Abstract
STAT6 interacts with PPARγ to elicit macrophage polarization towards an anti-inflammatory, insulin-sensitizing phenotype. Mice deficient in STAT6 display liver lipid accumulation (hepatosteatosis). Rosiglitazone (RSG), a PPARγ agonist, ameliorates hepatosteatosis and enhances insulin sensitivity. To elucidate the role of STAT6 in PPARγ action on hepatosteatosis we compared liver proteomes of RSG-treated wild type and STAT6-deficient mice and we identified pyruvate kinase M2 (PKM2), a glycolysis and proliferation-regulating enzyme that displayed STAT6-dependent expression. RSG induced PKM2 within inflammatory cells in liver but suppressed its expression in adipose tissue. RSG diminished hepatosteatosis and oxidative stress, enhanced fat accumulation and improved insulin sensitivity in STAT6-deficient mice. Our data reveal a complex interaction between STAT6 and PPARγ in the regulation of liver and adipose tissue lipid depot distribution and design STAT6 as a novel link between inflammatory cell metabolism and adipocyte and hepatocyte function.
Similar content being viewed by others
Introduction
Signal Transducer and Activator of Transcription 6 (STAT6) is the signal mediator of interleukin 4 and 13 (IL-4 and IL-13) promoting an anti-inflammatory process by inducing the development of Th2 lymphocytes and M2 type macrophages1,2,3,4,5,6,7,8. In hepatocytes, in vitro, IL-4/STAT6 diminishes triglyceride accumulation9. Accordingly, STAT6-deficient mice display aggravated high fat diet-induced hepatic liver deposition in vivo10.
STAT6 interacts with the peroxisome proliferator-activated receptor γ (PPARγ) to induce a metabolic switch from glycolysis towards fatty acid oxidation through direct interaction between STAT6 and PPARγ at the promoter regions of fatty acid β-oxidation genes6,7,11,12,13,14. This metabolic switch is tightly linked to the development of M2 type, anti-inflammatory macrophages7. Increased inflammatory state of adipose tissue is a hallmark of obesity and insulin resistance12. Macrophages activated by inflammatory cytokines via the classical (M1) pathway acquire a pro-inflammatory phenotype, release Monocyte Chemotactic Protein (MCP-1) and Interleukin 1β (IL-1β), thus enhancing adipose tissue inflammation and promoting insulin resistance12,15. By contrast, M2 type macrophage activation induced by IL-4/IL-13 or by PPARγ agonists is associated with improved insulin sensitivity in both human subjects and rodents11,16. Hepatosteatosis is one of the major metabolic complications related to obesity and insulin resistance17. Treatment with Rosiglitazone (RSG), a potent and specific PPARγ agonist ameliorated liver homeostasis in obese, insulin resistant patients and in divers mouse and rat models of hepatosteatosis18,19,20,21,22,23.
Taken together, these studies indicated an important beneficial role for PPARγ-mediated signaling in the modulation of liver lipid homeostasis. In immune cells, PPARγ-dependent modification of lipid metabolism entails an intertwined action with the STAT6 signaling network7. However, whether PPARγ-mediated modulation of hepatic lipid metabolism in vivo requires the presence of STAT6, has not yet been explored.
To gain insight into the interactions between PPARγ and STAT6 in hepatic homeostasis in vivo, we employed a comparative proteomic analysis and evaluated liver proteomes of wild type (WT) and STAT6-deficient mice after RSG treatment using an isobaric labeling strategy24. We identified pyruvate kinase isomer M2 (PKM2), a glycolytic and proliferation-regulating enzyme with attenuated RSG-induced expression in STAT6-deficient mice. In adipose tissue, RSG decreased PKM2 expression and this effect was absent in mice with STAT6 deletion. Immunohistochemistry localized PKM2 expression in inflammatory cells both in liver and adipose tissue, while hepatocytes and adipocytes were negative for PKM2 staining. RSG diminished hepatosteatosis and oxidative stress, enhanced fat accumulation and improved insulin sensitivity in STAT6-deficient mice. Taken together, our data suggest that STAT6-mediated signals play an important modulatory role in PPARγ-induced inflammatory cell metabolism and activation and that this action of STAT6 is a key component in the adjustment of PPARγ adipogenic and insulin-sensitizing effects.
Results
Rosiglitazone Normalizes Hepatic Triglyceride Content and Decreases Hepatocellular Stress in STAT6-deficient Mice
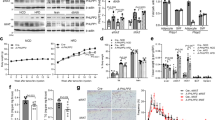
Confirming our previously published results10, quantification of hepatic triglyceride content demonstrated increased lipid accumulation in control STAT6-deficient mice compared to wild type (WT) mice. RSG treatment normalized triglyceride levels in STAT6-deficient mice but exerted no significant effect in WT mice (Figure 1a). Parallel to increased lipid accumulation livers of control STAT6-deficient mice showed elevated levels of malonyl-aldehyde (MDA), a hallmark of lipid peroxidation and oxidative stress. MDA amount was significantly decreased after RSG treatment in STAT6-deficient mice but remained unchanged in WT mice (Figure 1b). In parallel, expressions of two main anti-oxidant proteins, Glyoxilase1 (GLO1) and superoxide dismutase 1 (SOD1) displayed increased expressions in livers of control STAT6-deficient mice, but were normalized following RSG treatment (Figure 1c).
Rosiglitazone treatment normalizes liver triglyceride levels and hepatocellular stress in STAT6-deficient mice.
(a) Hepatic triglyceride content, (n = 5–10/group). (b) Thiobarbituric acid reactive substances (TBARS)/Malonaldehyde (MDA) levels in liver tissue, (n = 5–8/group). (c) Protein expressions of oxidative stress-related proteins: glyoxalase 1 (GLO1) and superoxide dismutase 1 (SOD1), (n = 3–8/group). Actin is used as loading control. All graphs represent mean ± S.E.M expressed as arbitrary units normalized to the mean of the WT control mice. * = p ≤ 0.05, ** ## = p ≤ 0.01, *** ### = p ≤ 0.001 (* WT vs. STAT6-deficient mice; #Control vs. RSG-treated mice) determined by one way analysis of variance (ANOVA) test. All runs were done in parallel with the same experimental conditions.
