Abstract
Mutually exclusive cell fate determination of CD4 helper or CD8 killer T cells occurs in the thymus. These T-cell subsets are not believed to redirect other lineages. Here we showed that retinoic acid and transforming growth factor-β1 promoted the differentiation of CD8αα T cells from CD4 T cells in a Runx3-dependent manner. These cells were inferred to belong to immunoregulatory populations because subpopulations of CD8αα+TCRαβ T cells are known to suppress activated T cells and mice with Runx3−/− T cells showed defects during recovery from experimental allergic encephalomyelitis. Our results demonstrate that CD4 T cells play fundamental roles in controlling immune reactions through promotion and attenuation. We accordingly anticipate that clarifying the mechanisms underlying this process will provide insights leading to autoimmune and immunodeficiency disease therapies.
Similar content being viewed by others
Introduction
During T cell maturation, T-cell receptor (TCR) αβ-bearing cells express both CD4 and CD8 (double-positive thymocytes) molecules on their surface. Transition from double to single positive (CD4+CD8− or CD4−CD8+) requires the selection of TCRαβ with intrathymic ligands that are presented by major histocompatibility complexes (MHCs). CD4 and CD8 coreceptors interact with MHC class II and I molecules, respectively, thereby resulting in the interaction of TCRαβ with ligands/MHC complexes and stabilizing CD4 or CD8 expression on maturing TCRαβ T cells. Simultaneously, thymocytes diverge into functionally distinct CD4 helper and CD8 killer cells1. A correlation between the mechanisms of antigen recognition and functional divergence of T cells suggests that the commitment of these cells is irreversible. The redirection of CD4 T cells to the CD8 lineage and vice versa is not believed to occur in the periphery.
Establishment of central and peripheral tolerance is important for maintaining immunological homeostasis. In addition to naturally occurring CD4+CD25+Foxp3+ regulatory T cells (nTregs), several phenotypically and functionally distinct regulatory T-cell populations have been suggested2,3,4,5,6. To date, at least four CD8+ Treg subsets have been identified and these include CD8+CD28−, CD8+CD25+, CD8+CD122+ and CD8αα T cells. Among them, two CD8+ T-cell subsets exhibit a special property in suppressing activated, but not naïve, T cells7,8,9. CD8αβ+CD122+CD44+ inducible costimulator ligand (ICOSL)+ TCR αβ+ T cells attenuate immune responses by inhibiting follicular T helper cell (TFH) via recognition of Qa-1, which is expressed on TFH cells in an activation-dependent manner7. The CD8αα+CD122+TCRαβ+ T-cell subset was identified during the screening of Treg cells that inhibit experimental autoimmune encephalomyelitis (EAE) in mice10,11. This CD8αα T-cell subset recognizes the pathogenic TCR-derived peptide that is present on the Qa-1 molecule of pathogenic T cells in an activation-dependent manner and inhibits pathogenic T cells. In addition, an immunoregulatory role of CD8αα+TCRαβ+ T cells has been shown in two other autoimmune disease models. First, transfer of CD8ααTCRαβ T cells inhibits colitis, induced by the adoptive transfer of naïve CD4 T cells into severe combined immunodeficient (SCID) mice6. Second, non-obese diabetic (NOD) mice are defective in the generation of CD8ααTCRαβ T cells, suggesting a regulatory role of these cell populations12. In addition, genetic control elements, which are required for clonal deviation to CD8αα T cells, also regulate rescue from clonal deletion of nTregs in the thymus13, suggesting that these different subsets of immunoregulatory T cells share a common thymic selection mechanism.
The developmental pathway of CD8αα+TCRαβ+ T cells is controversial because of the following findings. First, the accumulation of autoreactive TCRs was observed in the repertoire of CD8αα+TCRαβ+ T cells14. Second, CD8αα+TCRαβ+ T cells comprise only a small portion of T cells in the lymph node and spleen (<1% of T cells) but a large portion (approximately 40% of T cells) in the intraepithelium of the gut. For this reason it was previously believed that CD8αα+TCRαβ+ T cells were of extrathymic origin and differentiated locally in the gut; however, they are now considered to originate in the thymus15. In the thymus, CD8αα+TCRαβ+ precursor T cells were selected by high-affinity self-antigens, such as nTregs12,16,17. The selected precursor T cells were CD4- and CD8-double-negative cells. The final maturation of these cells, including the expression of CD8αα, occurs in the gut18. Transforming growth factor (TGF)-β1 plays a key role in the generation of nTregs and CD8αα T cells during selection in the thymus19,20. Thus, the regulatory factors that control development of CD8αα+TCRαβ+ intestinal intraepithelial lymphocytes (IELs) and the molecular pathways of this cell population have been elucidated. However, the origin of CD8αα+TCRαβ+ T cells outside intestines and the factors required for their generation are incompletely understood.
In this study, we showed that immune reactions promote the differentiation of CD8αα+TCRαβ+ T cells from naïve CD4 T cells within peripheral lymphoid tissues. Among the factors induced by inflammation, TGF-β1, all-trans retinoic acid (atRA) and interleukin (IL)-2 are indispensable for the differentiation of CD8αα T cells. Striking similarities were observed between induced Tregs (iTregs) and CD8αα T cells, with the same signals required for the differentiation of iTregs. In addition, Runx3-deficient CD4 T cells have lost their ability to become CD8αα T cells, indicating that Runx3 plays a critical role in this differentiation. Furthermore, mice whose T cells were Runx3-deficient showed defects in recovery from EAE. Thus CD8αα Tregs were confirmed to be generated in the thymus and periphery, like CD4+Foxp3+ Tregs and the signals required for CD8αα Tregs were the same as those required for CD4+Foxp3+ Tregs. We thus demonstrated that the newly identified Treg subset, namely CD8αα Tregs that are derived from CD4 T cells, plays an immunoregulatory role in EAE.
