Abstract
The evolution of peptidergic signaling has been accompanied by a significant degree of ligand-receptor coevolution. Closely related clusters of peptide signaling molecules are observed to activate related groups of receptors, implying that genes encoding these ligands may orchestrate an array of functions, a phenomenon known as pleiotropy. Here we examine whether pleiotropic actions of peptide genes might influence ligand-receptor coevolution. Four test groups of neuropeptides characterized by conserved C-terminal amino acid sequence motifs and their cognate receptors were examined in the red flour beetle (Tribolium castaneum): 1) cardioacceleratory peptide 2b (CAPA); CAPAr, 2) pyrokinin/diapause hormone (PK1/DH); PKr-A, -B, 3) pyrokinin/pheromone biosynthesis activating hormone (PK2/PBAN); PKr-C and 4) ecdysis triggering hormone (ETH); ETHr-b. Ligand-receptor specificities were established through heterologous expression of receptors in cell-based assays for 9 endogenous ligands. Based on ligand-receptor specificity analysis, we found positive pleiotropism exhibited by ETH on ETHR-b and CAPAr, whereas PK1/DH and CAPA are more highly selective for their respective authentic receptors than would be predicted by phylogenetic analysis. Disparities between evolutionary trees deduced from receptor sequences vs. functional ligand-receptor specificities lead to the conclusion that pleiotropy exhibited by peptide genes influences ligand-receptor coevolution.
Similar content being viewed by others
Introduction
Diffusible peptide signaling molecules acting via cell-surface receptors are evolutionary innovations for functional coordination of organ systems in animals. Partnerships between ligand and receptor persist through a process of ligand-receptor coevolution1. Among the diversity of peptides are closely related ligand groups that mediate distinct physiological functions.
Genome analysis reveals that acquisition and/or specialization of phenotypic traits accompanying speciation often includes appearance of new neuropeptide signaling pathways (i.e., tachykinin and natalisin signals2). Stability of a newly evolved ligand-receptor signaling pathway may require its rapid sub- or neo-functionalization3,4 for minimization of functional redundancy, for example through changes in spatial or temporal expression patterns. However, such a role for ligand-receptor coevolution in diversifying evolution is only one possible outcome. For example, actions of the neuropeptide FMRFamide on multiple receptors, including the receptor for the related neuropeptide FLRFamide, known as myosuppressin, in the silkworm Bombyx mori hints at likely evolutionary advantages associated with “cross-talk” or pleiotropism inherent in signaling strategies5. Pleiotropism occurs when a single gene influences multiple phenotypic traits. In the context of peptide signaling, a single peptide gene product signaling via distinct receptors may lead to multiple physiological and hence phenotypic outcomes. Such pleiotropic peptide action therefore could be an important evolutionary force. It may be time to recognize that evolution selects not just single ligand-receptor partners, but webs of interdigitating peptide signaling networks that orchestrate physiological states involving multiple organ systems and diverse functions.
Here we describe possible roles of pleiotropism in evolution of a neuropeptide cluster characterized by the C-terminal amino acid sequence motif “-PRXamide” and corresponding receptors in the red flour beetle, Tribolium castaneum. Our data support a role for pleiotropism in evolution of ligand-receptor interactions.
Results and Discussion
We investigated influences of pleiotropism in ligand-receptor coevolution by comparing evolutionary history according to alternative criteria: 1) amino acid sequence similarity and 2) ligand potency. According to the hypothesis of neutral evolution, evolutionary patterns should be similar whether gauged by molecular divergence of genes or by functional specificities of ligand-receptor interactions.
The PRXamide Peptide Cluster
The PRXamide cluster of neuropeptides has diversified considerably in insects, while its mammalian counterpart is confined to neuromedin U (F-x-P-R-x-amide) and possibly thyrotropin releasing hormone, based on molecular phylogeny of their corresponding receptors1. Hereafter, consensus sequences are represented according to Prosite syntax rules, where amino acids are separated by a hyphen (-), x denotes wild card amino acids and a square bracket denotes a small number of possible amino acid assignments at a given position.
The insect PRXamide cluster includes four families of neuropeptides (Fig. 1a): cardioacceleratory peptide 2b (hereafter named as CAPA), pyrokinin/diapausing hormone (PK1/DH or PK1), pyrokinin/pheromone biosynthesis activating hormone (PK2/PBAN or PK2) and ecdysis triggering hormone (ETH). Functions of this peptide group have been intensively studied over the past several decades, although information remains fragmented in some insect species and is limited to results from a few specific physiological assays, often chosen for ease of use rather than authentic physiological function(s) that are often unknown. It is notable that PRXamide peptides are predominantly hormonal signaling molecules, which allows for interactions with a broad diversity of receptors and for possible evolutionary selection based on adaptive influences of pleiotropism.