Analysis of Differential Protein Expression
Isobaric-labeling based quantitative proteomics using Tandem Mass Tag (TMT) technology identified differentially expressed liver proteins between RSG-treated WT and STAT6-deficient mice (four biological replicates in two different experiments)25. After liver protein extraction, tryptic digestion and off-gel electrophoresis, a total number of 1440 proteins were identified and quantified (Supplemental dataset). Among them, three were found differentially expressed in both experiments displaying lower expression levels in STAT6-deficient mice. In specific, these three proteins were the pyruvate kinase isozymes M1/M2, the hematopoietic system regulatory peptide (short isoform) (Thymosine beta 4, Tβ4) and the Histone H1.5 (chain 0) (Table 1). Further experiments were focused on PKM2 due to its dual interest as a cancer-specific metabolic enzyme and a recently identified PPARγ target gene in liver26,27,28.
Western Blot and Real-Time PCR Analyses of Pyruvate Kinase M2
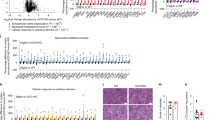
To confirm STAT6-dependent regulation of PKM2 we performed Western blot and real-time PCR experiments to analyze PKM2 protein and mRNA levels, respectively. Western blot examination demonstrated significant down-regulation of PKM2 expression in STAT6-deficient mice when data were analyzed by Student's t-test (p = 0.003) though the difference did not reach statistical significance when analyzed by one-way ANOVA test (p = 0.075). In line with these data, RSG treatment resulted in impaired up-regulation of PKM2 protein expression in STAT6-deficient mice compared to RSG-treated WT mice (Figure 2a). Similar STAT6-dependent upregulation was observed in PKM2 mRNA expression as well, though the difference between RSG-treated WT and STAT6-deficient mice did not reach statistical significance (Figure 2b).
RSG-induced PKM2 protein and mRNA up-regulation is STAT6-dependent.
(a) Western blot and (b) real-time PCR analysis of PKM2 expressions in livers of control and RSG-treated mice, (n = 5/group). Graphs represent mean ± S.E.M expressed as arbitrary units normalized to the mean of control WT mice. # = p ≤ 0.05, ** = p ≤ 0.01, ### = p ≤ 0.001 (* WT vs. STAT6-deficient mice; #Control vs. RSG-treated mice) determined by one way analysis of variance (ANOVA) test. (PKM2 protein Student's t-test p = 0.003, control WT vs. STAT6-deficient mice). All runs were done in parallel with the same experimental conditions.
Liver Histology and Immunohistological Localization of PKM2
Hematoxylin-eosin (HE) stained liver sections demonstrated the presence of several inflammatory foci of heterogenous cellular composition in RSG-treated WT mice that were absent from STAT6-deficient mice (Figure 3a). Hepatocytes of WT and STAT6-deficient mice were normal in appearance with physiological cytoplasm/nucleus ratio and with no sign of malignant transformation. Immunohistochemical analysis of liver sections from RSG-treated WT mice showed PKM2 labeling of numerous non-parenchymal cells located between hepatocytes but hepatocytes were clearly devoid of any PKM2 labeling (Figure 3a upper panels). The PKM2-labelled non-parenchymal cells were localized most abundantly in association with inflammatory foci. Some PKM2-positive cells showed a particular arrangement by surrounding healthy-appearing hepatocytes. In accordance with the diminished PKM2 expression detected by Western blot analysis, PKM2-labelled cells were substantially less abundant and scattered in RSG-treated STAT6-deficient mice than in WT mice (Figure 3a lower panels). In order to identify the nature of the non-parenchymal PKM2-positive cells in RSG-treated WT mice we performed immunohistochemistry on separate sets of three serial sections derived from RSG-treated WT mice; in each experiment the middle section was used for PKM2 immunohistochemistry while the adjacent sections on each side were subjected to immunolabeling for markers of macrophages and hepatic stellate cells, or to a standard staining procedure for fibrosis detection (Masson staining). In the first set of experiments we used two different macrophage markers, the F4/80 protein as a marker of M1-type activated macrophages and the CD206 antigen for M2-type activated macrophages, respectively29,30. Both F4/80 and CD206-positive cells could be distinctively identified, although with differential distributions, but without any substantial co-localized labeling with the PKM2-positive cells (Figure 3b, upper panels). In the second set of experiments we performed either an immunolabeling against the glial fibrillary acidic protein (GFAP), a marker of activated hepatic stellate cells (HSC), or a Masson staining to detect accumulation of fibrotic tissue31. GFAP-labeled HSC displayed typical star-like morphology and their position was also compatible with their specific location within the space of Disse between liver sinusoidal endothelial cells and hepatocytes (Figure 3b, lower panels)32. Some of these GFAP positive cells were also located within the inflammatory granulomas but without any significant colocalized labeling with the PKM2-positive cells (Figure 3b, lower panels). Upon activation, stellate cells differentiate into myofibroblasts to produce extracellular matrix, thus contributing to liver fibrosis. A recent case report described the development of granulomatosus hepatitis with fibrosus tissue accumulation in a patient treated with RSG33. In our study, however, collagen-specific Masson staining did not revealed fibrotic tissue build-up in the livers of RSG-treated WT or STAT6-deficient mice.
Immunohistochemical localization of liver PKM2 expression.