Results
Generation of CD8αα T cells in the spleen by immunization
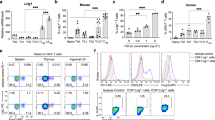
In addition to CD4+CD25+Foxp3+ T cells21,22,23, several populations have been identified as immunoregulatory cells7,10,24,25. CD8 Treg subsets negatively regulate activated, but not naïve, T cell functions7,10,24,25. Among these cell subsets, CD8ααTCRαβ T cells have been reported to play important roles in the gut mucosal immunity and prevent autoimmune diseases6,12,13. However, the origin of this subset and mechanisms of its development remain unclear. The initial commitment of CD8αα T cells occurs in the thymus following exposure to agonist self-peptides16,17,18 and their generation requires a TGF-β1 signal19. When positively selected CD8ααTCRαβ precursor T cells reach the gut, they express CD8αα molecules in an IL-15-dependent manner18. Importantly, the expression of CD8αα is induced on CD8αβ T cells by immunization and is required for the generation of memory CD8αβ T cells26,27. We accordingly examined the effect of immunization on the generation of CD8αα single-positive cells in the spleen. As anticipated, immunization induced the generation of CD8αα single-positive T cells (6.5% of CD4−CD8αβ−CD8αα+TCRβ+ T cells within CD4− T cells in immunized spleen compared with 2.5% in non-immunized spleen; Fig. 1A).
Generation of CD8αα T cells in Rag2−/− mice.
(A) Fluorescence-activated cell sorting (FACS) analysis of CD8αα+CD8αβ−CD4−TCRαβ T cells in the spleen after immunization of WT (C57BL/6) mice with NP-CGG/Alum. Horizontal axis indicate percentage of CD8αα T cells within CD4− T cells. *p < 0.05; n = 3. TCR, T-cell receptor. (B) FACS analysis of lymphocytes from Rag2−/− mice (CD45.1) that had received naïve CD4 T cells (CD45.2) 6 months previously. TCRαβ+CD45.2+ cells were examined for CD4 and CD8α expression. CD4−CD8α+ (I) and CD4+CD8α+ (II) T cells were further examined for CD8α and CD8β expression. IELs, intestinal intraepithelial lymphocytes, PP, Peyer's patches. CD8αα T cell differentiations from CD4 T cells in Rag2−/− mice were confirmed in two of four mice analyzed and representative data are shown.
Generation of CD8αα T cells from CD4TCRαβ T cells in Rag2−/− mice
With the aim of identifying the subsets that potentially differentiate into CD8αα T cells, their generation was examined at day 20 after naïve CD4 and CD8αβ T cells were adoptively transferred into Rag2−/− mice. Whereas CD8αα T cells were not found in either case after 20 days, in some Rag2−/− mice that received naïve CD4 T cells they were found after approximately 6 months. A few CD8αα-positive cells were found in all tissues examined, with a significantly larger number observed in Peyer's patches (PPs; Fig. 1B). These observations indicate that naïve CD4 T cells can differentiate into CD8αα T cells, at least under these experimental conditions.
TGF-β1, atRA and IL-2 promote differentiation of CD8ααTCRαβ Tregs from naïve CD4 T cells in vitro
To investigate the mechanisms of differentiation from CD4 into CD8αα T cells, we first focused on the factors required for the generation of iTregs or CD4+Foxp3+ cells. We added the factors to a conventional in vitro CD4 T-cell differentiation assay. IL-2, TGF-β1 and atRA generated CD8αα T cells from CD4 T cells (Fig. 2A). Although atRA played only a supporting role in the differentiation of iTreg cells, the generation of CD8αα T cells depended on all three factors (Fig. 2A and data not shown). To exclude the possibility that CD8αα T cells were derived from contaminated CD8αβ cells, CD8αβ T cells were cultured under the same conditions as those used for the differentiation of CD8αα from CD4 T cells and no CD8αα T cells were observed (Fig. 2B). Moreover, CD8αβ T cells have the ability to inhibit CD4-derived CD8αα T cell differentiation, as shown in Fig 2C. These observations indicate that CD4 T cells and not CD8αβ T cells are the major source of peripheral CD8αα T cells. Because ILs play critical roles in the determination of helper T-cell subsets, we further examined the effects of various ILs on the differentiation of CD8αα. Our results showed that IL-2 exerted the strongest effect on the generation of CD8αα T cells (Fig. 2D). Antigen dose has been shown to affect the differentiation of CD8αα T cells. In the thymus, a low concentration of an agonist induces CD8αβ T cells, whereas a high concentration induces CD8αα T cells. atRA concentration influences the differentiation of iTreg and Th17 cells28,29. Thus, antigen dose and atRA concentration influence various processes in cell fate determination. We accordingly examined the effects of TCR signal strength and atRA concentration on differentiation of CD8αα T cells. A high percentage of CD8αα T cells was found after culture with strong TCR stimulation and high atRA concentration (Fig. 2E).
TGF-β1, atRA and IL-2 promote differentiation of CD8ααTCRαβ regulatory T cell (Treg) from naïve CD4 T cells in vitro.
(A) Naïve CD4+ T cells were stimulated with α-CD28, IL-2, TGF-β1 and atRA on α-CD3ε-coated plates. CD8αα+CD4− T cells were analyzed by FACS. (B) CD8αβ+ T cells were stimulated under conditions mentioned in A. (C) Total spleen T cells (41% CD8αβ+ T cells and 56% CD4 T cells) were stimulated under conditions mentioned in A. (D) Naïve CD4+ T cells were cultured with various ILs and TGF-β1+atRA. (E) Naïve CD4+ T cells were stimulated with TGF-β1, IL-2 and various atRA concentrations on plates coated with indicated concentrations of α-CD3ε. Cell differentiation was analyzed by FACS. All experiments were performed at least thrice; representative data are shown.