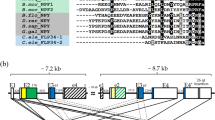
Three genes encoding –PRXamide peptide in the T. castaneum (a) and abdominal segments of a larva showing endocrine organs for ETH and CAPA visualized by immunohistochemistry (b).
(a) Schematic diagrams for CAPA, PBAN and ETH genes are shown on the top line of each gene product. Mature peptides are shown as color coded boxes for each peptide family. Empty boxes to the left in each diagram indicate signal peptides. Sequences for each putative mature peptide are aligned according to conserved C-terminal amino acid sequence motifs marked in bold. The italic used in the N-terminus of sequence ETH-2 indicates uncertainty with the canonical cleavage signal in the middle of the ETH-2 as GK. This study used the shorter version of ETH-2 starting with FFM in the alternative predictions. (b) Immunostaining of –PRXamide-expressing neurons. The top inset is a schematic diagram depicting stained elements visible below. Ventral larval abdominal segments are shown for the CNS and trachea containing the cells for CAPA and for ETH (Inka cell), respectively and for the perisympathetic organ, which serves as a neurohemal release site for the neuropeptide CAPA.
The CAPA family, named by virtue of cardioacceleratory activity, includes periviscerokinin, which shows myotropic activities through neurohemal release from segmental perisympathetic organs6. This family of peptides is characterized by the general C-terminal consensus sequence [LI]-x(2)-F-P-R-[VI]-amide (where x(2) means any two amino acids), with minor variations7. PK1/DH, known for its role in embryonic diapause in B. mori8,9 and for breaking pupal diapause in heliothine moths10,11,12,13, has consensus sequence W-F-G-P-R-L-amide with minor variations. PK2/PBAN is the well-known pheromone biosynthesis activating neuropeptide in Lepidoptera14,15 with consensus sequence F-x-P-R-L-amide. Notably, this sequence lacks the PK1/DH sequence motif. Finally, ETH targets numerous central peptidergic neuronal ensembles for activation of sequential preecdysis and ecdysis behaviors16,17,18. ETH has the C-terminal consensus sequence K-x(2)-P-R-[IVLM]-amide with minor variations. The nomenclature of neuropeptides described in this study follows that proposed by Coast and Schooley19 with minor modifications.
An interesting phenomenon in the PRXamide peptide group that possibly relates to functional pleiotropism involves two different genes, capa and pban, which encode mixed ligand arrays consisting of CAPA, PK1/DH and PK2/PBAN sequences (Fig. 1a). In most insects, the capa gene encodes a propeptide precursor containing CAPA and PK1/DH, while the pban gene encodes a precursor containing the related PK1/DH sequence and multiple PK2/PBAN-like peptides20. For example, the pban precursor gene of Helicoverpa zea encodes five mature peptides, including one copy of DH, one copy of PBAN and three PBAN-like peptides14. In this case, peptide precursors containing multiple peptides may undergo differential postranslational processing in different cell types, generating a diversity of peptide signals and physiological outcomes.
By way of example, the T. castaneum capa gene encodes authentic CAPA (also known as periviscerokinin) with a characteristic C-terminal FPRIa sequence motif (CAPA-2), but also encodes two additional peptides with SLRVamide and SPRLamide C-terminal sequence motifs (CAPA-1 and CAPA-3, respectively), along with one PK1/DH peptide (Fig. 1a). The pban gene encodes a PK1/DH peptide together with three peptides belonging to the PK2/PBAN group (PK2/PBAN-1, -2 and -3). The eth gene encodes two closely related ETH peptides: ETH-1 and ETH-2 (Fig. 1a). This evolutionary radiation of PRXamide peptides raises the possibility of pleiotropic actions on multiple receptors.
We also investigated proximity of PRXamide peptide release sites through immuno staining using an antiserum that cross-reacts with all members of the group. This suggests that the perisympathetic organ and Inka cell function as CAPA and ETH release sites, respectively in larval T. castaneum (Fig. 1b). The fact that these two release sites are in close proximity raises the possibility that potential functional interactions exist between these two closely related endocrine signals.