(a) Hematoxylin-eosin (HE) and PKM2 immunohistochemical stainings in livers of WT (upper panels) and STAT6-deficient (lower panels) mice. Images are representative of pictures obtained from five mice/genotype. Arrows point to PKM2 positive non-parenchymal cells in: 1) association with inflammatory foci, 2) individual localization and 3) surrounding hepatocytes. (b) Serial immunohistochemical or histological stainings from liver sections of WT RSG-treated mice. The middle section was used for PKM2 labelling and the two adjacent sections were used for markers for different non-parenchymal liver cell types. Upper row: the macrophage markers CD206 (M2 type) and F4/80 (M1 type); lower row: GFAP: Glial fibrillary acidic protein and Masson trichrome staining (collagen).
Western Blot Analysis of CD206
PPARγ induces M2-type polarization of macrophages and interaction with STAT6 is a necessary requirement for this action7,14,34. In line with the essential role of STAT6 in M2-type differentiation, RSG-treated STAT6-deficient mice displayed lower expression levels of CD206, the hallmark of M2-type macrophages (Figure 4).
Decreased alternative macrophage activation in livers of RSG-treated STAT6-deficient mice.
Western blot analysis of CD206 expression in livers of control and RSG-treated mice, (n = 5/group). Graphs represent mean ± S.E.M expressed as arbitrary units normalized to the mean of control WT mice. * # = p ≤ 0.05, (* WT vs. STAT6-deficient mice; #Control vs. RSG-treated mice) determined by one way analysis of variance (ANOVA) test. Actin is used as loading control. All runs were done in parallel with the same experimental conditions.
Enhanced Rosiglitazone Adipogenic and Insulin-sensitizing Effect in STAT6-deficient Mice
Liver steatosis and inflammation is closely related to alterations in adipose tissue function, the primary target tissue of PPARγ agonist drugs. In line with previous reports, control STAT6-deficient-mice had lower body fat mass than their WT littermates9. RSG treatment increased epididymal (WATe) and inguinal (WATi) fat pad accumulation in STAT6-deficient mice while this effect was not yet observed in WT type mice (Figure 5 a). Rosiglitazone induces adipocyte hyperplasia (cell number increase) and hypertrophy (cell size increase)35,36. In line with their lower adipose tissue mass, control STAT6-deficient mice displayed slightly but significantly smaller adipocyte size compared to control WT mice. RSG-treatment led to a shift in adipocyte distribution towards smaller sizes in both WT and STAT6-deficient mice but this effect was mitigated in the absence of STAT6 signaling. In line with the larger adipocyte size (a sign of adipocyte differentiation) RSG-treated STAT6-deficient mice demonstrated an elevation in the serum levels of leptin, a cytokine specific of mature adipocytes (Figure 5 b, c and d).
Enhanced RSG-induced adipogenesis in STAT6-deficient mice.
(a) Epididymal (WATe) and intra-abdominal (WATi) white adipose tissue weights of control and RSG-treated mice expressed as percentage of total body weight, (n = 5–8/group). (b) Representative hematoxylin-eosin stained images of WATe derived from WT and STAT6-deficient mice (upper panels) and their computer-assisted transformation (lower panels). Adipocytes are colored according to their size. (c) Histogram of adipocyte size distribution and the bar graph of adipocyte diameter of control and RSG-treated mice. Data were obtained from 6 images/mouse derived from two sections of three different levels, n = 5 mice/group. (d) Serum leptin levels measured by ELISA analysis (n = 4–5/group). Graph represents mean ± S.E.M; ** = p ≤ 0.01, *** ### = p ≤ 0.001 (* WT vs. STAT6-deficient mice; #Control vs. RSG-treated mice) determined by one-way analysis of variance (ANOVA) test.
Stimulation of IL-4 in vivo has opposing effects on STAT6 phosphorylation in liver and in adipose tissue suggesting that STAT6-mediated signals might have diverging effects in these two insulin-responsive tissues9. Indeed, in WT mice, in liver, PKM2 protein expression was up-regulated by RSG. By contrast, in adipose tissue PKM2 levels were down-regulated. However, in both tissues regulation of PKM2 expression was STAT6-dependent (Figure 6a compare to Figure 2a).
Decreased inflammatory cytokine production and increased insulin sensitivity in RSG-treated STAT6-deficient mice.
(a) Western blot of PKM2 in WAT of control and RSG-treated mice, (n = 5/group). (b) Serial immunohistochemical and histological stainings from adipose tissue sections of RSG-treated STAT6-deficient mice. The middle section was used for PKM2 labelling and the two adjacent sections were used for markers for the macrophage markers CD206 (M2 type) and F4/80 (M1 type) (c) Real-time PCR analysis of MCP1and IL-1β expression (d) Intraperitoneal insulin tolerance test (ipITT). Mice (n = 4–5/group) were starved overnight and injected intraperitonelly by insulin (1 U/Kg) and glucose levels were monitored for a period of one hour. Graph represents glucose levels expressed as the percentage of initial level. (e) AUC = area under the curve. All graphs represent mean ± S.E.M expressed as arbitrary units normalized to the mean of control WT mice. Results were analyzed by one-way ANOVA test, * # = p ≤ 0.05, (* WT vs. STAT6-deficient mice; #Control vs. RSG-treated mice). All runs were done in parallel with the same experimental conditions.
In an attempt to gain further insight into the physiological relevance of this differential regulation of PKM2, we performed a serial immunohistochemical analysis of adipose tissue sections from RSG-treated STAT6-deficient mice. Similar to liver, we observed marked PKM2 labeling of numerous cells located in the stromal-vascular fraction, situated between adipocytes (Figure 6b). Several of these PKM2 positive cells were arranged surrounding adipocytes, forming “crown-like” structures. In the same stromal space we also observed F40/80 (M1 type macrophage) and CD260 (M2 type macrophage) positive cells, but they did not display any obvious co-localization with the PKM2 positive cells.
