Abstract
Most viruses in human skin are known to be human papillomaviruses (HPVs). Previous sequencing of skin samples has identified 273 different cutaneous HPV types, including 47 previously unknown types. In the present study, we wished to extend prior studies using deeper sequencing. This deeper sequencing without prior PCR of a pool of 142 whole genome amplified skin lesions identified 23 known HPV types, 3 novel putative HPV types and 4 non-HPV viruses. The complete sequence was obtained for one of the known putative types and almost the complete sequence was obtained for one of the novel putative types. In addition, sequencing of amplimers from HPV consensus PCR of 326 skin lesions detected 385 different HPV types, including 226 previously unknown putative types. In conclusion, metagenomic deep sequencing of human skin samples identified no less than 396 different HPV types in human skin, out of which 229 putative HPV types were previously unknown.
Similar content being viewed by others
Introduction
Analysis of metagenomes using deep sequencing has in recent years become a routine approach1. The bacterial community of the skin has been investigated through metagenomic sequencing targeting the 16S ribosomal DNA. The viral part of the microbiome has not been analyzed as thoroughly, because the viruses do not share a common consensus sequence that can be targeted by molecular methods2.
However, metagenomic sequencing has revealed that >95% of the viral sequences present in skin samples belong to the Papillomavirus family, mostly to the β- and γ-genera3. There are now 197 different HPV types established at the International HPV Reference Center (www.hpvcenter.se, accessed on 2014-06-05), but putative novel HPV types are continuously being discovered4,5,6,7,8,9. The cutaneous HPV types have been found in healthy skin as well as in different skin lesions such as squamous cell carcinoma (SCC), actinic keratosis (AK) and keratoacanthoma (KA), in both immunocompetent and immunosuppressive patients10,11,12,13,14,15,16,17,18. Some mucosal HPV types cause cervical cancer19, as well as vulvar, anal and penile cancers20, whereas some cutaneous HPV types cause skin warts and others are associated with SCC in patients with a rare immunosuppressive disease21,22. Virus-associated cancers have an increased incidence among immunosuppressed patients23. This fact has spurred intensive efforts to search for viruses in cancers that have an increased incidence among the immunosuppressed patients, but that are not known to have a viral etiology. The cancer form that is most highly increased in incidence among the immunosuppressed is non-melanoma skin cancer23.
We previously performed metagenomic sequencing of 142 skin samples amplified only with whole genome amplification (that is, without prior PCR) using 454 and Ion Torrent sequencing technologies, where most of the viral sequences mapped to the HPV family and 7 and 12 HPV types were identified, respectively13. Classification of HPV is based on the sequence of the major capsid protein gene L1. The L1 sequence of a new HPV type should be <90% similar to the L1 gene in any known HPV type24. The HPV consensus PCR primer pair FAP59/64 amplifies both mucosal and cutaneous types25. As HPVs have been reported to be the most common viruses in the human skin3,13, we have also described the use of deep sequencing of general primer HPV PCR amplimers as a method to detect additional HPV types9,26. In this study, we wished to perform analysis with deeper sequencing using the much more powerful Illumina sequencing technology to investigate if there might be additional viruses present in human skin.
Results
Metagenomic sequencing of whole-genome amplified skin lesions (without prior PCR)
Swab samples from 82 SCCs and 60 AKs skin lesions were subjected to whole genome amplification, pooled and sequenced using the Illumina MiSeq sequencing platform. We found a total of almost 100 000 HPV reads (>99% of all viral reads) comprising 21 known established HPV types, 2 known putative types and 3 novel putative HPV types (Table I). The taxonomic definition of an HPV type is that the L1 sequence of a new HPV type should be less than 90% similar to the L1 gene in any known HPV type. Known putative HPV types refers to sequences that are present in GenBank, but have not yet been cloned and hence, no official HPV type number have been assigned to them by the International HPV Reference Center (www.hpvcenter.se). In addition, human polyomavirus 6, Merkel cell polyomavirus, torque teno virus and human endogenous retrovirus were detected with between 7 and 205 reads each (Table I).
