Abstract
The accumulation of single nucleotide polymorphisms (SNPs) in the displacement loop (D-loop) of mitochondrial DNA (mtDNA) has been described in various types of cancers and their association with cancer risk and disease outcome has been extensively identified. In the present study, we investigated the association between age-at-onset and SNPs in the mitochondrial D-loop using a population-based series of renal cell carcinoma(RCC). The SNP sites of nucleotides 16293A/G were identified for their association with age-at-onset using the log-rank test. The age-at-onset of patients with the minor allele G genotype was significantly lower than that of patients with the A genotype at the 16293 site (p < 0.001). Genetic polymorphisms in the D-loop are predictive markers of age-at-onset in RCC patients. Accordingly, the analysis of genetic polymorphisms in the mitochondrial D-loop may help identify RCC patient subgroups at high risk of early onset.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) accounts for almost 3% of all cases of adult malignancies worldwide, with more than 270,000 new cases (2.1% new cases of all cancers) and 100,000 deaths annually1. The incidence of RCC has been increasing globally2. In China, RCC is the second leading malignancy in the genitourinary system, with an average 6% annual number of cases over the past 20y3. RCC is a “quiet” disease and typically diagnosed at an advanced stage4. A few studies have focused on genetic factors associated with the age-at-onset of cancer, for example esophageal squamous cell carcinoma5. However, these studies have only demonstrated the genetic prevalence of this disease.
The human mitochondrial genome is 16 kb circular double - stranded DNA molecule. It contains 12 coding gene for engaged in respiration and oxidative phosphorylation, 2 rRNAs and a set of 22 tRNAs that are essential for the protein synthesis in mitochondria6. mtDNA is believed to be more susceptible to DNA damage and acquires mutations at a higher rate than nuclear DNA, due to high levels of reactive oxygen species (ROS), a lack of protective histones and a limited capacity for DNA repair in the mitochondria7,8.
In cancer patients, sequence changes accumulate extensively in the mitochondrial D-Loop region, which is significant for regulating the replication and expression of the mitochondrial genome, as it contains the leading-strand origin of replication and the main promoter for transcription9,10,11,12. Using blood samples from the RCC patients, we sequenced the D-loop, which contains a length of 1122 bps (nucleotide 16024–16569 and 1–576) and referred to mitochondria database http://www.mitomap.org. A total of 146 single nucleotide polymorphisms(SNPs) were identified in the D-loop of mtDNA in RCC patients and healthy controls. We also identified cancer outcome associated with SNPs using the Kaplan-Meier method (mitochondrial DNA, in press). The aim of this study was to assess the relationship between germline SNPs of the D-loop and age-at-onset in RCC patients.
Results
Age-at-onset distribution in RCC patients and clinical characteristics
A total of 75 patients, including 59 diagnosed with ccRCC, 13 with papillary RCC and 3 with oncocytoma RCC ,were enrolled in this study. The age-at-onset distribution of the RCC patients is listed in Table 1. The patients analyzed included: 4 patients aged < 40, 17 patients aged 40–50, 24 patients 51–60, 22 patients aged 60–70 and 8 patients aged > 70 years old. None of these patients had received any adjuvant chemotherapy or radiation therapy following RCC resection. The age-at-onset was analyzed against clinical characteristics using the Kaplan-Meier method and these were compared using the log-rank test. Gender and TNM stage were not associated with age-at-onset at statistically significant levels (Table 2).
Correlation between mtDNA genotype and survival
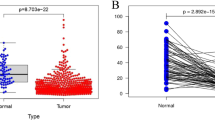
The correlation between the mtDNA genotype and survival was compared. SNPs with a minor allele frequency of <5% were excluded; thus, a remaining 14 SNPs were used for further analysis. The RCC patients were divided into two groups based on their genotype at each SNP site. For each site, the age-at-onset curve was plotted using the Kaplan-Meier method for all RCC patients. A noticeable difference in the age-at-onset appeared at nucleotide 16293, as revealed by the log-rank test (Fig. 1a). This SNP was previously identified in the mitochondrial database(www.mitomap.org). The age-at-onset for patients with the minor allele16293G genotype was significantly lower than that of patients with 16293A at the 16293 site (Fig. 1a). We randomly picked up 60 from all of the 75 RCC patients and repeated the processes of statistical analysis, the association for 16293 with age-at-onset of RCC is still exist (Fig. 1b).
