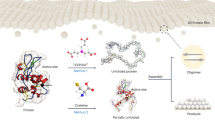
Abstract
Unlike physical patterning of materials at nanometer scale, manipulating soft matter such as biomolecules into patterns is still in its infancy. Self-assembled monolayer (SAM) with surface density gradient has the capability to drive biomolecules in specific directions to create hierarchical and discrete structures. Here, we report on a two-step process of self-assembly of the human serum albumin (HSA) protein into discrete ring structures based on density gradient of SAM. The methodology involves first creating a 2-dimensional (2D) polyethylene glycol (PEG) islands with responsive carboxyl functionalities. Incubation of proteins on such pre-patterned surfaces results in direct self-assembly of protein molecules around PEG islands. Immobilization and adsorption of protein on such structures over time evolve into the self-assembled patterns.
Similar content being viewed by others
Introduction
Self-assembly of soft-materials with functional groups into well-defined and discrete structures as a new form of nanofabrication has witnessed an unprecedented growth in the past decade1,2,3,4,5,6. Directed self-assembly of functional molecular groups into 2D and 3D structures to produce viable nanoscale devices and sensors is an attractive goal with many potential applications. Various techniques have been used to achieve micro- or nano- patterned 2D or 3D surfaces such as soft-lithography, nano-sphere lithography, photolithography, laser ablation, template-assisted patterning, dip-pen lithography and micro-contact printing7,8,9,10. Most of the current approaches focus on the initial modification of the structure and the chemistry of the surface to create patterns9,10,11. These techniques are typically used to produce sharp and distinct demarcations or boundaries of chemical and/or physical regions on a substrate. Recent advances in creating surface structures with material-gradient and spatio-temporal properties offer new opportunities in this field since it enables the fabrication of geometric structures with varying chemical and physical properties, which otherwise would be difficult to manufacture12.
Gradient assemblies may facilitate the accelerated screening of physio-chemical phenomena and are capable of creating complex geometries with directional soft-material transport13. For example, polymers and oligomers are materials with a functional gradient, allowing one to tune the geometry and functional character of the assembly. In the bottom-up approach, building blocks can be deposited gradually using a simple and cost-effective molecular self-assembly approach on the parent substrate. Position dependent continuous or discontinuous gradient patterns can be generated in a length scale using self-assembly. These patterns may lead to various directionalities of molecular distribution in radial, orthogonal or linear directions3,12,13,14. Molecular self-assembly is an attractive technique because of its simplicity and the potential efficiency for designing chemical “blueprints” of varied shapes and sizes with functionalities caused by the spontaneous assembly of molecules.
Recent advances in self-assembly have demonstrated the use of block-copolymers and nanoparticles in ordered arrays for different applications15,16,17,18. However, gradient self-assembly of biomolecules in ordered arrangements has not been widely explored. Tailoring the structure, as well as the physical and chemical properties of these patterns, paves the way towards a plethora of practical applications. A system containing molecules can generate gradient self-assembly if the constituents or building blocks can equilibrate to organize into stable dense arrays of nanoscale structures19,20. The chemical gradients of the self-assembly can be varied by tuning the functionalities of the building blocks and controlling the duration and kinetics of the self-assembly process to manipulate the spatio-temporal properties of the resultant patterns12,13,14,15,16,17,18,19,20. Bio-interfaces are essential for various bio-technological applications such as controlling the design of advanced bio-functional surfaces21,22,23,24,25. Regulating biomolecular immobilization is essential in the fabrication of biochips for biosensing or the engineering of surfaces for cell adhesion26,27,28,29,30. Gradient bio-molecular patterns with characteristic functionalities can produce decorated “smart surfaces” whose properties may change with variations in external physical and chemical stimuli such as pH, temperature, light, optical molecular switches, target analyte, charged molecules, electric field, etc.12,13,14.