Runx3-dependent generation of CD8ααTCRαβ Tregs in vitro
With the aim of characteristizing CD8αα T cells (CD8α positive fraction in Fig 3A or CD8αα cells in Fig 3B) that were generated in vitro, their gene expression profile was compared with iTregs (CD8 negative fraction shown in Fig 3A or iTreg cells in Fig 3B) derived from the same culture condition (as shown in Fig 3A) and previously well-characterized CD8 TCRαβ Tregs7,9,24,30. The expression of granzymes A and B increased in the CD8αα population, whereas that of Foxp3 decreased (Fig. 3A, B). The expression of CD122 and ICOSL on the surface was also specifically induced in CD8αα T cells (Fig. 3A). The expression pattern of these genes was similar to that of other CD8 Treg cells7,9,24,30. Recent studies have indicated that Runx3 is a major regulator of CD8αβ T cells, while ThPOK is a major regulator of CD4 T cells31,32,33. We accordingly examined the possibility that these factors were involved in the differentiation from CD4 to CD8αα T cells. Runx1 and ThPOK were expressed specifically in naïve CD4 T cells (Fig. 3B). After the generation of CD8αα T cells, only Runx3 expression was observed, while this was not found in the iTreg subset (Fig. 3B). Of note, we had previously found that Runx2 and Runx3 are required for TGF-β1 and atRA signals in IgA class switching in B cells34,35. Thus, it was conceivable that Runx3 contributed to the differentiation of CD8αα cells. As anticipated, Runx3−/−CD4 T cells were unable to differentiate into CD8αα T cells (Fig. 4). These observations support the hypothesis that CD4-derived CD8αα T cells function as regulatory cells in vivo and that Runx3 is essential for the generation of these lineage cells.
Gene expressions of CD8ααTCRαβ Tregs differentiated in vitro.
(A) Naïve CD4+ T cells were stimulated under conditions mentioned in Fig. 2(A). Gene expression was estimated by FACS. (B) Naïve CD4+ T cells were stimulated under conditions mentioned in Fig. 2(A). iTreg (CD4+T) and CD8αα T (CD8+ T) cells were sorted from the cultured cells (shown in Fig 3A) and gene expression was analyzed by reverse transcription–polymerase chain reaction. Naïve CD4 T cells (CD4+ CD62L+ T cells), CD8αβ T cells were used for control. Naive CD4, CD8αβ, iTreg and CD8αα indicate RNA source used for cDNA generation. Fivefold serial dilutions of cDNAs were amplified for the transcripts indicated. All experiments were performed at least thrice and representative data are shown.
Differentiation of CD8αα Tregs from CD4 T cells requires Runx3.
Naïve CD4 T cells from wild type (WT) or Runx3−/− FL-derived cells were stimulated under the conditions mentioned in Fig. 2(A). CD8αα+CD4− T cells were analyzed by FACS. All experiments were performed thrice; representative data are shown.
Generation of CD8αα Tregs from CD4 T cells in vivo requires Runx3
To determine whether CD4 T cells differentiate into CD8αα T cells in vivo, naïve CD4 T cells from C57BL/6 (CD45.2) mice were adoptively transferred into Rag2−/− mice with a C57BL/6 background (CD45.1) and the generation of CD8αα T cells was examined after atRA administration. Given that the homing location of transferred CD4 T cells cannot be controlled, absolute cell numbers of transferred T cells at PP, IEL and LPL varied substantially. Accordingly, CD8αα T cell differentiation was examined by assessing the percentages of CD8αα cells. CD8αα T cells appeared in an atRA-dependent manner in all tissues examined (Fig. 5). The percentage of CD8αα T cells was significantly higher in PPs and IELs than that in controls (12.2% of CD8αα+CD4−TCRαβ+ T cells in atRA-treated PPs compared with 0.05% in controls; 10.0% in the atRA-treated sIEL compared with 0.09% in controls; Fig. 5). These results indicate that the atRA-dependent differentiation of CD4 T cells into CD8αα T cells occurred in vivo. To exclude the possibility that CD8αα T cells were derived from contaminated CD8αβ T cells, we assessed the influence of CD8αβ T cells on the generation of CD8αα T cells by modulating the ratio of CD8αβ T cells within naïve CD4 T donor cells. Naïve CD4 T cells used for a series of cell transfer experiments contained fewer than 0.02% CD8αβ T cells. Purified naïve CD4 T cells were transferred into Rag2−/−mice with or without CD8αβ T cells and the generation of CD8αα T cells in vivo was assessed. There was negligible difference in the percentage of CD8αα T cells between mice carrying CD4 T cell populations containing 0.02% CD8αβ T cells and those carrying populations containing 0.12% CD8αβ T cells (data not shown). In addition, naïve CD4 T cells, contaminated less than 0.002% by CD8αβ T cells, differentiated as efficiently as CD4 T cell populations carrying 0.02% CD8αβ T cells in Rag2−/− mice. These results indicate that CD8αβ T cells are not the major source of CD8αα T cells differentiated in vivo.
Generation of CD8αα Tregs from CD4 T cells in vivo requires Runx3.
FACS analysis of lymphocytes from Rag2−/− mice (CD45.1) that received naïve CD4 T cells (CD45.2) from WT or Runx3−/− FL-derived cells after atRA administration. TCRαβ+CD45.2+ cells were examined for CD4 and CD8α expression (all samples). CD4-CD8α+ T cells were further examined for CD8α and CD8β expression in case of atRA-administrated samples. All experiments were performed at least thrice; representative data are shown. LPLs, lamina propria lymphocytes.