Discussion
Selected SNPs in the D-loop region have been examined for their ability to predict cancer risk and outcome in many types of tumor, including esophageal squamous cell carcinoma13, hepatocellular carcinoma14, oral squamous cell carcinoma15, lung cancer16. The present study has extended previous analyses to determine the relationship between age-at-onset and germline SNPs in a continuous sequence of mtDNA between nucleotides 16190 and 583 in RCC patients. The SNP 16293A/G was identified for its association with age-at-onset at a statistically significant level by the log-rank test.
We have performed a number of studies on D-loop SNPs in cancers of the genitourinary system. Apart from being a predictor for cancer risk and outcome, SNPs in the D-loop are also a predictor for age-at-onset in RCC patients. Based on the re-sampling method17, we also verified the robustness of the predictive power for association of 16293 with age-at-onset in RCC patients. Nucleotide 16293 is located in hypervariable (HV) segment region I.Although the functional of significance of this SNP is unclear, we have found it to be associated with the risk of RCC by conducting case-control study. Minor alleles of 16293G are associated with the early onset of RCC; the age-at-onset curve decreased precipitously in patients carrying these alleles (Fig. 1). We compared the distribution frequency of this SNP between RCC patients and healthy controls. Among the 68 age-matched controls, only two carried the 16293G allele, whereas 13 RCC patients carried the G allele. A much larger sample size is therefore required to perform a comparison for cancer risk at this site.
The mtDNA D-loop is important for regulation of mitochondrial genome replication and expression. SNPs in this region might affect mtDNA replication and lead to alteration of electron transport chain, which is responsible for the release of highly reactive oxygen species (ROS) and could contribute to nuclear genome damage as well as cancer initiation and promotion18,19,20. The SNP may alter transcription of mitochondrial genome and that the production of ROS is enhanced when the mitochondrial transcription is altered21, these ROS-mediated mechanism may accelerate the tumor development.
In conclusion, SNPs in the D-loop were found to be biomarkers for RCC age-at-onset. The analysis of genetic polymorphisms in the D-loop may help to identify patient subgroup at a high risk of an early onset in RCC patients, thereby helping to refine therapeutic decisions.
Methods
Tissue specimens and DNA extraction
Blood samples were obtained from 75 RCC patients who received nephrectomy in the Department of urinary surgery at the Fourth Hospital of Hebei University between 2002 and 2007. Total DNA was extracted using a Wizard Genomic DNA extraction kit (Promega, Madison, WI, USA) and stored at −20°C. 75 RCC patients ranging in age from 33 to 79 years were classified into five 10-year groups according to age at RCC onset. The study was approved by the Human Tissue Research Committee of the Fourth Hospital of Hebei Medical University. All patients provided written informed consent for the collection of samples and subsequent analysis.
PCR amplification and sequence analysis
The forward primer 5′-CCCCATGCTTACAAGCAAGT-3′(nucleotide 16,190–16,209) and reverse primer 5′-GCTTTGAGGAGGTAAGCTAC-3′(nucleotide 602–583) were used for amplification of a 982-bp product from the mtDNA D-loop region. PCR was performed according to the protocol of PCR Master Mix Kit (Promega) and purified prior to sequencing. The PCR condition consisted of one incubation of 2 min at 95°C,followed by 35 cycles of a 30 s denaturation at 95°C, a 30 s an annealing at 55°C and a 45 s extension at 72°C and a final extension at 72°C for 5 min. Cycle sequencing was performed with the Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystem, Foster City, CA, USA) and the products were then separated on the ABIPRISM Genetic Analyzer 3100 (Applied Biosystem). Polymorphisms were confirmed by repeated analyses from both strands.
Statistical analysis
The age-at-onset curve of RCC patients was calculated using the Kaplan-Meier method at each SNP site and compared with the log-rank test. All statistical analyses were performed using the SPSS17.0 software (SPSS Company, Chicago, IL, USA). For all the statistical tests, p < 0.05 was considered to indicate a statistically significant difference.
References
Remon, J., Lianes, P. & Martinez, S. Brain metastases from renal cell carcinoma. Should we change the current standard? Cancer Treat Rev 38, 249–257 (2012).