In this report, we describe a two-step strategy for designing and fabricating proteins into nanometer-scale structures. Using human serum albumin (HSA), we demonstrate a process for producing complex patterns by using gradual variation in chemistry of the surface. Polyethylene glycol (PEG) was used to generate a 2D-surface density gradient using bottom-up approach of self-assembly. Thin film gold surfaces grown on silicon were used as the parent substrate due to their significance in sensor biochips31. Surface ligand chemistry was employed to design 3D protein nano- and micro-patterns on a 2D self-assembled PEG surface. The variation in density of self-assembled PEG on the substrate is considered as the main driving factor for self-assembly of HSA into patterns. The pattern generation was dependent upon factors related to protein-protein and protein-ligand interactions. Previous reports have demonstrated the use of PEG polymers to produce protein nano-arrays by photo-modifying PEG32,33. In contrast, our strategy simply relies on controlling the self-assembly of the PEG and the incubation time of protein conjugation to create complex patterns of biomolecules.
Results
Growth of a PEG self-assembled monolayer (SAM)
A conventional self-assembly protocol was employed to achieve the gradient self-assembly of carboxylipoamide poly-ethylene glycol (CL-PEG12) on the gold substrate (see Methods). Atomic force microscopy (AFM) was used to characterize the self-assembly. The SAM formation was accompanied by PEG-island growth as observed in the AFM images (Supplementary Fig. S1). The PEG molecular conformation in a solution is affected by various factors such as the structure of PEG and the number of ethylene glycol (EG) units, inter- and intra- molecular hydrogen bonding, the polarity of solvent used and temperature34,35. The most viable structure of PEG is a gauche form because of the freely rotating EG unit, consisting of two methylene and one oxygen moiety34. During the growth of SAM on the substrate, PEG molecules are arranged parallel to the parent substrate. It is most likely that during the island growth period, the orientation of the PEG chains in the islands is towards the surface normal. Therefore, most of the surface is covered by PEG chains lying parallel to the surface and the areas with islands have the chains oriented in a crystalline phase towards the surface normal34. Distribution of discrete 2D PEG-islands on the parent substrate evolved over time creating a polymer surface density gradient. Growth of PEG on the parent substrate is considered as a “gradient self-assembly” because the density of PEG molecules at islands is higher than locations without islands. We have used the term “gradient” to address the chemical and/or physical property of PEG that changes with distance in respect to high and low density of molecules on the surface. The growth of PEG-islands was studied at various time points as presented in Fig. 1a–b. Generally, a constant height distribution of 18 ± 5 nm was observed among the PEG islands between incubation times of 30 min and 5 hours (Fig. 1a). This height is approximately three times the expected height of a single PEG molecule, indicating the possibility of multi-layer arrangement within the islands. Histograms for the width distribution of the islands show an increase in the range of width of the islands with increase in the duration of self-assembly time. The average width of the islands at 30 min was approximately 300 nm while it was 3 microns for 5 hour self-assembly. Increase in the width of the PEG islands during self-assembly may be due to surface diffusion phenomenon and is consistent with the diffusion limited aggregation (DLA) model35,36,37.
Analysis of topography of self- assembled monolayer formed using CL-PEG12.
(a) Variation in height of PEG islands as a function of number of events with variation in time of self-assembly. (b) Width histograms of PEG islands as a function of number percentage of islands at various time periods of self-assembly. (c) Height histogram of PEG islands as a function of number percentage of islands after 4 h (Inset: 3-D image showing islands of PEG formed at scale of 20 × 20 μm2). (d) Width histogram of PEG islands as a function of number of islands after 4 h (Inset: Topography of PEG islands after 4 h at a scale of 5 μm).
The PEG size distribution at 4 hours of self-assembly was chosen as the pre-patterned parent substrate for the protein incubation experiments. This specific growth time was selected because of the wide range of width distribution of the PEG islands (Fig. 1c–d) available for protein structure formation. Histograms for the height and the width distribution of PEG islands after 4 hours show a height distribution of 18 ± 5 nm (approximately 80%) and a width distribution of 1 ± 0.5 μm (approximately 70%).