The contribution of Runx3 to differentiation of CD4-CD8αα in vivo was further determined using Runx3-deficient naïve CD4 T cells as donor. No CD8αα T cells were observed in either atRA-treated or non treated mice (Fig. 5). However, the preferential differentiation of CD8αβ+ T cells, but not CD8αα+ T cells, was observed following treatment with atRA in high doses (Supplementary Fig. S1). Considering all results, Runx3 plays a special role in the differentiation of CD8αα T cells from CD4 T cells in vivo. In addition, several other signals that were not examined in this study contribute to CD8 T cell generation from CD4 T cells in vivo; Runx3−/− naïve CD4 T cells cannot differentiate CD8α+ T cells in vitro. In view of the functional redundancy of Runx family transcription factors, their contribution to the differentiation of CD4 T cells into CD8 T cells was investigated using Runx2−/−Runx3−/−CD4 T cells. Administration of high-dose atRA could not rescue the defect in the generation of CD8 T cells (data not shown), indicating that Runx2 and Runx3 play critical roles in the differentiation of CD4 T cells into CD8 T cells. However, the precise roles of each Runx factor in the differentiation into various CD4-derived CD8 T cell subsets remain to be investigated.
CD4 T cells differentiate into CD8αα Tregs during immune response and attenuate immune reactions
Accumulating evidence indicates that the gut is rich in atRA28,29,36,37 and RA-dependent events are believed to occur preferentially in the gut. However, the spontaneous differentiation of CD4 T cells into CD8αα T cells was not observed in the gut in the absence of atRA administration (Fig. 5). The physiological relevance of differentiation of CD4 T cells into CD8αα T cells in normal immunological reactions is thus debatable. Recent reports have demonstrated that RA is required to promote the effector responses of CD4 T cells and is more abundant in inflamed tissues than in the gut38,39. We accordingly investigated whether the differentiation of CD4 T cells into CD8αα T cells can be induced by immunization. Generation of CD8αα T cells in Rag2−/− mice that received naïve CD4 T cells was assessed after immunization. On days 1, 3 and 6 after immunization, approximately 1% of CD8αα T cells were observed in the spleen (Fig. 6A). This value was not very different from that observed after usual immunization. Approximately 2%–3% of the cells were CD8αα+ within TCRαβ+CD8αβ− T cells (Figure 1A and data not shown). Significantly larger populations of CD8αα+ T cells were observed in PPs, IELs and lamina propria lymphocytes on day 6 (Fig. 6B). In contrast, hardly any CD8αα+ T cells were observed in the gut on day 3 (Fig. 6B). Thus, these results suggested that differentiation of CD4 T cells into CD8αα T cells occurred at the site of immune reactions and that newly generated CD8αα T cells migrated into the gut through the expression of gut-homing receptors, as previously indicated26,36. In a previous study, we found that Runx3−/− mice showed defects in recovery from colitis and thereby increased the formation of inflammation-associated tumors40. We speculated that differentiated CD8αα T cells attenuate immune reactions and decrease the harmful effects of continuous inflammation, given that CD8αα Tregs are known to suppress activated T cells specifically. For a more detailed assessment of this hypothesis, we used mice with EAE as a model system. In this model, CD8 T cells play a key role in reducing the frequency of relapse after recovery from acute EAE41,42. This autoimmune disease is accordingly a suitable model for assessing the regulatory functions of CD8 Tregs. As expected, no differences in onset, clinical scores in the early phase, or EAE incidence were observed between genotypes. However, Runx3−/− mice exhibited defects in recovery from EAE, as indicated by the increased clinical scores after 6 weeks (P = 0.0010; Fig. 7A). To exclude effects derived from other lineage cells, we performed EAE-induction experiments using Rag2−/− mice, having transferred CD4 T cells from Runx3−/−and wild-type mice. Again, Rag2−/−mice having Runx3−/− CD4 T cells showed defects in recovery from EAE (P = 0.0048; Fig. 7B). These results indicate that Runx3 functions in CD4 T cells are important for recovery from EAE. Various CD4-derived cells are involved in EAE regulation: Th17, Th1 and CD4+Foxp3+ Treg cells. Th17 and Th1 are important for the induction of EAE disease. However, Runx family transcription factors are positive regulators for Th1 and Th17 differentiation, indicating that Runx3 deficiency will contribute to inhibiting EAE disease. Furthermore, Treg cell differentiation and function are not strongly affected by Runx3 deficiency, as shown previously40,48. In sum, the defect in recovery from EAE disease observed in Rag2−/− mice having Runx3-deficient CD4 T cells cannot be explained by previously known functions of Runx3 in CD4 T cells. For these reasons, we concluded that CD4-derived CD8αα T cells function as immune regulatory cells, at least in this EAE model. These observations support the hypothesis that differentiated CD8αα T cells play an important regulatory role in the establishment of a negative feedback loop that decreases adverse effects during immune reactions.
CD4 T cells differentiate into CD8αα Tregs during immune responses.
(A) Naïve CD4+ T cells (CD45.2) were injected into Rag2−/−mice. TCRαβ+CD45.2+ cells from spleen at 1, 3 and 6 days after immunization were analysed for expression of CD8α and CD4. (B) FACS analysis of lymphocytes from tissues of mice used for (A). TCRαβ+CD45.2+ cells were examined for expression of CD4 and CD8α. CD4-CD8α+ T cells were further examined for expression of CD8α and CD8β. All experiments were performed at least three times and representative data are shown.
Runx3−/− T cells cause defect in recovery from EAE.
Rag2−/− mice with (A) WT or Runx3−/− lymphocytes and (B) WT or Runx3−/− CD4 T cells were followed up for signs of neurological disease after experimental autoimmune encephalomyelitis (EAE) induction. Data shown are mean ± standard error of mean of EAE clinical score of 10 mice pooled from two independent experiments and percentage disease incidence.
This study revealed previously unknown CD8 T-cell subsets that were derived from activated peripheral CD4 T cells (Fig. 8) and a unique mechanism for generating activated CD8 T cells without other CD4 T cells. Furthermore, these T-cell subsets can likely inhibit immune reactions. The elucidation of the precise mechanism of these processes and the exact functions of these cell subsets will provide fundamental insights for future autoimmune disease therapies.