Cognate GPCRs of the PRXamide Peptide Cluster
The general consensus among receptor annotations for PRXamide neuropeptides across a wide range of insect orders places CAPA, PK and ETH receptors into distinct categories based on sequence homologies consistent with receptor specificities for their respective ligands. In a similar manner, PK receptors can be sub-divided into distinctive PK1/DH and PK2/PBAN receptors, but with larger degrees of ligand cross-activities1,21,22,23,24,25,26,27,28.
Information available from the genomic sequence of T. castaneum supports classification of five G protein-coupled receptors (GPCRs) in the PRXamide group29,30 (Fig. 2a): one CAPA receptor (CAPAr), three PK receptors denoted here as PKr–A, -B and -C and one ETH receptor that has alternatively spliced forms ETHr–a and ETHr–b. Based on our sequencing results, TcPKr-C and TcCAPAr match sequences predicted from the genome database. However, TcPKr-A has an eleven amino acid insertion and TcPKr-B has a three amino acid extension at the 5′ end not found in genome predictions. We updated the sequence information in GenBank on the NCBI website and corresponding accession numbers are provided in Table 1. Phylogenetic analyses clearly distinguish CAPAr, PKr and ETHr (Fig. 2a). Within the PKr group, TcPKr-A and TcPKr-B belong to the PK1/DH receptor cluster, whereas TcPKr-C forms an unstable branch on the basal lineage of the PK2/PBAN cluster with low bootstrapping supports (Fig. 2a).
Five T. castaneum GPCRs belonging to the cluster of receptors for –PRXamide peptides (a) and their expression patterns (b).
(a) A total of five receptors, TcPKr-A, -B and –C, TcCAPAr and TcETHr appear within a group of four by phylogenetic criteria: PK2/PBANr, PK1/DHr, CAPAr and ETHr. Arrowheads indicate bootstrapping values lower than 75, while all other nodes are supported by bootstrapping values higher than 75. Note that ETHr cluster is considerably separated from other clades. (b) Levels of mRNA for each receptor in different stages and tissues. Tissue specific expression data is for the late larval stage. The data show relative values for early egg in the stage specific data and for the CNS in the tissue specific data after normalization by rps3 expression.
Our results on stage- and tissue-specific expression patterns show that TcPKr-A and B, which are the most similar to each other in sequence, also show similar stage-specific expression patterns (Fig. 2b). TcPKr-B and –C exhibit similarly high expression in the central nervous system (CNS). TcPKr-A,-B,-C and TcCAPAr are all highly expressed in early larval stages. It is noteworthy that TcCAPAr is highly expressed in the hindgut, consistent with the hormonal diuretic function of CAPA previously documented in a number of insect species6,31,32. However, these expression data apparently are too limited in resolution to make inferences regarding evolutionary significance.
Ligand Specificities of PRXamide GPCRs
We characterized all PRXamide GPCRs through assays of 9 endogenous ligands. Ligand activities were quantified by detection of luminescence responses from CHO cells transiently transfected with GPCR (see Methods and Materials section for details). Dose-response curves were generated by logistic fitting of the log transformed ligand concentrations ranging from 0.01 nM to 10 μM (Fig. 3). All results, shown as averages, were replicated in three independent experiments (see Supplementary Data 1 for details). TcETHr-b, a splicing variant used in this study, showed relatively lower activity than those of other receptors, likely due to inefficient coupling to the reporter system.
Dose-response curves of five different PRXamide receptors of T. castaneum for nine endogenous ligands.
Left panel shows dose-response curves for each receptor with 9 putative endogenous ligands. Right panel shows names of ligands and their EC50s in rank order. Values of data points are found in Supplementary Data 1.
All GPCRs showed similar levels of sensitivity to their respective authentic ligands (low nanomolar EC50 values; Fig. 3), except for ETHr-b (EC50 values were 146.6 and 20.6 nM for TcETH1 and TcETH2, respectively; Fig. 3). Among three different Tribolium PK receptors, TcPKr-C was activated by only TcPK2/PBANs and activated by high concentrations of TcPK1/DH-2, but not by TcPK1/DH-1. Therefore, our results suggest that TcPKr-A and –B are activated by both TcPK/DH and TcPK/PBANs, while TcPKr-C is likely the authentic receptor specific for TcPK2/PBANs. TcCAPA and TcETH ligands had little or no activity on PK receptors. For TcCAPAr, the two TcCAPA ligands were highly potent with significant levels of cross activities of TcETHs. TcETHr-b was highly specific for TcETH ligands.