RSG treatment alleviated liver oxidative stress in STAT6-deficient mice. Similarly, in adipose tissue, expression of one of the major inflammatory cytokines, Monocyte Chemotactic Protein 1 (MCP1), was decreased in RSG-treated STAT6-deficient mice (Student's t-test p = 0.023) though this difference did not reach statistically significant value when analyzed by one-way ANOVA test (p = 0.055). RSG up-regulated levels of another inflammatory cytokine, Interleukin 1 β (IL-1β), in WT mice, but not STAT6-deficient mice. In fact, IL-1β expression tended to be decreased in RSG-treated STAT6-deficient mice compared to WT mice (Figure 6c). Decreased WAT pro-inflammatory cytokine expression is associated with enhanced insulin sensitivity37,38. Indeed, along with the attenuated adipose tissue MCP1 and IL-1β expression, RSG treatment led to increased insulin sensitivity in STAT6-deficient mice as demonstrated by intra-peritoneal insulin tests (ipITT). By contrast, no significant effect of RSG on insulin sensitivity was yet observed in WT mice (Figure 6d). The overall effect of RSG treatment was measured by calculating the Area Under the Curve (AUC) values from the ipITT graphs in the control and RSG treated WT and STAT6-deficient mice (Figure 6e).
Discussion
The goal of our study was to explore the association between PPARγ and STAT6-mediated signal transmission in the effect of Rosiglitazone, a potent antidiabetic and hypolipidemic drug on liver lipid homeostasis. To achieve this goal we applied short-term RSG treatment in WT and STAT6-deficient mice, compared their liver protein expression by a proteomic approach and validated the relevance of the identified proteins by various molecular biological and physiological methods. Our results imply an important interaction between PPARγ and STAT6 in vivo in inflammatory cells triggering alterations in their metabolic profile and modifying adipose tissue and liver metabolic homeostasis. Indeed, RSG treatment improved hepatosteatosis and decreased hepatocellular stress present in control STAT6-deficient mice. RSG treatment led to the development of inflammatory foci in livers of WT mice while STAT6-deficient mice were protected. Our proteomic analysis revealed that these alterations were accompanied by attenuated expression of three proteins, Histone H1.5, Thymosin4β and Pyruvate kinase M2 in RSG-treated STAT6-deficient mice. Histone H1.5 represses gene transcription through chromatin remodeling39. Tβ4 is secreted by macrophages and is one of their anti-inflammatory mediators40. Similar to PKM2, Tβ4 is also a target of the hypoxia-inducible factor 1α (HIF-1α) and its expression is up-regulated in cells with malignant transformation41. Pyruvate kinase (PK) is a glycolytic enzyme catalyzing the conversion of phosphoenolpyruvate (PEP) to pyruvate. It comprises of four isoforms (PKL, PKR, PKM1 and PKM2) that differ in their cellular distribution and enzyme kinetic properties. PKM2 is physiologically abundant only during embryogenesis but shows re-expression in adulthood in cells with enhanced mitogenesis and in tumors, among them hepatocellular cancer42. PKM2 promotes cell proliferation by providing energy preferentially through anaerobic glycolysis and thus diverting glucose metabolic products towards lipid, amino acid and nucleic acid synthesis26,43. These metabolic changes give advantage to cancerous cells to survive in a predominantly hypoxic condition associated with tumor environment and adapt cell metabolism to increased DNA synthesis and proliferation44. Similar metabolic changes are also present in non-tumor cells induced to proliferate, e.g. activated T lymphocytes45.
Adenovirus-mediated PPARγ overexpression has been shown to increase PKM2 expression in rodent livers28. Our results confirmed the stimulatory effect of the PPARγ agonist RSG on PKM2 protein expression. However, immunohistochemical analysis revealed that in the presence of physiological levels of liver PPARγ RSG-induced PKM2 protein is not located in hepatocytes but in a specific subset of non-parenchymal liver cells. Recent studies demonstrated that in macrophages PPARγ and STAT6 act in a cooperative fashion to induce the "alternative" pathway of activation and differentiation into M2-type activated cells, while another transcription factor HIF-1α mediates the "classical" activation pathway giving rise to M1-type cells. M1-type macrophages use glycolysis as their primary energy source while M2-type macrophages rely predominantly on fatty acid oxidation12,15. In our study, the distribution of PKM2 positive cells did not correspond to that of either fully differentiated F4/80-positive M1 or CD206-positive M2 type macrophage populations. Taken together, these data demonstrate that RSG induces high PKM2 expression in a subset of yet unidentified non-parenchymal cells. Current knowledge on PKM2 function in the modulation of cellular metabolism and proliferation implies that these PKM2-positive cells might represent inflammatory cells with metabolic alterations supporting increased proliferation and activation46,47. This explanation is compatible with the presence of PKM2-positive cells in inflammatory granulomas and would justify the lack of co-localization with any cell markers of already fully differentiated cell type. However, the importance of PKM2 is greatly reinforced by the finding that although RSG treatment caused only a 1.5-fold increase in PKM2 expression measured in whole liver lysate, our immunohistochemical analysis revealed that PKM2-positive cells represented just a small fraction of liver mass. Thus, the increase in PKM2 expression in specific cells largely surpasses the detected 1.5-fold increase. Of significance also, that Thymosin 4β, another protein with differential expression, also plays a role in immune cell proliferation40,48 (Supplementary Figure S2). Thus, taking the data of our proteomic analysis together, they suggest that STAT6 might act as a co-transcriptional regulator of PPARγ in inflammatory cell proliferation and/or activation in liver.