Salehipoor, M. et al. Role of viruses in renal cell carcinoma. Saudi J Kidney Dis Transpl 23, 53–57 (2012).
Zhao, P., Dai, M., Chen, W. & Li, N. Cancer trends in China. Jpn J Clin Oncol 40, 281–285 (2010).
Staehler, M., Rohrmann, K., Haseke, N., Stief, C. G. & Siebels, M. Targeted agents for the treatment of advanced renal cell carcinoma. Curr Drug Targets 6, 835–846 (2005).
Guo, Z., Yang, H., Zhang, F., Zhang, R. & Wang, C. Single nucleotide polymorphisms in the mitochondrial displacement loop and age-at-onset of esophageal squamous cell carcinoma. Oncol Lett 3, 482–484 (2012).
Lee, H. C. et al. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann N Y Acad Sci 1042, 109–122 (2005).
DiMauro, S. & Schon, E. A. Mitochondrial DNA mutations in human disease. Am J Med Genet 106, 18–26 (2001).
Beal, M. F. Mitochondria, free radicals and neurodegeneration. Curr Opin Neurobiol 6, 661–666 (1996).
Yoneyama, H. et al. Nucleotide sequence variation is frequent in the mitochondrial DNA displacement loop region of individual human tumor cells. Mol Cancer Res 3, 14–20 (2005).
Nishikawa, M. et al. Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma. Cancer Res 61, 1843–1845 (2001).
Sanchez-Cespedes, M. et al. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res 61, 7015–7019 (2001).
Taanman, J. W. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1410, 103–123 (1999).
Zhang, R. et al. Single nucleotide polymorphisms in the mitochondrial displacement loop and outcome of esophageal squamous cell carcinoma. J Exp Clin Cancer Res 29, 155 (2010).
Zhang, R. et al. Identification of sequence polymorphism in the D-Loop region of mitochondrial DNA as a risk factor for hepatocellular carcinoma with distinct etiology. J Exp Clin Cancer Res 29, 130 (2010).
Liu, S.-A., Jiang, R.-S., Chen, F.-J., Wang, W.-Y. & Lin, J.-C. Somatic mutations in the D-loop of mitochondrial DNA in oral squamous cell carcinoma. Eur Arch Otorhinolaryngol 269, 1665–1670 (2011).
Ding, C. et al. Identification of sequence polymorphisms in the D-loop region of mitochondrial DNA as a risk factor for lung cancer. Mitochondrial DNA 23, 251–254 (2012).
Li, J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun 1, 34 (2010).
Bandy, B. & Davison, A. J. Mitochondrial mutations may increase oxidative stress: implications for carcinogenesis and aging? Free Radic Biol Med 8, 523–539 (1990).
Gille, J. J. & Joenje, H. Cell culture models for oxidative stress: superoxide and hydrogen peroxide versus normobaric hyperoxia. Mutat Res 275, 405–414 (1992).
Shigenaga, M. K., Hagen, T. M. & Ames, B. N. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci U S A 91, 10771–10778 (1994).
Dement, G. A., Maloney, S. C. & Reeves, R. Nuclear HMGA1 nonhistone chromatin proteins directly influence mitochondrial transcription, maintenance and function. Exp Cell Res 313, 77–87 (2007).
Acknowledgements
This work was supported by the project of Hebei Natural Science Fund (H2012206157).
Author information
Authors and Affiliations
Contributions
J.S.X. and Z.J.G. analyzed the data and wrote the paper; J.X.Z. and L.W.C. carried out the experiments, analyzed the data and wrote the paper; S.L.Z. carried out the experiments; Y.L.B. designed the experiments, analyzed the data and wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Xu, J., Guo, Z., Zhang, J. et al. Single nucleotide polymorphisms in the mitochondrial displacement loop and age-at onset of renal cell carcinoma. Sci Rep 3, 2408 (2013). https://doi.org/10.1038/srep02408
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02408
This article is cited by
-
Renal Cell Carcinoma in Young Patients: a Review of Recent Literature
Current Urology Reports (2015)
-
Heterogeneity of Li-Fraumeni Syndrome links to unequal gain-of-function effects of p53 mutations
Scientific Reports (2014)
-
Next-generation sequencing of the mitochondrial genome and association with IgA nephropathy in a renal transplant population
Scientific Reports (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.