Compared to previous analysis of the same sample pool using 454GSFLX and Ion Torrent PGM technologies13, we obtained a 260-fold and 35-fold larger number of viral reads using the Illumina MiSeq, respectively. The MiSeq analysis detected 26 established or putative HPV types, compared to 7 and 14 detected by GSFLX and PGM, respectively. 11 of the 14 HPV types detected by any of the formerly used technologies, the GSFLX and the PGM, were detected by the Illumina MiSeq. Only 3 previously detected types (HPV45, HPV59, FA73) were not detected by MiSeq. However, these 3 HPV types were all only detected with a single read each during previous sequencing runs with the 454 or Ion Torrent technologies13.
For one of the known putative types, SE46, the complete sequence was obtained from a total of 4881 reads (maximum coverage of 273). From the previous GSFLX and PGM runs, SE46 was sequenced with 22 reads (maximum coverage of 5) and 132 reads (maximum coverage of 18), respectively with only a partial sequence obtained13. In this study, the Illumina MiSeq sequencing increased the sequencing depth approximately 220 times for this particular virus and the complete genome of the type SE46 was obtained (Figure 1). The genomic organization was similar to that of established γ-papillomaviruses.
Coverage plot of the putative HPV type SE46 genome.
Coverage is represented as percentages (maximum coverage/coverage at a particular position), comparing different sequencing methods with each other. Grey, green and red histograms correspond to genome coverage from the Roche 454GSFLX, Ion Torrent PGM and Illumina MiSeq platform, respectively. The data from GSFLX and PGM platforms are previously published13. Light blue lines represent putative open reading frames of SE46. The plot was generated using Circos visualization tool40.
In addition, partial sequences of the 3 previously unknown putative HPV types (herein named SE355, SE356, SE357), were found using the Illumina MiSeq platform (Table I). For SE355 we obtained a 7080 bp long contig, assembled from 369 reads, representing almost the complete HPV genome. The GenBank accession numbers for the 3 novel putative types (numbered SE355 to SE357) are KJ506870 to KJ506872.
Known and novel HPV types found after HPV consensus PCR
Sequencing of the HPV-amplimers in the 3 pools of in total 326 skin lesions identified 159 known HPV types, out of which 52 were known established HPV types and 107 were known putative types (Table II). In addition, the deep sequencing identified 226 sequences of previously unknown putative novel HPV types. 65% of the identified previously known HPV types (104/159) belonged to the γ-genus, whereas 63% of the novel putative types (142/226) belonged to the β-genus (Table II). There were only small differences between the 3 pools. They had similar distribution of different kinds of HPV types, both known and novel. For all 3 pools, more than half of all HPV sequences detected were novel putative types. The GenBank accession numbers for the 226 novel putative types (herein numbered SE129 to SE354) are KJ506873 to KJ507098.
The new HPV phylogenetic tree
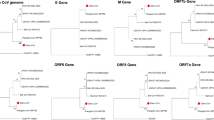
160 of the 226 novel putative HPV types identified by the MiSeq sequencing of the three pools of HPV amplimers contained >400 bp or >200 bp including the 3′-end of the HPV consensus PCR amplimers and are shown in the HPV tree together with all known established types and previously known putative types, labeled as SE-types (Figure 2). The novel putative types cluster together in the β- and γ-genera. One large group of 17 novel putative types and 2 known putative types forms a new branch in the tree, belonging to the γ-genus. Another group of 9 novel putative types forms a new branch belonging to the β-genus (Figure 2).
Bayesian phylogenetic tree based on the L1 part of the complete 164 established HPV types (+ bovine papillomavirus type 3 and type 5) and 160 putative novel HPV types (SE-types) that were >400 bp or contained >200 bp of the 3′-end of the amplimer.
SE-types discovered using 454GSFLX9,26 and using Illumina MiSeq are presented in blue and red colors, respectively.