RSG treatment alleviated liver steatosis with a concomitant increase in adipose tissue amount in STAT6-deficient mice suggesting a shift in lipid storage from liver towards adipose tissue. PPARγ induces white adipose tissue expansion and exerts an insulin-sensitizing effect on whole body metabolic homeostasis49,50. PPARγ-induced adipose tissue accumulation was enhanced in the absence of STAT6-mediated signaling evidenced by increased accrual of epididymal and inguinal fat pads compared to RSG-treated WT mice. RSG treatment leads to adipose tissue remodeling and enlargement by inducing hyperplasia and subsequent hypertrophy (lipid accumulation) of adipocytes35,36. In line with these previous studies RSG induced adipocyte hyperplasia in WT mice demonstrated by a significant shift towards smaller adipocyte sizes and a decrease in the levels of the differentiated adipocyte marker, leptin. RSG treatment led to a decrease in adipocyte size in STAT6-deficient mice as well, but this effect was mitigated resulting in larger adipocyte sizes compared to RSG-treated WT mice. In parallel, RSG-treated STAT6-deficient mice displayed elevated leptin levels indicating the presence of more differentiated adipocytes, thus, an acceleration of the adipocyte hyperplasia/hypertrophy sequential process. Taken together, these data imply a crucial negative effect of STAT6 on PPARγ-induced adipocyte differentiation. Increased adipocyte lipid storage capacity observed in the RSG-treated STAT6-deficient mice is also in line with the observed decrease in liver lipid content, oxidative stress and an improvement in insulin sensitivity as demonstrated by earlier studies51,52. The primary importance of RSG action on adipose tissue function is underlined by studies demonstrating that PPARγ-agonist treatment improves whole body metabolic homeostasis in liver- and muscle-specific PPARγ knock-out mice suggesting that these tissues contribute relatively little to the beneficial effects of PPARγ agonists50,53,54. An important role for inflammatory cell PPARγ in the positive effects in WAT function is underlined by the phenotype of the myeloid-specific PPARγ knock-out mice which display enhanced diet-induced insulin resistance11. Taken together, these data imply that the beneficial action of RSG treatment on hepatosteatosis and hepatic oxidative stress in STAT6-deficient mice is at least partially related to its positive effect on adipose tissue lipid storage capacity and changes in inflammatory cell metabolism associated with a decrease in WAT inflammatory cytokine secretion.
The exact physiological consequences of highly increased PKM2 expression and the related metabolic alterations in liver inflammatory cells and their relation to the observed changes in hepatic lipid homeostasis remain to be elucidated in further investigations. Of interest, however, that pathological activation of immune cells of different origins (macrophages, lymphocytes) and their crosstalk with hepatocytes are known to play important roles in the modulation of liver steatosis55. Our results demonstrate that STAT6, in cooperation with PPARγ, is an important component of this process.
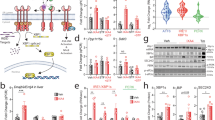
The summary of our current understanding of the role of STAT6 in the modulation of RSG action on liver and adipose tissue function and their crosstalk is depicted in Figure 7.
STAT6 modulates PPARγ action in adipose tissue and liver.
(a) Under normal conditions STAT6 cooperates with PPARγ to induce adipose tissue accumulation. In the absence of STAT6 adipose tissue mass is decreased and the excess fat is stored in the liver resulting in hepatic oxidative stress and inflammation. (b) Induction of PPARγ in the absence of STAT6 results in enhanced adipogenesis and adipose tissue lipid storage, resulting in decreased secretion of inflammatory cytokines (MCP1, IL-1β). In parallel, in liver, lipid accumulation decreases, along with diminished oxidative stress and inflammation. These changes are accompanied by attenuated immune cell PKM2 expression, indicating differences in metabolism and activation. The positive effect on hepatic homeostasis can be related to 1.) decreased adipose tissue inflammatory cytokine production, or 2.) a crosstalk between liver inflammatory cells and hepatocytes. In addition, 3.) immunmodulatory effects of adipose tissue cytokines on liver immune cells might results in STAT6-dependent upregulation of liver PKM2 expression indicating differences in activation profile. These three potential crosstalk events are indicated by dotted arrows.
In conclusion, we used a quantitative proteomic strategy to identify differences in liver protein expression induced by the PPARγ agonist drug RSG in the presence or absence of STAT6-mediated signaling. In particular, we demonstrated an attenuation of PPARγ-mediated up-regulation of PKM2 in a mixed population of non-parenchymal, inflammatory cells and explored adaptations in liver and adipose tissue metabolic homeostasis linked to differential inflammatory profile. Our data uncovered a novel aspect of the complex network between STAT6 and PPARγ in the modulation of anti-inflammatory and insulin-sensitizing actions of RSG and its potential relation to liver steatosis. Taken together, our data implicates STAT6 as a crucial signaling partner of PPARγ in vivo both in adipose tissue and liver and draws attention to its importance in the modification of the action of Rosiglitazone, a prominent anti-diabetic drug of its class.
Methods
Mice and Rosiglitazone Treatment
Wild type and STAT6 knock-out male mice with Balb/cJ genetic background were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were kept in conventional animal facility under controlled environmental conditions with free access to water and food. Rosiglitazone was administered by intraperitoneal injection with a dose of 10 mg/kg body weight for a period of 10 days. Control mice received injection of vehicle. Mice were anaesthetized and sacrificed by decapitation. Liver tissue was quickly removed, rinsed with ice-cold PBS solution, intersected and then frozen by liquid nitrogen and kept at −80°C until analysis. The Ethical Committee of the University of Geneva and the Veterinary Office of the Canton of Geneva accepted the experimental protocol.
Liver Lipid Extraction
Hepatic tissues were homogenized on ice in a mixture of Chloroform: Methanol (2:1) and lipids were left to extract in the organic phase overnight. After filtration, 0.9% NaCl (1/5 of volume) was added and the mixture was centrifuged at 1000 rpm. The lower organic lipid-containing phase was dried overnight at 4°C. Dried lipids were then resuspended in 250 μL of ethanol and stored at −20°C until triglyceride colorimetric assay was performed according to the manufacture's instructions (Roche Diagnostic).