Self-assembly of protein ring patterns
The second step in directed self-assembly involved a bio-conjugation approach to form protein ring patterns around PEG-islands (see Methods). The HSA was immobilized on the CL-PEG12 surface and allowed to incubate for 15 hours in order to generate the ring patterns. The size and structure of the ring pattern depends on the length of incubation. Moreover, as described in previous section, high density of PEG molecules at islands created more binding sites for protein molecules in comparison to the locations without islands. Hence “density gradient” of PEG on the surface is the driving force for protein self-assembly. Self-assembly of the ring pattern is illustrated by a schematic of protein binding on the gold substrate (Fig. 2a). Fig. 2b–e show the AFM and scanning electron microscopy (SEM) images of the self-assembled protein ring patterns on CL-PEG12 coated gold substrate. There are distinguishable components present in the self-assembled protein ring patterns (Fig. 2b). We will address these components here as a “nucleus” in the center and concentric rings of “beads” (protein aggregates) around the nucleus. These protein ring patterns are shown at different length scales in SEM and AFM images (Fig. 2c–e). Distinct protein ring patterns were observed on the overall surface of the substrate as seen in the SEM images (Fig. 2c). From the SEM images, it was clear that the ring patterns showed signs of degeneration after 3days of formation. Auger elemental analysis of the ring pattern showed the presence of C, O, N and S consistent with that of organic matter (Supplementary Fig. S2). Several sizes of ring patterns between 500 nm and 5.5 μm were observed using AFM images (Fig. 2d). An AFM image of a self-assembled ring pattern of 5.5 μm size is shown in Fig. 2e.
Images of distinct HSA ring patterns on the CL-PEG12 functionalized surface.
(a) Sketch showing HSA ring assembly on the CL-PEG12 coated substrate. (b) 3D-AFM image of HSA self-assembled protein ring pattern. (c) SEM image showing assembly of protein ring patterns at a scale bar of 10 μm. (d) AFM height (topography) image overlaid on AFM amplitude image showing a pattern with assembly at a scale of 5 μm. (e) AFM image (height overlaid on amplitude) showing a pattern with assembly of two chains of beads around the nucleus at a scale of 2 μm.
The size and shape of the self-assembled protein ring patterns were dependent on the size of the nucleus. From the AFM analysis, it was determined that the size of the nucleus corresponds to the size of the PEG islands observed in the pre-patterned parent substrate prior to immobilization of HSA. This implies that the PEG surface density gradient helped in systematic adsorption of HSA on the substrate. Fig. 3a–f show the size of PEG islands and the corresponding self-assembled protein ring patterns with a similar size of nucleus. The PEG islands with sizes of 0.7, 1 and 2 μm had corresponding approximate nucleus sizes of 0.7, 1 and 2 μm. The width of the nuclei after ring pattern formation varied from 300 nm to 3.6 μm and the complete self-assembled protein ring patterns had size distributions between 800 nm and 6 μm (Fig. 4a). As shown in Fig. 4b the size of self-assembled protein ring pattern grows linearly with the size of the nucleus. However, the linear growth of the ring size is restricted after the nucleus size reaches 1.5 μm and the limiting size of the pattern was about 5 ± 0.5 μm. It was observed that only a single ring of beads was formed around the nucleus when the size of the nucleus was between 500 nm and 2 μm. Two or more concentric rings were observed only when the nucleus size was above 2 μm. The growth in the width and the height of the beads seen in 3D was relative to the size of the nucleus (Supplementary Fig. S3). Growth of protein aggregates into beads was observed which might be convergent in direction, which means outer ring of beads may form initially followed by inner concentric rings.
It is interesting to note that no pattern developed under similar experimental conditions when an analogous ligand without any PEG chain was used (dihydrolipoic acid (DHLA) Supplementary Fig. S4). This reveals that the homogeneous surface grafting with responsive density gradient is required for bio-molecular pattern generation. These results also indicate that protein-protein interactions and protein-surface ligand interactions play a major role in self-assembled protein pattern generation.
Mechanism and growth analysis of the self-assembled protein ring patterns
In these experiments, the activation of PEG was accomplished using a 1-ethyl-3-(3-dimethyl aminopropyl)carbodiimide (EDC) and N-Hydroxysulfosuccinimide (Sulfo-NHS) coupling reaction. Carboxylate groups were activated and the conjugation of HSA and formation of amide bonds was accomplished through Arg or Lys side chains (HSA contains 58 Lys and 23 Arg residues) as an initial step. Incubation time was an important factor in the formation of ring patterns. The schematic of the bio-functionalization and self-assembly of HSA ring patterns is shown in Fig. 5a. Based on incubation time, the growth analysis of self-assembled protein ring patterns is presented in Fig. 5b. The conjugation of protein to the reactive complex of CL-PEG12 is a relatively slow and may take up to 2 hours. The growth of the self-assembled protein structure evolves over a period of time in a slow process as the height of structures increases from 20 to 240 nm over a period of 15 hours (Supplementary Fig. S5). No patterns were observed before 8 hours of incubation time. Therefore, ring pattern growth may consist of four major steps: conjugation, adsorption, condensation and growth. These steps revolve around the spatial distribution of PEG islands, the grafting density of polymer, the concentration gradients of protein and polymer and gradient dimensionality.