Pleiotropism
Evolutionary distances between PRXamide GPCRs were computed according to two criteria: 1) amino acid sequence similarity and 2) ligand specificity (Fig. 4a and b). Relative ligand activities were normalized to the strongest response of authentic partners as described in the Materials and Methods section (Fig. 4a). Overall, evolutionary distances computed by these two criteria (see Materials and Methods section for details) are well correlated, as the regression line shown in Fig. 4b indicates; this is consistent with the hypothesis of neutral evolution in determination of ligand-receptor evolutionary distances. However, we found a major deviation in the co-relationship of TcCAPAr and TcETHr. Although molecular evolution criteria predict considerable divergence between this receptor pair, functional ligand-receptor specificities predict a much smaller evolutionary distance (Fig. 4b). As a consequence, the evolutionary tree generated with ligand specificity as the determining criterion shows clustering of TcCAPAr and TcETHr (Fig. 4a), in contrast to the tree clustering TcCAPAr and TcPKrs according to amino acid sequence similarity.
Relative activities of endogenous ligands on five different PRXamide receptors (a), a plot showing relatedness of the sequence diversities and ligand specificities of PRXamide receptors (b) and a simplified diagram showing the asymmetric cross activities of ligands on target receptors (c).
(a) Values in the matrix are for log (EC50) normalized by the lowest EC50 for each receptor. The evolutionary tree based on GPCR amino acid sequence (top) is different from the tree inferred based on the ligand specificity (bottom). Color codes for each cell are low activity (green) to high activity (red). See Materials and Methods for further details. (b) Pairwise distances between receptors. Comparative distances between the distances based on ligand specificity (X-axis) and based on sequence similarity (Y-axis) for each pair of receptors are plotted. Error bars are for the standard error generated in 1000 bootstrapping in the sequence distances. The distance was normalized by the largest distance as 1. Regressed line is with 95% confidence interval (shaded area). Blue arrow implies directions of evolutionary forces for positive or negative pleiotropism. (c) Values in Figure 4a are used to depict a simplified view of asymmetry in pleiotropism. The line color indicates degree of activity; darker lines depict stronger activity.
The dual action of TcETH on TcETHr and TcCAPAr indicates positive pleiotropism and functional coupling of TcETHr and TcCAPAr (Fig. 4a). TcCAPA is released from segmental perisympathetic organs and likely promotes diuresis and myoactivity in vivo33, while TcETH triggers a downstream peptide signaling cascade that triggers sequential ecdysis behaviors and associated physiological events including diuresis, myoactivity and tracheal air-filling18. Targets of ETH include kinin neurons that promote diuresis and FMRFamide neurons that regulate myoactivity. Evidence presented here indicates that ETH, in addition to activating downstream peptidergic neurons, also activates related receptors such as CAPAr. Thus actions of ETH precursor gene products on both ETHr and CAPAr suggests that these two signaling pathways arose through concerted evolution through positive pleiotropism.
As shown in the matrices of ligand activities on each receptor (Fig. 4a), cross-activities are not necessarily reciprocal (Fig. 4c). For instance, the activity of TcETH on the TcCAPAr is not complemented by similar potency of TcCAPA on TcETHr. Another noticeable asymmetry occurs between PK1/DHs and PK2/PBANs. Strong activity of PK2/PBANs on all three PKr is asymmetric to the activity of PK1/DHs that has activities only to PKr-A and –B, but not to PKr-C (Fig. 4c). Therefore, CAPA and PK1/DHs act specifically only on their respective authentic receptor, but devoid of activating other receptors. This asymmetry supports the conclusion that divergence of ligand specificity is not random, but a consequence of selective pressure at the functional level for favoring pleiotropism or for high specificity.
We expanded our survey of PRXamide peptide pleiotropism to other species of insects, although only limited data were available to draw comprehensive conclusions. Data from two representatives of the Diptera (D. melanogaster34 and An. gambiae35) show that ETH is devoid of activity on CAPAr, in contrast to results of this study. Moderate levels of ETH activity on PKrs are commonly observed in many different insect species, including D. melanogaster21,27, An. gambiae35, H. virescens26 and H. zea22,23, in addition to the mild degree of positive pleiotropism we observe in the T. castaneum. Discrimination against PK1/DH by the PK2/PBAN receptor occurs in D. melanogaster21,27 and T. castaneum (i.e., TcPKr-C in this study), whereas moderate degrees of cross potency of PK1/DH on the PK2/PBAN receptor were observed in An. gambiae35, Ae. aegypti36, B. mori24,28, H. virescens26 and H. zea22,23. When more comprehensive data are available for expanded divergent taxa, evolutionary patterns of pleiotropism may reveal whether such activity is attributable to residual cross-activity (pleiotropism) in the duplicated system or whether pleiotropy was obtained secondarily in the evolutionary process.