Liver Protein Extraction
Mouse liver tissue was disrupted by freeze cracking in presence of liquid nitrogen. Then, tissues were homogenized in 20 mM Tris buffer (pH = 7.4) containing sucrose 250 mM, DTT 1 mM, EDTA 2 mM and a tablet of Complete protease inhibitor cocktail per 50 mL of buffer (Roche Diagnostic). The buffer volume was adjusted to obtain tissue amount of 50 mg/L. Protein concentration was measured using bicinchoninic acid method (BCA Protein Assay; Pierce).
Proteomic analysis by isobaric TMT labeling, off-gel electrophoresis and Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis
Liver protein extracts from four wild-type and four STAT6-deficient RSG treated mice were analyzed in two independent experiments, each comparing two wild-type and two STAT6-deficient mice. Altogether in this study we compared four biological individuals for each genotype. Details of liquid chromatography tandem mass spectrometry analysis are provided in Supplementary information. Briefly, in each experiment 100 μg proteins from each sample (two WT and two STAT6 KO) and two tubes of control mix (containing ¼ of each test sample) were separately prepared for digestion and submitted to the same procedure. This duplicate control mix allowed us to set the cut-off threshold for differential expression (the theoretical threshold ratio of all proteins in mix 1 and mix 2 should equal to one). The samples were digested overnight with 2 μg of porcine trypsin. The next day, all peptides from the four samples and the two mixes were separately labeled with six different Tandem Mass Tags (Thermo Scientific, San Jose, CA) according to the manufacturer's instructions. Next, the content of the six tubes was pooled, desalted on a Vidac C18 spin column (Harvard apparatus, Holliston, MA) and dried before off-gel electrophoresis. The 12 collected off-gel fractions were desalted and evaporated under speed-vacuum and stored at −20°C before LC-MS/MS analysis. Electrospray ionization linear ion trap quadrupole mass spectrometry (ESI LTQ-OT MS) was performed on a linear ion trap quadrupole-orbitrap (LTQ Orbitrap) Velos from Thermo Electron (San Jose, CA) equipped with a NanoAcquity system from Waters. Swiss-Prot database (2011_02 of 08-Feb-2011) specified to Mus musculus taxonomy was used for protein identification, where protein and peptide score were set up to maintain the false positive peptide ratio below 1%. For all analyses, only proteins matching two different peptide sequences were kept. Protein and peptide quantification was performed following the procedure described by Dayon et al.25. The protein ratio (Wild-type/STAT6 Knock-out) was calculated as the geometric mean of its corresponding peptide ratios. For each protein ratio, the geometric standard deviation (SD) was calculated as described by Tan et al.56 and the coefficient of variation (CV) determined as the SD divided by the protein ratio and multiplied by 100. To determine the proteins differentially expressed between the two stages (WT and STAT6-deficient), only proteins found to be above calculated thresholds in the experiment 1 (> 1.35) and experiment 2 (>1.53) were considered as ‘true’ differentially expressed proteins between two genotypes (Supplemental information MS).
Western blotting
Liver tissues were homogenized in lysis buffer (25 mM HEPES, 0.5% Triton X100, 65 mM NaCl, 2.5 mM EDTA, 25 mM sodium pyrophosphate, 50 mM NaF, 2 mM PMSF, 9 mM sodium orthovanadate, one tablet of Complete Inhibitor Cocktail in 20 ml (Roche Diagnostics), pH 7.5). Proteins were separated by SDS-PAGE, transferred onto nitrocellulose membranes and identified by immunoblotting using enhanced chemiluminescence. Signals were revealed in ChemiDoc machine and quantified by QuantityOne program (BioRad).
Immunohistochemistry
Tissue samples were fixed in 4% paraformaldehyde, embedded in paraffin and 4 μm thick liver serial sections were analyzed by immunohistochemistry using anti-PKM2, anti-CD206, anti-F4/80 and Glial fibrillary acidic protein (GFAP) antibodies with the Ventana Discovery automated staining system (Ventana Medical Systems, Tucson, AZ). Ventana reagents were used for the entire procedure. For both PKM2 and CD206 antigen retrieval, slides were heated with cell conditioning solution for 36 min (CC1; Tris-based buffer pH 8.4) using a standard protocol, whereas F4/80 and GFAP antigenicity was retrieved following P1 digestion during 8 min at 37°C. Primary antibodies were used at the dilution 1/100, 1/2500 and 1/50 in Dako diluent (S2022) for PKM2 (#3198, Cell-Signaling Technology), CD206 (#ab64693, Abcam), F4/80 (#ab6640, Abcam) and GFAP (Dako, Z0334), respectively and incubated 1 h at 37°C. Detection of primary antibodies was carried out using the anti-rabbit or anti-rat OmniMap-DAB kit (Ventana Medical Systems), based on conversion of diaminobenzidine to a dye with multimeric horseradish peroxidase (HRP).
Histology and Morphometric Measurements of Adipose Tissue
Epididymal adipose tissue samples were fixed in 4% paraformaldehyde, embedded in paraffin and serial sections were subjected to standard hematoxylin-eosin (HE) staining. Morphometric measurements were conducted on duplicate sections derived from three different levels of the tissue (6 images/mouse). Images were obtained by Axiocam Imaging System (Carl Zeiss AG, Feldbach, Switzerland). Cell diameter was calculated from computer-assisted image transformation using MetaMorph Image Analysis software (Molecular Devices, Union City, CA) as described previously57.
RNA Preparation and Quantitative Real-time RT-PCR
Total RNA was prepared by homogenizing approximately 100–200 mg liver tissue in TRIZOL Reagent (Invitrogen) and was purified by using RNAse free DNAse in combination with the RNeasy Mini Kit (Qiagen). cDNA was synthesized from 2 μg of DNA-free RNA by Superscript II Reverse Transcriptase (Invitrogen). Primers were designed by Primer Express software (Applied Biosystems) and are listed in the supplementary Table (Supplementary information). The results were quantified by the ΔΔCt method using cyclophillin A as the standard internal non-variable gene to compensate for differences in RNA input and efficiency of cDNA synthesis. Results were expressed as arbitrary units compared to the average expression levels in WT mice and are represented as mean ± S.E.M.