Discussion
CD4 T cells are known to differentiate into various helper and regulatory T-cell lineages, namely Th1, Th2, Th9, Th17, TFH, and CD4+Foxp3+ Tregs. In addition to these subsets, we identified a new cell population in this study called CD8αα Tregs, which have a regulatory function. Differentiation of CD8αα Tregs required the same signals as iTregs. However, the differentiation of CD8αα Tregs took more time than that of iTregs and stronger TCR stimulation and higher atRA concentrations were required for their generation. These data indicate that the determination of the cell fate of one of the two inducible Treg cell subsets was differentially controlled during immune reactions. Therefore, iTregs and CD8αα Tregs may have distinct functions in immune regulation. According to this concept, some CD8αα T-cell subsets have been shown to have the property of inhibiting activated, but not naïve, T cells. Qa-1 in mouse (or HLA-E in human), which is a nonclassical MHC class I molecule, has been believed to play a key role in restricting the activity of CD8αα Tregs only toward activated T cells because these molecules are transiently upregulated on the surface of activated T cells and antigen-presenting cells. The correlation between CD8 Tregs and Qa-1 was suggested on the basis of previous studies as follows. Immunization with Qa-1-expressing activated CD4 T cells induces Qa-1-restricted CD8 Tregs43,44; CD8αα Treg clones recognize a TCR-derived peptide presented on Qa-111; Qa-1−/− mice show augmentation of CD4 T-cell responses and increased susceptibility to EAE45; resistance to the re-induction of EAE was lost in CD8- and Qa-1-deficient mice41,42,45,46. In this study, we observed defects in recovery from EAE in Runx3-deficient mice in which CD4 T cells could not differentiate into CD8αα Tregs. These data indicate that CD8αα Tregs play a special role in the regulation of activated T cells. However, the molecular mechanisms underlying the suppressor functions of CD8αα Tregs are poorly understood. Furthermore, the exact target cells of CD8αα T cells and the relationship between CD8αα Tregs and Qa-1 in various immune reactions remain to be studied47.
We found striking similarities between signals for CD4 and CD8αα Tregs. In addition, differentiation of these two regulatory T-cell subsets requires Runx protein activities; CD8αα Tregs require Runx3 and CD4 Tregs require Runx148. Whether the different functions of Runx1 and Runx3 or the total amount of Runx proteins influence the determination of cell fate remains to be clarified.
CD8αα T cells are major populations of IELs and most of them are selected in the thymus. The commitment of CD8ααTCRαβ T cells occurs from both MHC class I- and class II-restricted T cells16,17. Moreover, CD8αα IELs are absent in β2-microglobulin-deficient mice but not in MHC class Ia-deficient mice49,50. From these observations, MHC class Ib molecules, including Qa-1, are believed to be required for the selection of CD8αα T cells. In this study, we showed that CD4 T cells differentiated into CD8αα T cells in the periphery, indicating that MHC class II-dependent primary selection of these cells occurred in the thymus. It is of interest to determine the exact roles of the Qa-1 molecule in the differentiation, maintenance, or expansion of CD4-derived CD8αα T cells.
In summary, we have identified a new subset of inducible Treg cells (Fig. 8). This differentiation system can be affected by various immune dysfunctions, including autoimmune diseases and tumor immunity. We anticipate that further elucidation of the precise mechanisms of this process will provide a basis for development of anti-cancer drugs and a rationale for treatment of autoimmunity.
Methods
Mice
This study received appropriate ethics approval from the Institutional Review Board of Kyoto University (Reference Number: Med Kyo12076). Wild-type (WT), Runx3+/−, Runx2+/− and Rag2−/− mice (CD45.1) in a C57BL/6 genetic background were maintained in a specific pathogen-free mouse facility. Procedures involving animals and their care were in accordance with the guidelines for animal treatment of the Institute of Laboratory Animals, Kyoto University.
Fetal liver transfer
Single-cell suspensions of 2–4 × 106 whole fetal liver mononuclear cells harvested from Runx3−/−, Runx2−/−Runx3−/−and WT (CD45.2) embryos at E14.5 were injected intravenously into sublethally irradiated (4 Gy) Rag2−/− recipient mice (CD45.1). At least 10 weeks after transplantation, mice were used for EAE induction or were killed and their naïve CD4 T cells were analyzed by flow cytometry or used for in vitro culture. In some experiments, naïve CD4 T cells were purified and adoptively transferred into other Rag2−/− recipient mice (CD45.1). All Rag2−/− (CD45.1) mice used for each experimental set were age- and sex-matched.
Cell preparation
Naïve CD4+TCRαβ+ was prepared by magnetic cell sorting (MACS) using a CD4+ CD62L+ T cell isolation kit (Miltenyi Biotec, Bergisch, Germany) in combination with various antibodies (BD Pharmingen). Briefly, lineage positive cells, but not CD4, were removed according to the manufacturer's instruction using biotin-conjugated lineage-specific antibodies and α-Biotin microbeads. After negative selection, we made a rough prediction of the final concentration of contaminated CD4-CD62L+ cells after purification of CD4+CD62L+ cells. For this purpose, a part of the negatively selected cells were stained with α-CD4 and α-CD62L antibodies. If more than 5% cells were CD4-CD62L+ among CD62L+ cells, we performed negative selection again using the following antibodies and magnetic beads; α-CD8α-FITC, α-CD25-Biotin, α-CD19-Biotin, α-B220-Biotin, α-CD11b-Biotin, α-CD11c-Biotin, α-Ia-Biotin, α-Dx5-Biotin, α-TCRγδ-Biotin, Ter119-Biotin (Becton Dickinson, Mountain View, CA), α-FITC microbeads and streptavidin microbeads (Myltenyi Biotech Bergisch, Germany). After the second round of negative selection was finished, CD62L+ cells were further selected according to the manufacturer's instructions. Purities were determined by staining with antibodies for CD8α, CD8β, CD4 and CD62L. Usually, more than 97% cells were CD4+CD62L+ and fewer than 0. 02% cells were CD8α+. CD8αα cells cannot be detected. If more than 0.02% cells were positive for CD8α expression or fewer than 97% cells were CD4+CD62L+, we further purified CD4+CD62L+ cells by flow cytometry using FACSAria. All the purified cells using the above method were used for both in vitro culture and cell transfer experiments into RAG2−/− mice. Essentially all the methods of purification were used for in vitro culture and cell-transfer experiments.