Comparison of results from different samples
The swabbing of the surface of human skin is intended to characterize the diversity of viruses present, but is less informative for making associations with skin diseases as viruses that are shed from other places on the body might be detectable in swabs of skin surfaces also on rather distant sites on the body. Skin surface swabs were used in the metagenomic analysis (pool D) as well is with deep sequencing of HPV general primer amplimers (pool C). The sequencing of amplimers was unquestionably more sensitive (352 different HPVs detected) compared to the metagenomic sequencing (26 different HPVs detected), but it is noteworthy that no less than 11 of the HPVs that were detected in the metagenomic sequencing were not detected when sequencing HPV general primer PCR amplimers (Tables I and II) implying that these viruses were not effectively amplified by the general primers used. The metagenomic sequencing had an average coverage of the human of 2.5-fold, suggesting that viruses that are WGA-amplified about as effectively as the human genome had been detectable if present at about 1 copy per cell and that viruses that are WGA-amplified about as effectively as the HPV control plasmid had been detectable if present at about 0.04 copies/cell.
Two analyzed samples contained biopsies from either SCC and its precursor AK (pool A) or from KA (pool B). Most of the detected HPVs were present in about equal abundance in all the pools tested, suggesting that they were not specifically associated with any particular skin disease. Viruses that were much more (>10-fold number of reads) abundant in the pool with SCC and AK were HPV158, FA9, GC05, KC45, SE126, SE253, SE279, SE337 and SE341. Viruses that were much more abundant in the KA pool were HPV4, HPV77, HPV148, FA89, FA199,SE205, SE242, SE 243, SE265, SE270, SE273,SE274 and SE325.
Discussion
The aim of the present study was to investigate if deeper sequencing using the Illumina platform would extend the diversity of HPVs found to be present in human skin. Indeed, our study found that human skin contains an extreme diversity of cutaneous HPV types, establishing that the HPV types are far more numerous than previously known. We have previously used first generation deep sequencing technologies to study the viral metagenome in skin, for example the 454GSFLX with original and titanium chemistries9,14,15,27 as well as Ion Torrent PGM13. The current, much deeper, sequencing using the Illumina platform resulted in an approximately thousand-fold increase in sequence depth and revealed 229 previously unknown putative HPV types, resulting in a fundamental change in our understanding of the diversity of HPVs.
Impressively, deep sequencing appears to be a reliable method for detection of viruses in skin samples. In the pool with 142 skin samples amplified only with whole genome amplification (but not with prior PCR), the deeper Illumina sequencing resulted in 200 times and 25 times more total number of reads compared to previous metagenomic sequencing using 454 and Ion Torrent technologies13 and generated 260 times and 35 times more number of viral sequences as well as 4 times and 2 times more HPV types, respectively. 3 previously unknown putative HPV types were only found using the Illumina technology and for one of them almost the complete sequence was obtained. These findings indicate that the Illumina deep sequencing technology is a useful method to obtain the complete picture of the viruses that are present in human skin. 9 out of 12 viral types that were detected with only a single read using 454GSFLX or Ion Torrent PGM were confirmed by the Illumina, with many more reads. Only 3 of the 12 viruses previously detected by only a single viral read were not confirmed by the deeper sequencing.
The whole genome amplification method used (GenomiPhiHighYield) is intended for a random amplification of all DNA in a sample and is thus routinely used for amplifying linear DNA. However, the GenomiPhi reaction has a preference for circular templates, which will have made it easier to detect circular genomes in the pool amplified with WGA. The WGA amplified the human genome 25 times and a circular HPV plasmid an additional 25 times. Thus, although the WGA will have made it easier to detect the circular HPV genomes, it is unlikely that we would have missed linear viruses unless they were present only in very small amounts.
Three of the pools were amplified with an HPV consensus PCR before sequencing. Although the amplimer length is 450 bp, some sequences obtained were longer or shorter than expected This could be due to the fact that these PCR primers are highly degenerate and may to some extent also bind unspecifically.