Pleiotropic interactions among closely related ligands and corresponding receptors appear to be important in fine tuning orchestrated multi-organ physiological outcomes and consequently may be subject to selection pressures in the evolutionary process. In conclusion, this study provides evidence for influences of pleiotropism in ligand-receptor co-evolution in the PRXamide-receptor cluster. It seems possible that this principle may be expanded and applied to algorithms with greater effectiveness and selectivity in drug design targeting GPCRs.
Methods
Insects and chemicals
T. castaneum was kept in a 30°C growth chamber under a 16 : 8 (L : D) photoperiod and fed on a diet of wheat flour and Brewer's yeast (10 : 1). Peptides from T. castaneum were synthesized by Genescript (Piscataway, NJ). For culture of Chinese Hamster Ovary cells (CHO), DMEM/F12 medium, fetal bovine serum (FBS), Fungizone and Penicillin/Streptomycin and coelenterazine for aequorin functional assay were purchased from Gibco cell culture at Life TechnologiesTM (Grand Island, NY). TransIT-LT1 Transfection Reagent (Mirus Bio LLC, Madison, WI) was used for transient transfections.
Molecular cloning
Total RNA was isolated from the whole body of six individuals of T. castaneum last larval instar using TRI reagent (Ambion), treated by DNase I (Ambion) to eliminate genomic DNA and followed by the Phenol-Chloroform extraction. First strand cDNA was synthesized by SuperScriptTMII First-Strand Synthesis System for RT-PCR with random hexamer in the total volume 20 μL, according to manufacturer's instructions (Invitrogen Life Technologies). The 1st strand cDNA was used as template to amplify target PRXamide receptors utilizing high fidelity DNA polymerase PrimeSTARTM HS (Takara). Primers for all five GPCRs were designed based on T. castaneum genome data. For CAPA and three pyrokinin receptors (TcPKr-A, -B and -C), we started with four predictions including TC007170, TC011171, TC011320 and TC011318, respectively. Meanwhile, we started with the experimentally confirmed sequence (GenBank accession number is EF222294) for ETHr-b. Functional expression of ETH-a failed for unknown reasons. All primer information is provided in Table 1. Reactions were conducted in a volume of 50 μL and included ~50 ng cDNA, 10 μL 5x PrimeSTAR buffer with Mg2+, 0.32 mM for each dNTP, 0.2 μM of each primer. The PCR program included 35 cycles: 98°C for 10 sec, 58°C for 10 sec and 72°C for 90 sec with a final extension of 6 min at 72°C. The PCR product was cloned into a pGEMT Easy vector (Promega) and sequenced.
Sequence analysis and tree construction
Nucleotide sequences and deduced amino acid sequences of the PRXamide receptors were analyzed using DNAMAN7 (LynnonBioSoft). Searches of similar sequences were done with BlastP in the non-redundant protein sequences (nr) database of the NCBI website (http://www.ncbi.nlm.nih.gov/). Sequence alignments were made by ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Transmembrane helices were predicted using the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM) and only sequences covering transmembrane segments 1 to 7 and their linkers, excluding N- and C-termini of highly diversified regions, were used for the tree (Fig. 4a). A phylogenetic tree was constructed by MEGA537 by applying the Neighbor-Joining method with a bootstrap test of 1000 replications and complete deletion of gaps produced by the alignment. The distances between receptors obtained in the phylogenetic tree were.normalized by the maximum distance as 1 and used the plot value for “relative distance based on the amino acid sequence” (Fig. 4b).
The ligand activity matrix shown in Fig. 4A depicts log10-transformed EC50s normalized to the most potent authentic ligand for a given receptor using the formula: log{[EC50(test ligand)]/log[EC50(lowest)]}, where EC50(lowest) is the EC50 for endogenous partner ligand. In order to avoid artifacts introduced by extremely high EC50 values, we set 4 as the maximum value (e.g., EC50 of 104 nM) for log transformed EC50s. The ligand specificity tree was constructed in Cluster 3.019. The distances based on ligand specificity were obtained by using the R Stats Package Euclidean method in Distance Matrix Computation (http://stat.ethz.ch/R-manual/R-patched/library/stats/html/00Index.html). Relative distances were obtained by normalization to the maximum distance as 1.