Determination of liver oxidative stress
Oxidative stress was assessed by measuring tissue concentrations of malonyl-aldehide (MDA) as a thiobarbituric acid reactive substance (TBARS) using a commercially available kit (ZeptoMetrix, Buffalo, NY).
Intraperitoneal Insulin Tolerance Test (ipITT)
Mice were fasted overnight and were administered intraperioneally with 1 U/kg insulin. Glucose levels were monitored during a period of one hour using a glucometer (Accu-check, Roche Diagnostics GmbH, Mannheim, Germany).
References
Hebenstreit, D., Wirnsberger, G., Horejs-Hoeck, J. & Duschl, A. Signaling mechanisms, interaction partners and target genes of STAT6. Cytokine Growth Factor Rev 17, 173–188 (2006).
Takeda, K., Kishimoto, T. & Akira, S. STAT6: its role in interleukin 4-mediated biological functions. J Mol Med 75, 317–326 (1997).
Kaplan, M. H., Schindler, U., Smiley, S. T. & Grusby, M. J. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 4, 313–319 (1996).
Tomkinson, A. et al. The failure of STAT6-deficient mice to develop airway eosinophilia and airway hyperresponsiveness is overcome by interleukin-5. Am J Respir Crit Care Med 160, 1283–1291 (1999).
Goenka, S. & Kaplan, M. H. Transcriptional regulation by STAT6. Immunol Res (2011).
Odegaard, J. I. & Chawla, A. Mechanisms of macrophage activation in obesity-induced insulin resistance. Nat Clin Pract Endocrinol Metab 4, 619–626 (2008).
Szanto, A. et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity 33, 699–712 (2010).
Mishra, B. B., Gundra, U. M. & Teale, J. M. STAT6(-/-) mice exhibit decreased cells with alternatively activated macrophage phenotypes and enhanced disease severity in murine neurocysticercosis. J Neuroimmunol 232, 26–34 (2011).
Ricardo-Gonzalez, R. R. et al. IL-4/STAT6 immune axis regulates peripheral nutrient metabolism and insulin sensitivity. Proc Natl Acad Sci U S A 107, 22617–22622 (2010).
Iff, J. et al. Differential proteomic analysis of STAT6 knockout mice reveals new regulatory function in liver lipid homeostasis. J Proteome Res 8, 4511–4524 (2009).
Odegaard, J. I. et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 447, 1116–1120 (2007).
Olefsky, J. M. & Glass, C. K. Macrophages, inflammation and insulin resistance. Annu Rev Physiol 72, 219–246 (2010).
Bensinger, S. J. & Tontonoz, P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature 454, 470–47 (2008).
Chawla, A. Control of macrophage activation and function by PPARs. Circ Res 106, 1559–1569 (2010).
Odegaard, J. I. & Chawla, A. Alternative macrophage activation and metabolism. Annu Rev Pathol 6, 275–297 (2011).
Hotamisligil, G. S. & Spiegelman, B. M. Tumor necrosis factor alpha: a key component of the obesity-diabetes link. Diabetes 43, 1271–1278 (1994).
Yeh, M. M. & Brunt, E. M. Pathology of nonalcoholic fatty liver disease. American journal of clinical pathology 128, 837–84 (2007).
Mayerson, A. B. et al. The effects of rosiglitazone on insulin sensitivity, lipolysis and hepatic and skeletal muscle triglyceride content in patients with type 2 diabetes. Diabetes 51, 797–802 (2002).
Juurinen, L., Kotronen, A., Graner, M. & Yki-Jarvinen, H. Rosiglitazone reduces liver fat and insulin requirements and improves hepatic insulin sensitivity and glycemic control in patients with type 2 diabetes requiring high insulin doses. The Journal of clinical endocrinology and metabolism 93, 118–124 (2008).
Neuschwander-Tetri, B. A., Brunt, E. M., Wehmeier, K. R., Oliver, D. & Bacon, B. R. Improved nonalcoholic steatohepatitis after 48 weeks of treatment with the PPAR-gamma ligand rosiglitazone. Hepatology 38, 1008–101 (2003).
Sanchez, J. C. et al. Effect of rosiglitazone on the differential expression of obesity and insulin resistance associated proteins in lep/lep mice. Proteomics 3, 1500–1520 (2003).
Ackerman, Z. et al. Hepatic effects of rosiglitazone in rats with the metabolic syndrome. Basic Clin Pharmacol Toxicol 107, 663–668 (2010).
Hockings, P. D. et al. Rapid reversal of hepatic steatosis and reduction of muscle triglyceride, by rosiglitazone: MRI/S studies in Zucker fatty rats. Diabetes Obes Metab 5, 234–243 (2003).
Thompson, A. et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75, 1895–1904 (2003).
Dayon, L. et al. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem 80, 2921–2931 (2008).
Mazurek, S. Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol, (2010).
Mazurek, S. Pyruvate kinase type M2: a key regulator within the tumour metabolome and a tool for metabolic profiling of tumours. Ernst Schering Found Symp Proc, 99–124 (2007).
Panasyuk, G. et al. PPARgamma contributes to PKM2 and HK2 expression in fatty liver. Nat Commun 3, 672 (2012).
Fujisaka, S. et al. Regulatory mechanisms for adipose tissue M1 and M2 macrophages in diet-induced obese mice. Diabetes 58, 2574–2582 (2009).
Nishimura, S. et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med 15, 914–920 (2009).
Baratta, J. L. et al. Cellular organization of normal mouse liver: a histological, quantitative immunocytochemical and fine structural analysis. Histochem Cell Biol 131, 713–726 (2009).
Friedman, S. L. Hepatic stellate cells: protean, multifunctional and enigmatic cells of the liver. Physiol Rev 88, 125–172 (2008).