The majority of the HPV types and putative types detected in this study belonged to the β- and γ-genera, but 11 mucosal types from α-genus were also found, out of which one was a putatively novel HPV type. Both presence of anogenital oncogenic HPV types in skin samples28,29 and contamination of the skin by viruses originating from mucosal surfaces, mediated by the fingers, has been reported30. In this study, biopsies from SCCs and AKs were taken after tape-stripping of the skin surface15, to reduce the probability of detecting contaminating viruses.
Phylogenetically, the majority of the novel HPV sequences belonged to the β-genus (142/226) followed by the γ-genus (83/226). In previous studies, the majority of novel putative HPV types found belonged to the γ-genus. Possibly, the viruses in the γ-genus may exist at higher viral loads, making them easier to detect, wheras viruses of β-genus might be biologically different and be present in lower viral loads. It might intuitively be surmised that viruses present in high amounts are more pathogenic, but there is no data to support this notion. Alternative scenarios are possible where the many different viruses present may interact. A first step that will enable investigations of this issue is the basic knowledge that these viruses are present - the basic and fundamental discovery of the paper. Although 22 of the HPVs detected appeared to be more common in either the SCC/AK or KA sample, the massive number of viruses detected suggests that the association may have been due to chance and that it needs to be investigated in further studies.
The β-genus is already diverse with 45 completely sequenced HPV types established (www.hpvcenter.se). The γ-genus has been growing rapidly and has surpassed the β-genus, with now 61 completely sequenced γ types established (www.hpvcenter.se). It appears that some of the new HPV types detected in the present study form clusters outside the previously defined species. Two of them are particularly noticeable, a new branch in the β-genus formed from 9 novel putative HPV types and a new branch in the γ-genus formed from 17 novel putative HPV types. The second branch also includes 2 previously detected putative HPV types26. However, whether these new branches do indeed represent new HPV species needs confirmation by cloning, sequencing and deposition at the international HPV reference center (www.hpvcenter.se).
With increasing sensitivity and throughput of the sequencing technology, the possibility always exists that assembly algorithms may construct erroneous “chimeric” sequences by the assembly of two different sequences from different viruses. Genomic recombination has been described for cetacean papillomaviruses31. Even though this has not been described for HPVs, multiple co-infections of related microorganisms can result in recombination32. Both naturally occurring genomic recombination and PCR-mediated recombination may mislead phylogenetic analysis32. We have previously developed a bioinformatics pipeline, which detects and removes putative chimeras that may have resulted from PCR-mediated recombination between related templates26,33. The bioinformatics in this study was even stricter regarding chimera checking also for known viruses. Hence, some sequences reported in the previous studies13,26 did not pass the chimera checking step in the data analysis pipeline in this study. The phylogenetic tree was constructed only with the sequences that had passed our most strict chimera checking algorithm.
In conclusion, we demonstrate that the diversity of HPVs in human skin is far greater than previously known. Deep sequencing using the Illumina platform is effective to detect a plethora of HPVs that are present in skin samples. As deep sequencing technologies are continuously evolving with increasing throughput and decreasing cost per base pair, it is likely that an extraordinary and expanding diversity of cutaneous HPVs will continue to be revealed. As the HPVs are much more diverse than previously known, this needs to be taken into consideration in the design of detection methods and when designing studies of HPV biology or epidemiology.