For purification of CD4 T cells, spleen cells were subjected to negative selection using the following antibodies and magnetic beads; α-CD8α-FITC, α-CD19-Biotin, α-B220-α-CD11b-Biotin, α-CD11c-Biotin, α-Ia-Biotin, α-Dx5-Biotin, α-TCRγδ-Biotin, Ter119-Biotin (Becton Dickinson), α-FITC microbeads and streptavidin microbeads.
CD8αβ+TCRαβ+ T cells was prepared by MACS using a CD8α+ T cell isolation kit (Miltenyi Biotec) in combination with various antibodies (BD Pharmingen). Briefly, lineage positive cells, but not CD8, were removed according to the manufacturer's instruction using biotin-conjugated lineage-specific antibodies and α-Biotin microbeads. Then we further performed second round of negative selection using the following antibodies and magnetic beads; α-CD4-Biotin, α-CD19-Biotin, α-B220-Biotin, α-CD11b-Biotin, α-CD11c-Biotin, α-Ia-Biotin, α-Dx5-Biotin, α-TCRγδ-Biotin, Ter119-Biotin (Becton Dickinson) and Streptavidin microbeads. After a second round of negative selection, CD8β cells were positively selected by MACS. Usually more than 98% of populations were CD8αβ+ cells and fewer than 0.5 % were CD8αα+ cells. If fewer than 96% cells were CD8αβ+, or more than 1.0% were CD8 αα+ cells, we further purified CD8αβ+ cells by flow cytometry using FACSAria. All the purified cells using above method were used for both in vitro culture or cell transfer experiments into RAG2−/− mice.
Transfer of CD4 T cells or naïve CD4 T cells
Naïve CD4 T cells were prepared as described above; 0.5–2.5 × 106 naïve CD4 T cells (CD45.2) were transferred into Rag2−/− (CD45.1) mice. At least 4 weeks after transplantation, the mice were used for various experiments. For EAE experiments, 2.5 x106 CD4 T cells (CD45.2) were transferred into Rag2−/− (CD45.1) mice. Several days after transplantation, mice were used for EAE induction.
Cell cultures
In all experiments, the percentages of CD62L+CD4+ T cells or CD8αβ+ T cells were above 97. Naïve CD4+ T cells (1 × 105 cells /mL) were activated with plate-bound anti-CD3 (5 μg/mL; BD Pharmingen), soluble anti-CD28 (1 μg/mL; BD Pharmingen), anti-interferon-γ (3 μg/mL; BD Pharmingen), anti-IL-4 (3 μg/mL; BD Pharmingen), TGF-β1 (3 ng/mL; R&D) and IL-2 (20 ng/mL; R&D) in the presence or absence of atRA (10 nM–10 μM; Sigma-Aldrich) for 7–9 days.
RA injection
atRA (Sigma-Aldrich, Saint Louis, MO) was dissolved in soybean oil at 6 mg/mL and aliquots were stored at −80°C. A working solution was prepared by diluting the atRA aliquots in soybean oil and an equal volume of dimethyl sulfoxide (DMSO). atRA of 200 μg/100 μL, 600 μg/300 μL or vehicle was delivered by intraperitoneal injection 1, 3 and 6 d before analysis.
Flow cytometric analysis
The following antibodies were used for staining: PECy7 anti-CD45.2 (BD Pharmingen); allophycocyanin (APC) anti-mouse TCRαβ (BD Pharmingen); APCCy7 anti-mouse CD8α (BD Pharmingen); fluorescein isothiocyanate (FITC) anti-mouse CD8β (BD Pharmingen); phycoerythrin (PE) anti-mouse CD4 (BD Pharmingen); PE anti-mouse CD8α (BD Pharmingen); APC anti-mouse CD4 (BD Pharmingen); FITC anti-mouse CD8α (BD Pharmingen); FITC anti-mouse CD4 (BD Pharmingen); PE anti-mouse CD44 (BD Pharmingen); Biotin anti-mouse CD122 (BD Pharmingen); APC anti-mouse CXCR5 (BD Pharmingen); Biotin anti-mouse α4β7 (BD Pharmingen); FITC anti-mouse CCR9 (R&D); PE anti-mouse Granzyme A (Santa Cruz CA); Alexa 647 anti-Foxp3 (BD Pharmingen); Alexa 647 anti-Granzyme B (BD Pharmingen); PerCP–streptavidin (Molecular Probes, Eugene, OR); APC–streptavidin (Molecular Probes, Eugene, OR); APCCy7–streptavidin (BD Pharmingen); PECy7–streptavidin (BD Pharmingen); PE–streptavidin (Molecular Probes); FITC–streptavidin (Molecular Probes).
For cytoplasmic staining, BD Cytofix/Cytoperm solution and BD Perm/Wash solutions (BD Pharmingen) were used.
All analyses were performed with FACSCalibur or FACSAria (Becton Dickinson).