Heterologous expression and functional assay
cDNAs for full GPCR ORFs were inserted into the expression vector pcDNA3.1(+) (Invitrogen) or pcDNA5/FRT (Invitrogen). Methods for expression in Chinese Hamster Ovary (CHO) cell supplemented with aequorin and G-α16 and the assays were performed as previously described38. Briefly, cells were transfected using the TransIT-LT1 transfection reagent (Mirus Bio LLC). Thirty hours after the transfection, the cells were collected and preincubated with the coelenterazine (Invitrogen) prior to the functional assay. A serial dilution (10 folds) of the 9 endogenous ligands were applied to the cells. The luminescence caused by the intracellular calcium mobilization were measured in continuous 20 seconds in every half-second interval. The authentic ligands showing most potent activity on the receptors were selected as model ligand for normalization of the luminescence. Based on which, a dose response curve was generated for each receptor by logistic fitting in Origin 8.6 (OriginLab).
Quantitative reverse transcriptase PCR (Q-PCR)
Expression patterns of GPCRs were examined by Q-PCR. Total RNA was collected from insects at different developmental stages and prepared as described previously: early egg (<24 h), late egg (>24 h), early larva (<24 h post-hatching), late larva (older than 5th instar including prepupa), early pupa (<24 h post-pupation), late pupa (>72 h post-pupation), early adult (<24 h post-eclosion) and late adult (one week old)39. To quantify tissue-specific expression, we collected total RNA from each of the following dissected tissues: CNS (central nervous system including brain and ganglia), midgut, hindgut and carcass excluding the aforementioned tissues. A pool of ten last-instar larvae was used for preparation of each tissue. Total RNA was treated by DNaseI (Ambion). Primers used in Q-PCR were listed in Table 1. 1st strand cDNA for qPCR was synthesized using ImProm-II™ Reverse Transcription System (Promega). Real-time RT-PCR was performed by using the iTaqTM Universal SYBR Green supermix (Biorad) on a CFX Connect real time detection system (Biorad). Data expressed here as relative mRNA levels were normalized to a reference gene ribosomal protein S3 (rpS3, GenBank accession number is CB335975), using the ΔΔCT method40. For samples collected from each developmental stage or tissue, we performed three biological replicates. For data analysis, relative expression of the target transcripts in early eggs (EE) served as the calibrator for developmental expression profiling, while target transcript expression in the CNS was employed as the calibrator for tissue specific expression profiling.
Immunohistochemistry
The antibody (anti-DrmETH1) used for recognition of–PRXamide motifs was raised in a rabbit. The central nervous system from last instar larvae was dissected in ice-cold phosphate-buffered saline (PBS: 137 mM NaCl, 1.45 mM NaH2PO4, 20.5 mM Na2HPO4, pH 7.2), freed of remaining fat and fixed in 4% paraformaldehyde at 4°C overnight. Fixed samples were washed in PBS containing 1% Triton X-100 (PBST). Tissues were then preadsorbed with 5% normal goat serum (Sigma) in PBST for 10 minutes and subsequently incubated with anti-DrmETH1 (1:500 dilution) for 2 days. After three washes with PBST (4 times for 15 minutes), tissues were incubated overnight in goat anti–rabbit IgG antibody conjugated with Alexa Fluor 488 (Molecular Probes). Images were captured in an epifluorescence microscope (Nikon E800).
References
Park, Y., Kim, Y.-J. & Adams, M. E. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin and AKH supports a theory of ligand-receptor coevolution. Proc Natl Acad Sci U S A 99, 11423–11428 (2002).
Jiang, H. et al. Natalisin, a tachykinin-like signaling system, regulates sexual activity and fecundity in insects. Proc. Natl. Acad. Sci. 110, E3526–E3534 (2013).
Conant, G. C. & Wolfe, K. H. Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet 9, 938–950 (2008).
Lynch, M. & Conery, J. S. The Evolutionary Fate and Consequences of Duplicate Genes. Science 290, 1151–1155 (2000).
Yamanaka, N. et al. Neuropeptide Receptor Transcriptome Reveals Unidentified Neuroendocrine Pathways. PLoS ONE 3, e3048 (2008).
Predel, R. & Wegener, C. Biology of the CAPA peptides in insects. Cell Mol Life Sci 63, 2477–2490 (2006).
Predel, R. et al. Identification of the first neuropeptides from the CNS of Hemiptera: CAPA peptides of the southern green stinkbug Nezara viridula (L.). Peptides 27, 2670–2677 (2006).