HSA has a globular structure that consists of 585 residues, composing three main domains loosely joined together by physical forces and six subdomains enfolded by disulfide bonds. Seventeen disulfide bridges and one free SH group in a single protein molecule may facilitate dimerization or a higher level of aggregation38. Under physiological conditions, native HSA lack any properties which suggest aggregation. In our work, due to perturbation of the environment of the protein on the substrate, aggregation may be observed. Generally, in proteins or high molecular weight biomolecules, the Nernst diffusion layer (approximately 10 μm)39 surrounding the biomolecule gets depleted due to high rate of binding at the surface40. Uniform aggregation of beads may be caused by the local high concentration of ions around the protein causing the shrinkage of the protein Nernst layer as well as the salting out effect, which may allow the HSA molecules to aggregate40.
The initial step is the conjugation of proteins on the surface of carboxyl terminated PEG moieties over a period of 30 min to 2 hours. Surface-anchored PEG island gradients with responsive carboxyl functionalities oriented towards the surface normal facilitate the binding of HSA in large numbers creating a 3D chemical gradient. After conjugation on the surface the overall charge on the protein may change, generating a new surface charge gradient. Protein molecules have a net charge (Znet), which is the sum of the charge without (Zown) and with (Zbound) ions bound on them. The isoelectric point of HSA is 5.4 and the net charge on HSA at pH 7.4 is negative41. The binding of protein molecules to the ligand on the substrate may affect the surface charge density on the protein. Therefore, a surface charge gradient introduced on the substrate enhances electrostatic interactions which may be dominated by salt bridge formation and hydrogen bonding42. Physiochemical properties, such as the pH of the solution and the interface propagates the charge density gradient thereby draining the protein molecules from the bulk solution and adsorption on the conjugated substrate. The higher the density of bound-protein molecules on a pre-patterned parent substrate, the greater the protein adsorption from the solution which initiates self-assembly. Circular dichroism investigations in solutions conducted over a period of 15 hours show no change in the protein conformation (Supplementary Fig. S6). In the case of no large conformational changes being observed in the protein solution, the association rate is less than or equal to the diffusion limited rate (k ≤ kdiff)42,43.

where ka is rate of association of transition state, kao is the basal rate in the absence of electrostatic interactions, kB is Boltzmann's constant, ΔG++ is Gibb's free energy change for transition state and T is temperature. For free energy minimization, the linear arrangement tries to bend into a circular shape depending on the potential gradient44. The protein molecules carry some charge and the charged entities or aggregates on the substrate group together depending on the repulsive and attractive forces around the nucleus. Depending on the charge, the coupling may break creating a potential gradient and the charge per unit area may determine the pattern formation44. It is a possibility that the entropy change and electronic charge on the protein molecules are the guiding forces for self-assembly of HSA.
It has also been observed in the case of radial mesoporous silica growth, that the surface tension constant (Δ) is proportional to the radius of the crystal45. Therefore, in order to maintain constant free energy, the crystal acquires a smooth round shape. This shape corresponds to an angle (ψ) defined as the angle at the point of the director-field near the ray to be at ψo = π/2 due to increase in Δ during the growth mechanism45. In general however, contraction geometry in protein self-assembly may be condensation induced and an additional factor of charge and thickness of the electrical double layer around the nucleus may affect the distance between the concentric rings. Consequently, self-assembly begins at the nano-scale level to create micro-scale ring patterns. The dynamic gradient in the physiochemical properties such as charge, concentration, surface energy, surface potential and surface tension, which evolve over time, is the driving force for generation of complex HSA geometric patterns.