Methods
Samples
Swabs and biopsies of non-melanoma skin cancer lesions (SCC, AK and KA) from immunocompetent patients attending Swedish and Austrian hospitals27 and biopsies of KA lesions from both immunosuppressed and immunocompetent patients attending the Norwegian National Hospital in Norway14 were used. The swab samples had been collected both from the top of the lesion and from normal adjacent skin by a pre-wetted (0.9% NaCl) cotton-tipped swab that was rolled on the lesion (within margins of the lesion) or on the normal skin respectively and suspended in 1 mL of saline. In addition, 2 mm diameter punch-biopsies had been taken after tape-stripping from each lesion as well as from normal adjacent skin15. Biopsies from SCC and AK had been HPV-positive in a previous study27. The DNA was extracted with a phenol-free method25 for the Swedish/Austrian biopsies and with the QIAamp DNA Minikit (Qiagen, Germany)14 for the Norwegian biopsies. Swab samples were not extracted, only frozen and thawed. Informed consent was obtained from participants. The study adhered to the declaration of Helsinki and was approved by the Ethical Review Committees of Karolinska Institutet and of Lund University (Sweden), Medical University Vienna (Austria) and Institutional Review Board in Oslo (Norway). All methods were carried out in accordance with the approved guidelines.
DNA amplification and pooling
The samples were mixed into 4 pools. Pool A, B and C were used in a previous study26, where pool A constituted fresh frozen biopsies from 29 SCC lesions and 31 AK lesions, pool B fresh frozen biopsies from 91 KA lesions and pool C top of lesion swabs of 84 SCCs and 91 AKs. Prior pooling into A, B and C, the samples were PCR amplified with the HPV consensus primer pair FAP59/64, targeting the L1 gene of α-, β- and γ-HPVs, as described previously25, using 5 μL of each sample per reaction. For SCC and AK biopsies, only samples that were HPV positive by PCR27 were used (SCC, n = 29 and AK, n = 31). A plasmid including the sequence of HPV type 12 was used as a positive control and a detection limit of one copy per microliter was found. The PCR amplimers from the three pools were purified using the MinElute spin column kit from Qiagen according to manufacturer's guidelines. Two columns per sample pool were used with a sample volume of 120 μL per column. The elution volume was 10 μL of EB-buffer. The two elutions from each sample were mixed, generating a purified PCR product of 20 μL. Pool D, also used in a previous study13, was a mix of swab samples from 82 SCC lesions and 60 AK lesions. The 142 swab samples were subjected to random whole genome amplification using the GenomiPhi High Yield kit (GE Healthcare, UK) and combined into pool D, where upon the DNA was ethanol precipitated and dissolved in 100 uL water. The 4 pools were quantified using QuantiFluor-ST (Promega, US), a fluorometric assay quantifying dsDNA, according to manufacturer's user guide. The concentrations were 22.4 ng/μL, 17.8 ng/μL, 19.6 ng/μL and 56.4 ng/μL for pool A, B, C and D respectively.
Sample library preparation
DNA libraries for the 3 HPV consensus PCR amplified pools, A, B and C, were prepared using the TruSeq Nano DNA Sample Preparation kit according to the user guide revision A (Illumina) with the following modifications: as the samples consisted of approximately 450 bp long PCR products, the fragmentation, end-repair and size selection steps were omitted and hence, the library preparation started with adenylation of 3′-ends. As Illumina's recommended input DNA is 100 ng and 200 ng for 350 bp and 550 bp insert size respectively, we used 150 ng of the 450 bp long PCR product as input in a volume of 17.5 μL, diluted in resuspension buffer, for pool B (first pool to be sequenced). Due to low cluster density in the sequencing flow cell for pool B, we adjusted the input DNA for library preparation to 50 ng for pool A and C. Having too much DNA during adapter ligation will result in more fragments containing only one adapter, which will be part of library quantification, but not be able to generate clusters in the sequencing flow cell, leading to low cluster density. The index adapters AD007, AD002 and AD019 were used for pool A, B and C respectively.
DNA library of the random amplified pool D was prepared using the Nextera DNA Sample Preparation kit according to the user guide revision B (Illumina), starting with 50 ng DNA in the tagmentation reaction. A MinElute spin column was used in the clean-up step according to the MinElute user guide (Qiagen, US) instead of the Zymo spin plate.