IELs and LPLs preparation
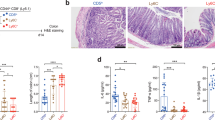
To prepare IELs and LPLs, the intestines were removed and PPs and mononuclear lymphocytes were isolated. The intestines were opened longitudinally, washed with phosphate-buffered saline and cut into 5-mm pieces. To obtain IELs, pieces were shaken in Hank's balanced salt solution (+) containing 5 mM ethylenediaminetetraacetic acid for 20 min at 37°C and passed through a cell strainer twice. The remaining tissue was cut into smaller pieces and digested with RPMI 1640 medium containing 4% fetal calf serum, 1 mg/mL collagenase II, 1 mg/mL Dispase (Gibco) and 40 μg/ml DNase I (TaKaRa) at 37°C and stirred for 20 min. Whole intestinal cell suspensions, which included LPL or IEL, were passed through a cell strainer and loaded on a 40/80% discontinuous Percoll gradient (GE Healthcare) and cells at the interface between 40% and 80% were collected and used as IELs or LPLs.
Immunization
100 μg NP- chicken gamma globulin (CGG) in alum or complete Freund's adjuvant (incomplete Freund's adjuvant with Mycobacterium tuberculosis H37RA; Difco) was intraperitoneally injected.
EAE induction and clinical scoring
Age- and sex-matched mice were immunized subcutaneously with 100 μg MOG35-55 (Genway, China) in IFA (incomplete Freund's adjuvant) supplemented with 500 μg Mycobacterium tuberculosis on day 1. On days 1 and 3, mice were injected with 300 ng pertussis toxin (List Biological Laboratories). Clinical signs were scored on a scale of 1–5: 0, no clinical sign; 0.5, partially limp tail; 1, paralyzed tail; 2, loss of coordinated movement, hind limb paresis; 2.5, one hind limb paralyzed; 3, both hind limbs paralyzed, 3.5, hind limbs paralyzed, weakness in fore limbs; 4, fore limbs paralyzed; and 5, moribund.
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNAs were extracted from sorted T cells using TRIzol reagent (Gibco-BRL, Gaithersburg, MD). Oligo (dT)-primed cDNAs were prepared by reverse transcription. For semiquantitation, 50 ng cDNA was serially diluted and subjected to PCR. All PCR products were resolved electrophoretically on 2% agarose gels and visualized by ethidium bromide staining.
Statistical analysis
Proportions of cells were compared with Student's t test (Fig. 1A). To see the effect of Runx3 in EAE recovery, the mean clinical scores were compared between genotypes using repeated measures analysis of variance (ANOVA). Greenhouse–Geiser correction was applied to adjust the degree of freedom for deviation from the sphericity assumption (Fig. 7A). All reported P values were two-tailed and P values lower than 0.05 were considered to indicate statistical significance.
References
Singer, A., Adoro, S. & Park, J. H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol 8, 788–801 (2008).
Menager-Marcq, I., Pomie, C., Romagnoli, P. & van Meerwijk, J. P. CD8+CD28- regulatory T lymphocytes prevent experimental inflammatory bowel disease in mice. Gastroenterology 131, 1775–1785 (2006).
Rifa'i, M. et al. CD8+CD122+ regulatory T cells recognize activated T cells via conventional MHC class I-alphabetaTCR interaction and become IL-10-producing active regulatory cells. Int Immunol 20, 937–947 (2008).
Suzuki, H., Shi, Z., Okuno, Y. & Isobe, K. Are CD8+CD122+ cells regulatory T cells or memory T cells? Hum Immunol 69, 751–754 (2008).
Cosmi, L. et al. Th2 cells are less susceptible than Th1 cells to the suppressive activity of CD25+ regulatory thymocytes because of their responsiveness to different cytokines. Blood 103, 3117–3121 (2004).
Poussier, P., Ning, T., Banerjee, D. & Julius, M. A unique subset of self-specific intraintestinal T cells maintains gut integrity. J Exp Med 195, 1491–1497 (2002).
Kim, H. J., Verbinnen, B., Tang, X., Lu, L. & Cantor, H. Inhibition of follicular T-helper cells by CD8(+) regulatory T cells is essential for self tolerance. Nature 467, 328–332 (2010).
Jiang, H. & Chess, L. The specific regulation of immune responses by CD8+ T cells restricted by the MHC class Ib molecule, Qa-1. Annu Rev Immunol 18, 185–216 (2000).
Tang, X. et al. Regulation of immunity by a novel population of Qa-1-restricted CD8alphaalpha+TCRalphabeta+ T cells. J Immunol 177, 7645–7655 (2006).
Kumar, V. Homeostatic control of immunity by TCR peptide-specific Tregs. J Clin Invest 114, 1222–1226 (2004).
Tang, X., Maricic, I. & Kumar, V. Anti-TCR antibody treatment activates a novel population of nonintestinal CD8 alpha alpha+ TCR alpha beta+ regulatory T cells and prevents experimental autoimmune encephalomyelitis. J Immunol 178, 6043–6050 (2007).
Zucchelli, S. et al. Defective central tolerance induction in NOD mice: genomics and genetics. Immunity 22, 385–396 (2005).
Holler, P. D. et al. The same genomic region conditions clonal deletion and clonal deviation to the CD8alphaalpha and regulatory T cell lineages in NOD versus C57BL/6 mice. Proc Natl Acad Sci U S A 104, 7187–7192 (2007).
Rocha, B., Vassalli, P. & Guy-Grand, D. The V beta repertoire of mouse gut homodimeric alpha CD8+ intraepithelial T cell receptor alpha/beta + lymphocytes reveals a major extrathymic pathway of T cell differentiation. J Exp Med 173, 483–486 (1991).
Eberl, G. & Littman, D. R. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science 305, 248–251 (2004).
Yamagata, T., Mathis, D. & Benoist, C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol 5, 597–605 (2004).
Leishman, A. J. et al. Precursors of functional MHC class I- or class II-restricted CD8alphaalpha(+) T cells are positively selected in the thymus by agonist self-peptides. Immunity 16, 355–364 (2002).