Spectroscopic analysis of the binding energy and conformation of protein ring patterns
The entropy constraint of the reaction pathway, which is essentially leading towards the formation of patterns, is linked to the binding energy of the reaction. The anchoring of the protein on the support has an effect on the hydration of the protein molecule. Progressive attachment of the protein on the support can change the conformation. By remaining in its native state, the protein attachment on the surface minimizes the entropy gain46.
X-ray photoelectron spectroscopy (XPS) analyses were performed on the surface to examine CL-PEG12 and HSA modified self-assembled protein ring pattern surfaces. The survey spectra showed main peaks corresponding to elemental species C, N, O and S. The peak intensity for all elements in the case of PEG modified substrates was very low compared to the peaks observed on the ring pattern substrate (Supplementary Fig. S7). A characteristic peak for the C 1s core was observed at 284 eV assigned to C-C and C-H aromatic and aliphatic bonds in the protein (Fig. 6a). A prominent component at 285.3 eV represented the corresponding C-O environment in the protein and was faintly observed in PEG modified substrate. A strong component at a high binding energy of 287.2 eV was evident due to the C = O backbone of the protein molecules present on the substrate.
Spectroscopic characterization of HSA Protein self-assembly on gold substrate.
(a) High resolution XPS peak fitted C1s region for self-assembled protein ring patterns surface composition is indicated in the integrated areas. Fitting curves are shown in blue and pink colors (b) Variable angle FTIR spectroscopy of human serum albumin ring patterns and PEG modified on a gold substrate.
The variable angle Fourier Transform Infrared (FTIR) spectroscopy shows a distinct protein conjugation on the PEG-modified surface (Fig. 6b) after incubation. After 15 hours, the peak position at 1662 cm−1 suggests there is a shift in position for α-helix to β-turns or random coil in protein molecules47. A small but distinct peak at 1624 cm−1 shows the aggregated state of protein or β-sheet formation. After 2 hours, a broad peak was observed between 1700 and 1500 cm−1. This demonstrates the appearance of an amide I band at 1657 cm−1 on the PEG modified surface which represents a major part of the protein as an α-helix structure. Moreover, the intensity of peaks is low after 2 hours of incubation indicating a low protein density on the surface. Characteristic bands for pure HSA show a typical α-helical structure with peak positions at 1644 and 1533 cm−1 (Supplementary Fig. S8). It can be inferred from this that protein conjugation may lead to aggregated structures over a longer period of incubation time.
Discussion
Our work has explored the interesting field of chemical gradient variations for the fabrication of complex “smart biomolecular” patterns. We have demonstrated the switching of a 2D array of discrete islands of PEG on a substrate into discrete 3D protein patterns in a two-step process. The utilization of physiochemical gradients to generate HSA self-assembled ring formations has also been demonstrated. The molecular density gradient of PEG islands might have provided directionality for the binding and symmetrical adsorption of protein molecules. The biomolecules have arranged themselves in a symmetric and discrete radial pattern spreading out from the nucleus instead of in a continuous arrangement observed with conventional diffusion. Three major factors were responsible for controlling and generating the spontaneous self-assembled ring patterns. The first was the gradual variation of responsive PEG island width in length scale; the second was the variation of physiochemical gradients in space; and the third was the incubation time. Not much has been reported regarding the creation of spatiotemporal biomolecular geometries and much still needs to be accomplished in this direction. Similar approaches can be employed to create distinct geometric patterns by using different protein molecules or varying the constituents of the surface ligand or varying the charge on the protein subunits. More studies are in progress to understand aggregation of HSA triggered by PEG islands to produce functional sensing assemblies. In summary, “density gradient” of pre-patterned surfaces offer an opportunity to design complex “biomolecular blueprints” with spatiotemporal characteristics. Such protein nano- and micro-patterns which have a straightforward approach of generation may find huge application potential in the field of biosensors, biomaterials, bio-Micro-Electro-Mechanical-Systems (Bio-MEMS), cell adhesion and bacteria detection.