The library pools were quantified with the QuantiFluor system as above and the library sizes were checked using the Bioanalyzer (Agilent). Pool A contained fragments between 200 to 1370 bp. The main peaks for pool B and C was at 566 bp and 560 bp respectively, corresponding to the FAP-PCR amplimer size. Pool D contained fragments between 300 to 7000 bp with the main peak at 1200 bp. For pool A, B and C the mean DNA fragment sizes were estimated to be 500 bp and the conversion formula 1 ng/μL = 3 nM DNA was used. For pool C the mean fragment size was estimated to be 1–1.5 kb and the conversion formula 1 ng/μL = 1.5 nM DNA was used.
Sequencing on the Illumina MiSeq
Denatured libraries at 20 pM from the FAP-PCR amplified pool A, B and C were spiked with 5% PhiX control and individually sequenced by paired-end 301 + 301 cycles on the MiSeq instrument using version 3 reagent kit (Illumina, US). For the multiple displacement amplified pool D, 10 pM denatured library was spiked with 1% PhiX control and sequenced by paired-end 251 + 251 cycles on the MiSeq using version 2 reagent kit. The sequencing preparations were made according to the user guides Preparing Libraries for Sequencing on the MiSeq revision C, Reagent Preparation Guide revision A and MiSeq System User Guide revision K.
The sequencing flow cell cluster density for pool B, first to be sequenced, was 422 K/mm2, the total yield was 6.2 Gb, 68% >Q30 and 98% of reads were passing filter. The cluster densities for pool A and C were 1113 K/mm2 and 1339 K/mm2, the total yields were 15.6 Gb and 18.6 Gb, 66% and 67% >Q30, 95% and 94% of reads passing filter - an improvement compared to pool B presumably due to the adjusted DNA input for library preparation. Pool D had a cluster density of 627 K/mm2, the total yield was 5.9 Gb, 86% were >Q30 and 97% of reads were passing filter.
Analysis of sequences
Sequences were obtained from the MiSeq (Illumina) instrument. Indices, included in the Illumina adaptors, were used to assign the sequences obtained to the originating sample. The bioinformatic analysis started with quality checking, where sequences were trimmed according to their Phred quality scores34. Quality checked reads were then screened against the human reference genome hg19 using BWA-MEM35 and SOAP aligner (http://soap.genomics.org.cn) and reads with >95% identity over 75% of their length to human DNA were removed from further analysis. The rest of the sequences were normalized (http://ged.msu.edu/papers/2012-diginorm) to discard redundant data and reduce sampling variation and sequencing errors. The normalized dataset was then processed for assembly using the Trinity36, SOAPdenovo and SOAPdenovo-Trans (http://soap.genomics.org.cn/) assemblers into contiguous sequences (contigs). Reads before assembly were re-mapped to assembled contigs and the result was used to calculate number of reads for each assembled contig. The use of several assembly algorithms and re-mapping of all singleton reads to assembled contigs were used to validate assembly results13,37. Assembled contigs were then subjected to taxonomic classification by comparing them against GenBank nucleotide database using paracel blast (www.strikingdevelopment.com) blastn to classify them as i) previously known sequences, ii) related to previously known sequences, or iii) unrelated to any previously known sequences. To identify possible artifactual “chimeric” sequences, contigs containing sequences originating from different viruses, all papillomavirus-related contigs and singletons were checked as described38. Shortly, the sequence that aligned to its most closely related sequence in GenBank was divided into three equal segments. If at least one of the segments differed in similarity to the corresponding overlapping parts with more than 5% (for example, if segment 1 was 88% similar and segment 2 was 94% similar) the sequence was considered as a “possible chimera”. All analyses were performed using in-house R (www.R-project.org/) and python (www.python.org) scripts that run on a high performance (40 core, 2 TB RAM) Linux server.
Phylogenetic analysis
BEAST v1.8.039 was used to construct a Bayesian Phylogenetic tree. 4 independent Markov chain Monte Carlo (MCMC) tests were run under a coalescent tree prior and relaxed uncorrelated lognormal molecular clock for 100 000 000 generations, with a tree logged every 1000th generation. A maximum clade credibility tree was constructed by a tree annotator39 after discarding the initial 25% generations as a conservative generalization of the “burn-in” phase. The analyses were restricted to sequences that were >400 bp or contained >200 bp of the 3′-end of the amplimer. This restriction was necessary in order to avoid the possibility that non-overlapping sequences might derive from the same virus.