Yamashita, O. Diapause hormone of the silkworm, Bombyx mori: Structure, gene expression and function. J Insect Physiol 42, 669–679 (1996).
Sato, Y. et al. Precursor polyprotein for multiple neuropeptides secreted from the suboesophageal ganglion of the silkworm Bombyx mori: characterization of the cDNA encoding the diapause hormone precursor and identification of additional peptides. Proc Natl Acad Sci U S A 90, 3251–3255 (1993).
Zhang, Q., Zdarek, J., Nachman, R. J. & Denlinger, D. L. Diapause hormone in the corn earworm, Helicoverpa zea: Optimum temperature for activity, structure–activity relationships and efficacy in accelerating flesh fly pupariation. Peptides 29, 196–205 (2008).
Zhang, T. Y., Sun, J. S., Zhang, L. B., Shen, J. L. & Xu, W. H. Cloning and expression of the cDNA encoding the FXPRL family of peptides and a functional analysis of their effect on breaking pupal diapause in Helicoverpa armigera. J Insect Physiol 50, 25–33 (2004).
Xu, W. H. & Denlinger, D. L. Molecular characterization of prothoracicotropic hormone and diapause hormone in Heliothis virescens during diapause and a new role for diapause hormone. Insect Mol Biol 12, 509–516 (2003).
Zhao, J. Y., Xu, W. H. & Kang, L. Functional analysis of the SGNP I in the pupal diapause of the oriental tobacco budworm, Helicoverpa assulta (Lepidoptera: Noctuidae). Regul Pept 118, 25–31 (2004).
Ma, P. W. K., Knipple, D. C. & Roelofs, W. L. Structural organization of the Helicoverpa zea gene encoding the precursor protein for pheromone biosynthesis-activating neuropeptide and other neuropeptides. Proc Natl Acad Sci U S A 91, 6506–6510 (1994).
Raina, A. K. et al. Identification of a Neuropeptide Hormone That Regulates Sex Pheromone Production in Female Moths. Science 244, 796–798 (1989).
Žitňan, D. et al. Conservation of ecdysis-triggering hormone signalling in insects. J Exp Biol 206, 1275–1289 (2003).
Roller, L. et al. Ecdysis triggering hormone signaling in arthropods. Peptides 31, 429–441 (2010).
Kim, Y.-J., Žitňan, D., Galizia, C. G., Cho, K.-H. & Adams, M. E. A Command Chemical Triggers an Innate Behavior by Sequential Activation of Multiple Peptidergic Ensembles. Curr Biol 16, 1395–1407 (2006).
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95, 14863–14868 (1998).
Jurenka, R. & Nusawardani, T. The pyrokinin/pheromone biosynthesis-activating neuropeptide (PBAN) family of peptides and their receptors in Insecta: evolutionary trace indicates potential receptor ligand-binding domains. Insect Mol Biol 20, 323–334 (2011).
Cazzamali, G., Torp, M., Hauser, F., Williamson, M. & Grimmelikhuijzen, C. J. P. The Drosophila gene CG9918 codes for a pyrokinin-1 receptor. Biochem Biophys Res Commun 335, 14–19 (2005).
Choi, M. Y., Fuerst, E. J., Rafaeli, A. & Jurenka, R. Identification of a G protein-coupled receptor for pheromone biosynthesis activating neuropeptide from pheromone glands of the moth Helicoverpa zea. Proc Natl Acad Sci U. S. A. 100, 9721–9726 (2003).
Choi, M. Y., Fuerst, E. J., Rafaeli, A. & Jurenka, R. Role of extracellular domains in PBAN/pyrokinin GPCRs from insects using chimera receptors. Insect Biochem Mol Biol 37, 296–306 (2007).
Homma, T. et al. G protein-coupled receptor for diapause hormone, an inducer of Bombyx embryonic diapause. Biochem Biophys Res Commun 344, 386–393 (2006).
Jiang, H., Wei, Z., Nachman, R. J. & Park, Y. Molecular cloning and functional characterization of the diapause hormone receptor in the corn earworm Helicoverpa zea. Peptides 53, 243–249 (2014).
Kim, Y. J., Nachman, R. J., Aimanova, K., Gill, S. & Adams, M. E. The pheromone biosynthesis activating neuropeptide (PBAN) receptor of Heliothis virescens: Identification, functional expression and structure-activity relationships of ligand analogs. Peptides 29, 268–275 (2008).
Rosenkilde, C. et al. Molecular cloning, functional expression and gene silencing of two Drosophila receptors for the Drosophila neuropeptide pyrokinin-2. Biochem Biophys Res Commun 309, 485–494 (2003).