Methods
Materials
All chemicals used including NaCl, Na2PO4, 1-ethyl-3-(3-dimethyl aminopropyl)carbodiimide (EDC),N-Hydroxysulfosuccinimide (Sulfo-NHS) and 2-(N-morpholino) ethanesulfonic acid (MES), lipoic acid, human serum albumin (30% in 0.85% sodium chloride) were purchased from Sigma Aldrich (Oakville, Ontario, CA). The ligands carboxy-PEG12-lipoamide (CL-PEG12) and solvents such as ethanol were purchased from Fisher Scientific. Deionized (DI) water with resistivity of 18 MΩ·cm−1 from Milli-Q-water purification system was used in all the experiments.
Pattern generation
Silicon substrate (1 × 1 cm2) was cleaned in piranha (H2SO4:H2O2 (3:1)) (Caution) and rinsed with plenty of water and coated with Ti/Au (5/50 nm) layers using e-beam evaporation. The substrate smoothness and cleanliness plays an important role in pattern generation. The substrate was cleaned again with piranha and ethanol before functionalization with 1 mM CL-PEG12 or 1 mM dihydrolipoic acid (DHLA). PBS buffer (20 mM Na2PO4, 0.15 M NaCl) at pH 7.4 was used for SAM formation. Homogeneous SAM was formed after 4 h. The substrates were rinsed in the buffer several times. After SAM formation, the carboxyl groups on PEG were activated using EDC (0.2 M) and NHS-sulfo (0.05 M) in MES buffer (0.1 M MES, 0.5 M HCl) at pH 6 by stirring for 30 min. Then 0.2 mg/ml protein was functionalized on the activated substrate in PBS buffer at pH 7.4 for 15 hrs.
Atomic force microscopy (AFM)
MFP-3D AFM (Asylum Research, Santa Barbara, CA, USA) with tapping mode (in air) was used for topography of the self-assembled patterns. A silicon probe with Al reflex coating (spring constant: 42 N/m, resonant frequency: 300 kHz) was used as tip for AFM imaging. A scan rate of 1 Hz was used for imaging, while SPIP 6.0.9 software was used for the in-depth analysis of AFM images.
Infrared spectroscopy (IR)
NEXUS 670 FTIR (Thermo Nicolet, Madison, WI, USA) was used for ATR-IR analysis of HSA samples equipped with ZnSe crystal. ATR-IR for the protein was taken after spreading the HSA solution at pH 7.4 on the ZnSe crystal and dried under nitrogen until the surface was dried without visible water on the crystal. Variable angle FTIR was accomplished at an angle of 70 degrees for all the functionalized substrates with p-polarized light.
X-ray photoelectron spectroscopy (XPS)
Chemical composition was investigated using Axis 165 X-ray Photoelectron Spectrometer (Kratos Analytical).
Scanning electron microscope (SEM)
JAMP 9500F (JEOL) Scanning Electron Microscope for SEM was used in High vacuum mode. The substrates were mounted on SEM Stubs.
Circular dichroism (CD) spectropolarimetery
The CD spectra were measured on an OLIS DSM 17 Circular Dichroism instrument (OLIS Inc. Bogart, Georgia, USA). Quartz cell of a 0.02 cm path length was used to contain the sample and the spectra were recorded in the far-UV region with wavelength between 190 and 260 nm. The spectrum was recorded with five scan accumulations.
References
Gates, B. D. et al. New approaches to nanofabrication: Molding, printing and other techniques. Chem. Rev. 105, 1171–1196 (2005).
Love, J. C., Estroff, L. A., Kriebel, J. K., Nuzzo, R. G. & Whitesides, G. M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 105, 1103–1169 (2005).
Dhar, B. M. et al. Field-effect tuned lateral organic diodes. Proc. Natl. Acad. Sci. USA 107, 3972–3976 (2010).
Genzer, J., Fisher, D. A. & Efimenko, K. Combinatorial near-edge x-ray absorption fine structure: Simultaneous determination of molecular orientation and bond concentration on chemically heterogeneous surfaces. Appl. Phys. Lett. 82, 266–268 (2003).
Wang, B. et al. Size selected monodispersed clusters on supported grapheme: Bonding, isomerism and mobility. Nano Lett. 12, 5907–5912 (2012).
Steiner, J., Schafer, R., Wieczoreck, H., McCord, J. & Otto, F. Formation and coarsening of the concertina magnetization pattern in elongated thin-film elements. Phys. Rev. B 85, 104407/1–104407/25 (2012).