Amplification of viral and human DNA by whole genome amplification (WGA)
Because the samplesin pool D were amplified by WGA before sequencing, we quantified the amount of amplification of human DNA and of HPV DNA. To samples with human DNA (human placental DNA at 1 ng/ul), we added 20 copies/ul of HPV16 plasmid and amplified the samples with WGA using the GenomiPhi HighYield Ready-to-go kit (GE Healthcare) in the same manner as for the clinical samples. The amounts of human DNA and viral DNA were quantified using real-time PCR for Betaglobin and for HPV16, respectively. The human DNA was found to be amplified 26-fold, whereas the HPV16 DNA was amplified 679-fold.
References
Cheval, J. et al. Evaluation of high-throughput sequencing for identifying known and unknown viruses in biological samples. J Clin Microbiol 49, 3268–75 (2011).
Grice, E. A. & Segre, J. A. The skin microbiome. Nat Rev Microbiol 9, 244–53 (2011).
Foulongne, V. et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One 7, e38499 (2012).
Chen, Z., Schiffman, M., Herrero, R. & Burk, R. D. Identification and characterization of two novel human papillomaviruses (HPVs) by overlapping PCR: HPV102 and HPV106. J Gen Virol 88, 2952–5 (2007).
Vasiljevic, N. et al. Characterization of two novel cutaneous human papillomaviruses, HPV93 and HPV96. J Gen Virol 88, 1479–83 (2007).
Bernard, H. U. et al. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401, 70–9 (2010).
Ekstrom, J., Forslund, O. & Dillner, J. Three novel papillomaviruses (HPV109, HPV112 and HPV114) and their presence in cutaneous and mucosal samples. Virology 397, 331–6 (2010).
Bottalico, D. et al. The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. J Infect Dis 204, 787–92 (2011).
Ekstrom, J., Bzhalava, D., Svenback, D., Forslund, O. & Dillner, J. High throughput sequencing reveals diversity of Human Papillomaviruses in cutaneous lesions. Int J Cancer 129, 2643–50 (2011).
Bzhalava, D., Guan, P., Franceschi, S., Dillner, J. & Clifford, G. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology 445, 224–31 (2013).
Antonsson, A. et al. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J Gen Virol 84, 1881–6 (2003).
Chen, A. C., McMillan, N. A. & Antonsson, A. Human papillomavirus type spectrum in normal skin of individuals with or without a history of frequent sun exposure. J Gen Virol 89, 2891–7 (2008).
Bzhalava, D. et al. Unbiased approach for virus detection in skin lesions. PLoS One 8, e65953 (2013).
Forslund, O., DeAngelis, P. M., Beigi, M., Schjolberg, A. R. & Clausen, O. P. Identification of human papillomavirus in keratoacanthomas. J Cutan Pathol 30, 423–9 (2003).
Forslund, O. et al. High prevalence of cutaneous human papillomavirus DNA on the top of skin tumors but not in “Stripped” biopsies from the same tumors. J Invest Dermatol 123, 388–94 (2004).
Kohler, A. et al. Genomic characterization of ten novel cutaneous human papillomaviruses from keratotic lesions of immunosuppressed patients. J Gen Virol 92, 1585–94 (2011).
Li, J. et al. Nine complete genome sequences of cutaneous human papillomavirus genotypes isolated from healthy skin of individuals living in rural He Nan province, China. J Virol 86, 11936 (2012).
Vasiljevic, N., Hazard, K., Dillner, J. & Forslund, O. Four novel human betapapillomaviruses of species 2 preferentially found in actinic keratosis. J Gen Virol 89, 2467–74 (2008).