Watanabe, K. et al. FXPRL-amide peptides induce ecdysteroidogenesis through a G-protein coupled receptor expressed in the prothoracic gland of Bombyx mori. Mol Cell Endocrinol 273, 51–58 (2007).
Hauser, F. et al. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front Neuroendocrinol 29, 142–165 (2008).
Li, B. et al. Genomics, transcriptomics and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res 18, 113–122 (2008).
Paluzzi, J. P., Russell, W. K., Nachman, R. J. & Orchard, I. Isolation, cloning and expression mapping of a gene encoding an antidiuretic hormone and other CAPA-related peptides in the disease vector, Rhodnius prolixus. Endocrinology 149, 4638–4646 (2008).
Kean, L. et al. Two nitridergic peptides are encoded by the gene capability in Drosophila melanogaster. Am J Physiol Regul Integr Comp Physiol 282, R1297–R1307 (2002).
Eckert, M., Herbert, Z., Pollák, E., Molnár, L. & Predel, R. Identical cellular distribution of all abundant neuropeptides in the major abdominal neurohemal system of an insect (Periplaneta Americana). J Comp Neurol 452, 264–275 (2002).
Iversen, A., Cazzamali, G., Williamson, M., Hauser, F. & Grimmelikhuijzen, C. Molecular cloning and functional expression of a Drosophila receptor for the neuropeptides capa-1 and -2. Biochem Biophys Res Commun 299, 628–633 (2002).
Olsen, S. S., Cazzamali, G., Williamson, M., Grimmelikhuijzen, C. J. P. & Hauser, F. Identification of one capa and two pyrokinin receptors from the malaria mosquito Anopheles gambiae. Biochem Biophys Res Commun 362, 245–251 (2007).
Choi, M.-Y., Estep, A., Sanscrainte, N., Becnel, J. & Vander Meer, R. K. Identification and expression of PBAN/diapause hormone and GPCRs from Aedes aegypti. Mol Cell Endocrinol 375, 113–120 (2013).
Tamura, K. et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance and Maximum Parsimony Methods. Mol Biol Evol 28, 2731–2739 (2011).
Park, Y. & Adams, M. E. Insect G protein-coupled receptors. in Insect Pharmacology (eds. L.I. Gilbert & S.S. Gill) 331–359 (Academic Press, London; 2010).
Begum, K., Li, B., Beeman, R. W. & Park, Y. Functions of ion transport peptide and ion transport peptide-like in the red flour beetle Tribolium castaneum. Insect Biochem Mol Biol 39, 717–725 (2009).
Livak, K. J. & Schmittgen, T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408 (2001).
Acknowledgements
This paper is contribution no. 15-099-J from the Kansas Agricultural Experiment Station. HJ and YP were supported in part by National institute of health; Grant Number: R01AI090062; RJN, was supported in part by grants from BARD (4205-09C) and USDA/DOD DWFP (60-0208-4-001). This project was supported in part by the National Nature Science Foundation of China (31201508) and the Specialized Research Fund for the Doctoral Program of Higher Education of China (20120182120019). Publication of this article was funded in part by the Kansas State University.
Author information
Authors and Affiliations
Contributions
H.J., Z.W. performed the experiments. R.J.N. provided the reagents. H.J., M.E.A., Y.P. analyzed the data. Y.P., H.J., R.J.N. and M.E.A. wrote the paper. Y.P. supervised the project.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Dataset 1
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Jiang, H., Wei, Z., Nachman, R. et al. Functional Phylogenetics Reveals Contributions of Pleiotropic Peptide Action to Ligand-Receptor Coevolution. Sci Rep 4, 6800 (2014). https://doi.org/10.1038/srep06800
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06800
This article is cited by
-
Bee-safe peptidomimetic acaricides achieved by comparative genomics
Scientific Reports (2022)
-
CAPA neuropeptides and their receptor form an anti-diuretic hormone signaling system in the human disease vector, Aedes aegypti
Scientific Reports (2020)
-
Desiccation, thermal stress and associated mortality in Drosophila fruit flies induced by neuropeptide analogue treatment
Journal of Pest Science (2019)
-
The ecdysis triggering hormone system is essential for successful moulting of a major hemimetabolous pest insect, Schistocerca gregaria
Scientific Reports (2017)
-
Ligand selectivity in tachykinin and natalisin neuropeptidergic systems of the honey bee parasitic mite Varroa destructor
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.