Gooding, J. J. Advances in interfacial design for electrochemical biosensors and sensors: aryl diazonium salts for modifying carbon and metal electrodes. Electroanalysis 20, 573–582 (2008).
Gilroy, J. B. et al. Monodisperse cylindrical micelles by living crystallization-driven self-asssembly. Nat. Chem. 2, 566–570 (2010).
Chen, C. L., Bromley, K. M., Oldak, J. M. & DeYoreo, J. J. A multistage pathway for human prion protein aggregation in vitro: from multimeric seeds to β-oligomers and nonfibrillar structures. J. Am. Chem. Soc. 133, 8586–8593 (2011).
Singh, G. et al. Highly ordered nanometer-scale chemical and protein patterns by binary colloidal crystal lithography combined with plasma polymerization. Adv. Funct. Mater. 21, 540–546 (2011).
Piner, R. D., Zhu, J., Xu, F., Hong, S. & Mirkin, C. A. “Dip-Pen” Nanolithography. Science 283, 661–663 (1999).
Genzer, J. & Bhat, R. J. Surface-bound soft matter gradients. Langmuir 24, 2294–2317 (2008).
Genzer, J. Surface-bound gradients for studies of soft materials behavior. Annu. Rev. Mater. Res. 42 435–468 (2012).
Leng, L., McAllister, A., Zhang, B., Radisic, M. & Gunther, A. Mosaic hydrogels: One step formation of multiscale soft materials. Adv. Mater. 24, 3650–3658 (2012).
Dri, C., Peters, M. V., Schwarz, J., Hecht, S. & Grill, L. Spatial periodicity in molecular switching. Nat. Nanotechnol. 3, 649–653 (2008).
Hong, S. W. et al. Unidirectionally aligned patterns driven by entropic effects on faceted surfaces. Proc. Natl. Acad. Sci. USA 109, 1402–1406 (2012).
Bhat, R. R., Fischer, D. A. & Genzer, J. Fabricating planar nanoparticle assemblies with number density gradients. Langmuir 18, 5640–5653 (2002).
Efimenko, K. et al. Nestling self-similar wrinkling patterns in skins. Nat. Mater. 4, 293–297 (2005).
Otero, R., Gallego, J. M., Parga, A. L. V. D., Martín, N. & Miranda, R. Molecular self-assembly at solid surfaces. Adv. Mater. 23, 5148–5176 (2011).
Cohen Stuart, M. A. et al. Emerging applications of stimuli-responsive polymer materials. Nat. Mater. 9, 101–113 (2010).
Agheli, H. et al. Large area protein nanopatterning. Nano Lett. 6, 1165–1171 (2006).
Valesesia, A. et al. Immobilization of antibodies on biosensing devices by nanoarrayed self-assembled monolayers. Langmuir 22, 1763–1767 (2006).
Willner, I. Biomaterials for sensors, fuel cells and circuitry. Science 298, 2407–2408 (2002).
Choi, H. J., Kim, N. H., Chung, B. H. & Seong, G. H. Micropatterning of biomolecules on glass surfaces modified with various functional groups using photoactivable biotin. Anal. Biochem. 347, 60–66 (2005).
Gooding, J. J. Advances in interfacial design for electrochemical biosensors and sensors: aryl diazonium salts for modifying carbon and metal electrodes. Electroanalysis 20, 573–582 (2008).
Gilroy, J. B. et al. Monodisperse cylindrical micelles by living crystallization-driven self-sssembly. Nat. Chem. 2, 566–570 (2010).
Burns, J. W. Biology takes centre stage. Nat. Mater. 8, 441–443 (2009).
Singh, G., Griesser, H. J., Bremmell, K. & Kingshott, P. Highly ordered nanometer-scale chemical and protein patterns by binary colloidal crystal lithography combined with plasma polymerization. Adv. Funct. Mater. 21, 540–546 (2011).
Mann, S. Self-assembly and transformation of hybrid nano-objects and nanostructures under equilibrium and non-equilibrium conditions. Nat. Mater. 8, 781–792 (2009).
Sinclair, J. C. Self-assembly: Proteins on parade. Nat. Chem. 4, 346–347 (2012).