Gangadharan, D. et al. Identification of pre- and postselection TCRalphabeta+ intraepithelial lymphocyte precursors in the thymus. Immunity 25, 631–641 (2006).
Konkel, J. E. et al. Control of the development of CD8alphaalpha+ intestinal intraepithelial lymphocytes by TGF-beta. Nat Immunol 12, 312–319 (2011).
Ouyang, W., Beckett, O., Ma, Q. & Li, M. O. Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity 32, 642–653 (2010).
Rudensky, A. Y. Regulatory T cells and Foxp3. Immunol Rev 241, 260–268 (2011).
Sakaguchi, S., Miyara, M., Costantino, C. M. & Hafler, D. A. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol 10, 490–500 (2010).
Littman, D. R. & Rudensky, A. Y. Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 (2010).
Rifa'i, M., Kawamoto, Y., Nakashima, I. & Suzuki, H. Essential roles of CD8+CD122+ regulatory T cells in the maintenance of T cell homeostasis. J Exp Med 200, 1123–1134 (2004).
Smith, T. R. & Kumar, V. Revival of CD8+ Treg-mediated suppression. Trends Immunol 29, 337–342 (2008).
Huang, Y. et al. Mucosal memory CD8(+) T cells are selected in the periphery by an MHC class I molecule. Nat Immunol (2011).
Madakamutil, L. T. et al. CD8alphaalpha-mediated survival and differentiation of CD8 memory T cell precursors. Science 304, 590–593 (2004).
Uematsu, S. et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol 9, 769–776 (2008).
Mucida, D. et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317, 256–260 (2007).
Fanchiang, S. S. et al. Global expression profiling of peripheral Qa-1-restricted CD8alphaalpha+TCRalphabeta+ regulatory T cells reveals innate-like features: Implications for immune-regulatory repertoire. Hum Immunol (2011).
Wang, L. et al. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity 29, 876–887 (2008).
Setoguchi, R. et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825 (2008).
Taniuchi, I. et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002).
Sugai, M., Watanabe, K., Nambu, Y., Hayashi, T. & Shimizu, A. Functions of Runx in IgA class switch recombination. J Cell Biochem (2010).
Watanabe, K. et al. Requirement for Runx proteins in IgA class switching acting downstream of TGF-beta 1 and retinoic acid signaling. J Immunol 184, 2785–2792 (2010).
Iwata, M. et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity 21, 527–538 (2004).
Tezuka, H. et al. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature 448, 929–933 (2007).
Hall, J. A. et al. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity 34, 435–447 (2011).
Pino-Lagos, K. et al. A retinoic acid-dependent checkpoint in the development of CD4+ T cell-mediated immunity. J Exp Med 208, 1767–1775 (2011).
Sugai, M. et al. Runx3 is required for full activation of regulatory T cells to prevent colitis-associated tumor formation. J Immunol 186, 6515–6520 (2011).
Koh, D. R. et al. Less mortality but more relapses in experimental allergic encephalomyelitis in CD8−/− mice. Science 256, 1210–1213 (1992).
Jiang, H., Zhang, S. I. & Pernis, B. Role of CD8+ T cells in murine experimental allergic encephalomyelitis. Science 256, 1213–1215 (1992).
Panoutsakopoulou, V. et al. Suppression of autoimmune disease after vaccination with autoreactive T cells that express Qa-1 peptide complexes. J Clin Invest 113, 1218–1224 (2004).
Jiang, H. et al. T cell vaccination induces T cell receptor Vbeta-specific Qa-1-restricted regulatory CD8(+) T cells. Proc Natl Acad Sci U S A 95, 4533–4537 (1998).
Hu, D. et al. Analysis of regulatory CD8 T cells in Qa-1-deficient mice. Nat Immunol 5, 516–523 (2004).
Kumar, V. & Sercarz, E. E. The involvement of T cell receptor peptide-specific regulatory CD4+ T cells in recovery from antigen-induced autoimmune disease. J Exp Med 178, 909–916 (1993).
Das, G. & Janeway, C. A., Jr MHC specificity of iIELs. Trends Immunol 24, 88–93 (2003).
Kitoh, A. et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity 31, 609–620 (2009).
Gapin, L., Cheroutre, H. & Kronenberg, M. Cutting edge: TCR alpha beta+ CD8 alpha alpha+ T cells are found in intestinal intraepithelial lymphocytes of mice that lack classical MHC class I molecules. J Immunol 163, 4100–4104 (1999).
Das, G. & Janeway, C. A., Jr Development of CD8alpha/alpha and CD8alpha/beta T cells in major histocompatibility complex class I-deficient mice. J Exp Med 190, 881–884 (1999).
Acknowledgements
We thank Dr K. Ikuta (Kyoto University, Virus Institute) for discussion of the findings and for CD45.1 mice, Dr F. Alt (Harvard University) for Rag2−/− mice and Dr Y. Yokota (Fukui University) for critically reading the manuscript. This work was supported in part by Grants-In-Aid for Science Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Contributions
Y.N. and M.S. performed the experiments; T.H., K.-J.J., K.N., H.M., K.A. and K.T. assisted with experiments; M.S. designed and conceptualized the study; Y.I. and T.K. provided critical materials; S.T. performed statistical analysis; Y.N., K.A., M.O., K.I. and J.F. made comments; A.S. , Y.N. and M.S. interpret the data; and M.S. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Figure 1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Nambu, Y., Hayashi, T., Jang, KJ. et al. In situ differentiation of CD8αα Τ cells from CD4 T cells in peripheral lymphoid tissues. Sci Rep 2, 642 (2012). https://doi.org/10.1038/srep00642
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00642
This article is cited by
-
Non-canonicaly recruited TCRαβCD8αα IELs recognize microbial antigens
Scientific Reports (2018)
-
Changing course by lymphocyte lineage redirection
Nature Immunology (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.