Walboomers, J. M. et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189, 12–9 (1999).
IARC. Human Papillomaviruses. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans 90, 1–636 (2007).
Jablonska, S., Dabrowski, J. & Jakubowicz, K. Epidermodysplasia verruciformis as a model in studies on the role of papovaviruses in oncogenesis. Cancer Res 32, 583–9 (1972).
Jablonska, S., Majewski, S., Obalek, S. & Orth, G. Cutaneous warts. Clin Dermatol 15, 309–19 (1997).
Vajdic, C. M. & van Leeuwen, M. T. Cancer incidence and risk factors after solid organ transplantation. Int J Cancer 125, 1747–54 (2009).
de Villiers, E. M., Fauquet, C., Broker, T. R., Bernard, H. U. & zur Hausen, H. Classification of papillomaviruses. Virology 324, 17–27 (2004).
Forslund, O., Antonsson, A., Nordin, P., Stenquist, B. & Hansson, B. G. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol 80 (Pt 9), 2437–43 (1999).
Ekstrom, J. et al. Diversity of human papillomaviruses in skin lesions. Virology 447, 300–11 (2013).
Forslund, O. et al. Cutaneous human papillomaviruses found in sun-exposed skin: Beta-papillomavirus species 2 predominates in squamous cell carcinoma. J Infect Dis 196, 876–83 (2007).
Asgari, M. M. et al. Detection of human papillomavirus DNA in cutaneous squamous cell carcinoma among immunocompetent individuals. J Invest Dermatol 128, 1409–17 (2008).
Iftner, A. et al. The prevalence of human papillomavirus genotypes in nonmelanoma skin cancers of nonimmunosuppressed individuals identifies high-risk genital types as possible risk factors. Cancer Res 63, 7515–9 (2003).
Alam, M., Caldwell, J. B. & Eliezri, Y. D. Human papillomavirus-associated digital squamous cell carcinoma: literature review and report of 21 new cases. J Am Acad Dermatol 48, 385–93 (2003).
Robles-Sikisaka, R. et al. Evidence of recombination and positive selection in cetacean papillomaviruses. Virology 427, 189–97 (2012).
Liu, W. et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 467, 420–5 (2010).
Johansson, H. et al. Metagenomic sequencing of “HPV-negative” condylomas detects novel putative HPV types. Virology 440, 1–7 (2013).
Bokulich, N. A. et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10, 57–9 (2013).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–95 (2010).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8, 1494–512 (2013).
Meiring, T. L. et al. Next-generation sequencing of cervical DNA detects human papillomavirus types not detected by commercial kits. Virol J 9, 164 (2012).
Johansson, H. et al. Metagenomic sequencing of “HPV-negative” condylomas detects novel putative HPV types. Virology (2013).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214 (2007).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639–45 (2009).
Acknowledgements
We thank Carina Eklund for excellent technical assistance.
Author information
Authors and Affiliations
Contributions
E.H. and J.D. conceived the project. E.H., J.E. and O.F. designed the experiments. E.H. performed the experiments. D.B. performed bioinformatic analyses and prepared figures. D.B., E.H., L.S.A.M. and J.E. interpreted the data. E.H., D.B. and J.D. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Bzhalava, D., Mühr, L., Lagheden, C. et al. Deep sequencing extends the diversity of human papillomaviruses in human skin. Sci Rep 4, 5807 (2014). https://doi.org/10.1038/srep05807
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05807
This article is cited by
-
The routine use of LCD-Array hybridisation technique for HPV subtyping in the diagnosis of penile carcinoma compared to other methods
BMC Urology (2022)
-
A broadly protective vaccine against cutaneous human papillomaviruses
npj Vaccines (2022)
-
Metatranscriptome analysis in human papillomavirus negative cervical cancers
Scientific Reports (2022)
-
Using HPV-meta for human papillomavirus RNA quality detection
Scientific Reports (2022)
-
Explainable deep neural networks for novel viral genome prediction
Applied Intelligence (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.