van de Water, J. Breaking boundaries in neural-immune interactions. Neuron 64, 9–12 (2009).
Sagar, D. et al. Dendritic cell CNS recruitment correlates with disease severity in EAE via CCL2 chemotaxis at the blood–brain barrier through paracellular transmigration and ERK activation. J Neuroinflammation 9, 245 (2012).
Kawakami, N. & Flügel, A. Knocking at the brain's door: intravital two-photon imaging of autoreactive T cell interactions with CNS structures. Semin Immunopathol 32, 275–287 (2010).
Ahrens, E. T. & Bulte, J. W. M. Tracking immune cells in vivo using magnetic resonance imaging. Nat Rev Immunol 13, 755–763 (2013).
Wu, Y. L. et al. In situ labeling of immune cells with iron oxide particles: an approach to detect organ rejection by cellular MRI. Proc Natl Acad Sci USA 103, 1852–1857 (2006).
Mori, Y., Umeda, M., Fukunaga, M., Ogasawara, K. & Yoshioka, Y. MR contrast in mouse lymph nodes with subcutaneous administration of iron oxide particles: size dependency. Magn Reson Med Sci 10, 219–227 (2011).
Richards, J. M. J. et al. In vivo mononuclear cell tracking using superparamagnetic particles of iron oxide: feasibility and safety in humans. Circ Cardiovasc Imaging 5, 509–517 (2012).
Murray, P. J. & Wynn, T. A. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 11, 723–737 (2011).
Shi, C. & Pamer, E. G. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 11, 762–774 (2011).
Laskin, D. L., Weinberger, B. & Laskin, J. D. Functional heterogeneity in liver and lung macrophages. J Leukoc Biol 70, 163–170 (2001).
Hickey, W. F. Leukocyte traffic in the central nervous system: the participants and their roles. Semin Immunol 11, 125–137 (1999).
Luciani, A. et al. Adipose tissue macrophages: MR tracking to monitor obesity-associated inflammation. Radiology 263, 786–793 (2012).
Lawaczeck, R. et al. Magnetic iron oxide particles coated with carboxydextran for parenteral administration and liver contrasting. Pre-clinical profile of SH U555A. Acta Radiol 38, 584–597 (1997).
Kato, Y. & Pathak, A. P. Combined contrast and therapeutic nanocarriers for oncologic MRI. Nanoimaging [Goins, B. A. & Phillips, W. T. (ed)], 1–17 (Pan Stanford Publishing, America, 2011).
Rooijen, N. V. & Sanders, A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J Immunol Methods 174, 83–93 (1994).
Ohno, K., Akashi, T., Tsujii, Y., Yamamoto, M. & Tabata, Y. Blood clearance and biodistribution of polymer brush-afforded silica particles prepared by surface-initiated living radical polymerization. Biomacromolecules 13, 927–936 (2012).
Fulci, G. et al. Depletion of Peripheral Macrophages and Brain Microglia Increases Brain Tumor Titers of Oncolytic Viruses. Cancer Res 67, 9398–9406 (2007).
Jacquelin, S. et al. CX3CR1 reduces Ly6Chigh-monocyte motility within and release from the bone marrow after chemotherapy in mice. Blood 122, 674–683 (2013).
Wagner, D. D. & Frenette, P. S. The vessel wall and its interactions. Blood 111, 5271–5281 (2008).
Bendszus, M. & Stoll, G. Caught in the act: in vivo mapping of macrophage infiltration in nerve injury by magnetic resonance imaging. J Neurosci 23, 10892–10896 (2003).
Roth, T. L. et al. Transcranial amelioration of inflammation and cell death after brain injury. Nature 505, 223–228 (2014).
Greenwood, J. et al. Review: leucocyte-endothelial cell crosstalk at the blood-brain barrier: a prerequisite for successful immune cell entry to the brain. Neuropathol Appl Neurobiol 37, 24–39 (2011).
Hickey, W. F. Migration of hematogenous cells through the blood-brain barrier and the initiation of CNS inflammation. Brain Pathol 1, 97–105 (1991).
Martinez, F. O. & Gordon, S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000 Prime Rep 6 (2014).
Shapiro, E. M. et al. MRI detection of single particles for cellular imaging. Proc Natl Acad Sci USA 101, 10901–10906 (2004).
Wiart, M. et al. MRI monitoring of neuroinflammation in mouse focal ischemia. Stroke 38, 131–137 (2007).
Ebisu, T. et al. Neuroprotective effects of an immunosuppressant agent on diffusion/perfusion mismatch in transient focal ischemia. Magn Reson Med 51, 1173–1180 (2004).
Tofts, P. S. & Kermode, A. G. Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magn Reson Med 17, 357–367 (1991).
Taheri, S., Candelario-Jalil, E., Estrada, E. Y. & Rosenberg, G. A. Spatiotemporal correlations between blood-brain barrier permeability and apparent diffusion coefficient in a rat model of ischemic stroke. PLoS ONE 4, e6597 (2008).
Ahrens, E. T., Feili-Hariri, M., Xu, H., Genove, G. & Morel, P. A. Receptor-mediated endocytosis of iron-oxide particles provides efficient labeling of dendritic cells for in vivo MR imaging. Magn Reson Med 49, 1006–1013 (2003).
Acknowledgements
We appreciate the contributions of Prof. N. Takemura and Prof. S. Uematsu (Institute of Medical Science, The University of Tokyo, Tokyo, Japan) who provided technical assistance and discussion and of Ms. K. Mori who assisted in data processing. We also thank Dr. P. Karagiannis (RIKEN QBiC, Osaka, Japan) for carefully reading this manuscript and members of the Y.Y. laboratory for their suggestions. This study was supported by Grant-in-aid for scientific research 24791302 (to Y.M.), Grant-in-aid for scientific research 23228001 (to Y.Y.) and funding for World-Leading Innovative R&D in Science and Technology (Akira FIRST Program) from the Japan Society for the Promotion of Science (to Y.M. and Y.Y.).
Author information
Authors and Affiliations
Contributions
Y.M. and Y.Y. designed all experiments and interpreted the data. Y.M. and T.C. performed all MRI experiments and Y.M., T.C. and Y.K. performed the cell culture and microscopic experiments. T.F., S.K. and Y.H. designed the automatic post-processing program and provided the image registrations. K.O., S.Y. and Y.T. synthesized and provided the SPIO particles. Y.M. and Y.Y. wrote the manuscript with input from the co-authors.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Supplementary Information
Supplementary Video 1
Supplementary Information
Supplementary Video 2
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Mori, Y., Chen, T., Fujisawa, T. et al. From cartoon to real time MRI: in vivo monitoring of phagocyte migration in mouse brain. Sci Rep 4, 6997 (2014). https://doi.org/10.1038/srep06997
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06997
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.