Vericat, C., Vela, M. E., Benitez, G., Carro, P. & Salvarezza, R. C. Self-assembled monolayers of thiols and dithiols on gold: new challenges for a well known system. Chem. Soc. Rev. 39, 1805–1834 (2010).
Lee, B. K. Stepwise self-assembly of a proteinnanoarray from a nanoimprinted poly(ethylene glycol) hydrogel. Small 4, 342–348 (2008).
Jeyachandran, Y. L. & Zharnikov, M. Comprehensive analysis of the effect of electron irradiation on oligo(ethylene glycol) terminated self-assembled monolayers applicable for specific and nonspecific patterning of protein. J. Phys. Chem. C 116, 4950–14959 (2012).
Rundqvist, J., Hoh, J. H. & Haviland, D. B. Poly(ethylene glycol) self-assembled monolayer island growth. Langmuir 21, 2981–2987 (2005).
Malysheva, L. et al. Spectroscopic characterization and modeling of methyl- and hydrogen-terminated oligo(ethylene glycol) self-assembled monolayers. J. Phys. Chem. C. 116, 12008–12016 (2012).
Amar, J. G. & Family, F. Critical cluster size: island morphology and size distribution in submonolayer epitaxial growth. Phys. Rev. Lett. 74, 2066–2096 (1995).
Amar, J. G. & Family, F. Dynamic scaling of the island-size distribution and percolation in a model of submonolayer molecular beam epitaxy. Phys. Rev. Lett. 50, 8781–8797 (1994).
Juarez, J., Taboda, P. & Mosquera, V. Existence of different structural intermediates on the fibrillation pathway of human serum albumin. Biophys. J. 96, 2353–2370 (2009).
Karlsson, R., Roos, H., Faegerstam, L. & Persson, B. Kinetic and concentration analysis using BIA technology. Methods, 6, 99–110 (1994).
Jennissen, H. P. & Zumbrink, T. A novel nanolayer biosensor. Biosens. Bioelectron. 19, 987–997 (2004).
Fogh-Andersen, N., Bjerrum, P. J. & Siggaard-Andersen, O. Ionic binding, net charge and Donnan effect of human serum albumin as a function of pH. Clin. Chem. 39, 48–52 (1993).
Dell'Orco, D. Fast prediction of thermodynamics and kinetics of protein-protein recognition from structures: from molecular design to system biology. Mol. BioSyst. 5, 323–334 (2009).
Alsallaq, R. & Zhou, H. X. Electrostatic rate enhancement and transient complex of protein-protein interaction. Structure 15, 215–224 (2007).
Bandopadhyay, A., Pati, R., Sahu, S., Peper, F. & Fujita, D. Massively parallel computing on an organic molecular layer. Nat. Phys. 6, 369–375 (2010).
Solokolov, I., Yang, H., Ozin, G. A. & Kresge, C. T. Radial Patterns in Mesoporous Silica. Adv. Mater. 11, 636–642 (1999).
Rialdi, G. & Battistel, E. Thermodynamics of proteins in unusual environments. Biophys. Chem. 126, 65–69 (2007).
Pribie, R., VanStokkum, I. H. M., Chapman, D., Haris, P. I. & Bolemendal, M. Protein secondary structure from Fourier transform infrared and/or circular dichroism spectra. Anal. Biochem. 214, 366–378 (1993).
Acknowledgements
This work was supported by Canada Excellence Research Chairs (CERC) Program.
Author information
Authors and Affiliations
Contributions
G.T. and K.P. planned, carried out the experiments, did AFM image processing, analyzed the data and wrote the manuscript; T.T. analyzed results and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Directed self-assembly of proteins into discrete patterns
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Thakur, G., Prashanthi, K. & Thundat, T. Directed self-assembly of proteins into discrete radial patterns. Sci Rep 3, 1923 (2013). https://doi.org/10.1038/srep01923
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01923
This article is cited by
-
Tug-of-war of microtubule filaments at the boundary of a kinesin- and dynein-patterned surface
Scientific Reports (2014)
-
Artificial photosynthesis of oxalate and oxalate-based polymer by a photovoltaic reactor
Scientific Reports (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.