Dhawan, M. et al. Rosiglitazone-induced granulomatous hepatitis. J Clin Gastroenterol 34, 582–584 (2002).
Vats, D. et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell metabolism 4, 13–24 (2006).
Johnson, J. A., Trasino, S. E., Ferrante, A. W., Jr & Vasselli, J. R. Prolonged decrease of adipocyte size after rosiglitazone treatment in high- and low-fat-fed rats. Obesity (Silver Spring) 15, 2653–2663 (2007).
MacKellar, J., Cushman, S. W. & Periwal, V. Differential effects of thiazolidinediones on adipocyte growth and recruitment in Zucker fatty rats. PloS one 4, e8196 (2009).
Shoelson, S. E., Lee, J. & Goldfine, A. B. Inflammation and insulin resistance. The Journal of clinical investigation 116, 1793–1801 (2006).
Foryst-Ludwig, A. et al. PPARgamma activation attenuates T-lymphocyte-dependent inflammation of adipose tissue and development of insulin resistance in obese mice. Cardiovascular diabetology 9, 64 (2010).
Murphy, K. M. & Reiner, S. L. The lineage decisions of helper T cells. Nature reviews. Immunology 2, 933–944 (2002).
Nemolato, S. et al. Different thymosin Beta 4 immunoreactivity in foetal and adult gastrointestinal tract. PloS one 5, e9111 (2010).
Yoon, S. Y. et al. Thymosin beta4 expression correlates with lymph node metastasis through hypoxia inducible factor-alpha induction in breast cancer. Oncol Rep 25, 23–31 (2011).
Mazurek, S., Boschek, C. B., Hugo, F. & Eigenbrodt, E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol 15, 300–308 (2005).
Cairns, R. A., Harris, I. S. & Mak, T. W. Regulation of cancer cell metabolism. Nat Rev Cancer 11, 85–95 (2011).
Pelicano, H., Martin, D. S., Xu, R. H. & Huang, P. Glycolysis inhibition for anticancer treatment. Oncogene 25, 4633–4646 (2006).
Munoz-Pinedo, C., El Mjiyad, N. & Ricci, J. E. Cancer metabolism: current perspectives and future directions. Cell Death Dis 3, e248 (2012).
Wang, R. & Green, D. R. The immune diet: meeting the metabolic demands of lymphocyte activation. F1000 biology reports 4, 9 (2012).
Shapiro, H., Lutaty, A. & Ariel, A. Macrophages, meta-inflammation and immuno-metabolism. TheScientificWorldJournal 11, 2509–2529 (2011).
Kannan, L., Rath, N. C., Liyanage, R. & Lay, J. O., Jr Effect of toll-like receptor activation on thymosin beta-4 production by chicken macrophages. Mol Cell Biochem 344, 55–63 (2010).
Sharma, A. M. & Staels, B. Review: Peroxisome proliferator-activated receptor gamma and adipose tissue--understanding obesity-related changes in regulation of lipid and glucose metabolism. The Journal of clinical endocrinology and metabolism 92, 386–395 (2007).
Kintscher, U. & Law, R. E. PPARgamma-mediated insulin sensitization: the importance of fat versus muscle. American journal of physiology. Endocrinology and metabolism 288, E287–291 (2005).
Bays, H., Mandarino, L. & DeFronzo, R. A. Role of the adipocyte, free fatty acids and ectopic fat in pathogenesis of type 2 diabetes mellitus: peroxisomal proliferator-activated receptor agonists provide a rational therapeutic approach. The Journal of clinical endocrinology and metabolism 89, 463–478 (2004).
Yki-Jarvinen, H. Thiazolidinediones. The New England journal of medicine 351, 1106–1118 (2004).
Gavrilova, O. et al. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance and regulation of body fat mass. J Biol Chem 278, 34268–34276 (2003).
Norris, A. W. et al. Muscle-specific PPARgamma-deficient mice develop increased adiposity and insulin resistance but respond to thiazolidinediones. The Journal of clinical investigation 112, 608–618 (2003).
Zhan, Y. T. & An, W. Roles of liver innate immune cells in nonalcoholic fatty liver disease. World journal of gastroenterology : WJG 16, 4652–4660 (2010).
Tan, H. T. et al. Quantitative and temporal proteome analysis of butyrate-treated colorectal cancer cells. Mol Cell Proteomics 7, 1174–1185 (2008).
Li, Y. et al. Deficiency in the NADPH oxidase 4 predisposes towards diet-induced obesity. Int J Obes (Lond), (2012).
Acknowledgements
We thank Prof. D. Hochstrasser (Geneva University and Geneva University Hospitals) for helpful discussions and critical reading of the manuscript and Nathalie Lin-Marq, Marie Ebrahim-Malek and Sarah Mouche (University of Geneva) for excellent technical help. This work was supported by COST in the frame of COST Action BM0602 (grant No C07.0080) to I.S.
Author information
Authors and Affiliations
Contributions
T.S., A.H. and A.S. performed proteomic analysis and analyzed the results and wrote part of the manuscript; A.W. supervised and analyzed immunohistological stainings, wrote part of the manuscript; F.N. analyzed immunohistological stainings, wrote part of the manuscript; J.C.S. planned proteomic experiments, supervised proteomic data analysis and wrote part of the manuscript; I.S. designed and directed the experiment, wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Supplementary Information
Supplementary dataset 1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Sajic, T., Hainard, A., Scherl, A. et al. STAT6 promotes bi-directional modulation of PKM2 in liver and adipose inflammatory cells in Rosiglitazone-treated mice. Sci Rep 3, 2350 (2013). https://doi.org/10.1038/srep02350
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02350
This article is cited by
-
STAT proteins in cancer: orchestration of metabolism
Nature Reviews Cancer (2023)
-
STAT6 contributes to renal fibrosis by modulating PPARα-mediated tubular fatty acid oxidation
Cell Death & Disease (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.