Abstract
Background
Genome, transcriptome and methylome-wide association studies have identified single-nucleotide polymorphisms (SNPs) or genes at 258 loci associated with colorectal cancer (CRC) risk. We studied the relationship between these and patient outcome.
Methods
We studied 1926 unrelated patients with advanced CRC from COIN and COIN-B. Of 205 CRC-risk SNPs, 19 were directly genotyped and 162 were imputed, and of 53 risk genes, 52 were tested. An additive Cox model for overall survival was adjusted for known prognostic factors. For nominally significant SNPs or genes, we considered a recessive model with a Bonferroni corrected threshold of P = 2.1 × 10−4. We examined SNPs as expression quantitative trait loci (eQTL) and the relationship between gene expression in colorectal tumours and survival in 597 unrelated patients.
Results
Eleven SNPs or genes were nominally associated with survival under an additive model. Only rs117079142 mapping to UTP23 and EIF3H (Hazard Ratio [HR] = 2.79, 95% Confidence Intervals [CI] = 1.70–4.58, P = 4.7 × 10−5) and rs9924886 mapping to CDH1 and CDH3 (HR = 1.24, 95% CI = 1.12–1.38, P = 5.2 × 10−5) passed the multiple testing threshold under a recessive model. rs117079142 was an eQTL for UTP23 and rs9924886 for CDH1, CDH3 and ZFP90. Decreased CDH1 expression in CRCs was associated with worse survival (HR = 2.18, 95% CI = 1.3–3.5, P = 1.8 × 10−3).
Conclusion
rs117079142 and rs9924886 may represent potential prognostic biomarkers for CRC.
Similar content being viewed by others
Introduction
Genome-wide association studies (GWAS) have identified single-nucleotide polymorphisms (SNPs) associated with risk of developing colorectal cancer (CRC) [1]. Some studies have suggested that a subset of these may also influence patient survival [2,3,4,5,6,7] although other studies have not supported these observations [8,9,10,11]. We previously studied the relationship between SNP genotype and patient outcome for 83 CRC-risk SNPs [12] by analysing patients with advanced CRC from the COIN and COIN-B clinical trials [13, 14]. A recent meta-analysis of all available GWAS augmented by transcriptome and methylome-wide association studies (TWAS and MWAS, respectively) has identified further loci taking the total number of CRC-risk loci to 258 [15].
To gain a more comprehensive understanding of the relationship between inherited genetic variation and patient survival, we assessed 233 of these risk loci for their prognostic role in 1926 patients from COIN and COIN-B.
Materials and methods
Patients and genotyping
Germline DNAs were extracted from EDTA venous blood samples from 2244 unrelated patients with metastatic or locally advanced colorectal adenocarcinoma participating in the MRC clinical trials COIN (NCT00182715) [13] and COIN-B (NCT00640081) [14]. All patients gave fully informed consent for bowel cancer research (approved by NHS Research Ethics Committee [04/MRE06/60]). COIN patients were randomised 1:1:1 to receive continuous oxaliplatin and fluoropyrimidine chemotherapy, continuous chemotherapy and cetuximab, or intermittent chemotherapy. COIN-B patients were randomised 1:1 to receive intermittent chemotherapy and cetuximab, or intermittent chemotherapy and continuous cetuximab. There was no heterogeneity in overall survival (OS; time from trial randomisation to death or end of trial) between patients when analysed by trial, trial arm, type of chemotherapy received, or cetuximab use [12], so we combined groups for survival analyses. Patient DNAs were genotyped using Affymetrix Axiom Genome-Wide CEU 1 Human Mapping Arrays [16].
Prediction of untyped SNPs was carried out using IMPUTE2 v2.3.0 [17] based on data from the 1000 Genomes Project as reference [18, 19]. After quality control (QC), SNP genotypes were available on 1950 patients. Two patients had no data on survival and a further 22 lacked clinicopathological data leaving 1926 for analysis (of which 1435 died at censorship).
SNPs and genes analysed
For the 205 CRC-risk SNPs, 19 were directly genotyped, 162 were imputed and 24 were not analysed (one because it was on the X-chromosome which was not genotyped, 19 had INFO scores <0.7 and 4 had minor allele frequencies [MAFs] <0.01). Therefore, in total, 181 CRC-risk SNPs were tested for an association with OS.
For the CRC-risk genes identified from TWAS and MWAS, we used data from a GWAS of COIN and COIN-B (2.9 million SNPs post-QC; [16]). SNPs were mapped to a region spanning 35 kilobases before and 10 kilobases after the transcription zone and analysed using MAGMA v1.07b [20]. Of the 53 genes, 52 were successfully analysed (one had insufficient SNPs in their annotation window).
Statistical analysis
The relationship between genotype and OS was determined using an additive Cox survival model adjusting for 11 prognostic covariates previously identified in COIN and COIN-B: WHO performance status (P = 3.1 × 10−23), resection status of the primary tumour (P = 1.8 × 10−21), WBC count (P = 1.2 × 10−31), platelet count (P = 1.7 × 10−29), alkaline phosphatase levels (P = 1.5 × 10−27), number of metastatic sites (P = 1.7 × 10−13), liver metastases (P = 1.3 × 10−4), site of primary tumour (P = 9.1 × 10−9), surface area of primary tumour (P = 1.1 × 10−5), time from diagnosis to metastases (P = 1.7 × 10−7), and metachronous versus synchronous metastases (P = 6.0 × 10−8) [21]. For gene level analysis in MAGMA, SNP P-values were assessed with the linkage disequilibrium (LD) between them using the multi=snp-wise option. This model takes advantage of the sum of the -log10(P) for all SNPs, as well as the top SNP associations within each gene, to assess the association of their constituent genes. For any SNPs or genes nominally associated with OS (P < 0.05), we also considered a recessive model to uncover associations potentially missed under additive analyses [22]. We used Bonferroni correction to address multiple testing with P < 2.1 × 10−4 being considered statistically significant (0.05/233 SNPs or genes tested). Based on the number of patients analysed, our analysis provided over 70% power to demonstrate a HR of 1.2 for SNPs with MAFs >0.30. Power was calculated using the ‘survSNP.power.table’ function from the ‘survSNP’ package in R [23].
Bioinformatic analyses
We queried the GTEx [24] database to examine SNPs as potential expression quantitative trait loci (eQTLs) for neighbouring genes. Significance for tissue association was set at P < 1.0 × 10-3 (Bonferroni correction for 49 tissues [0.05/49]). We correlated gene expression with survival by analysing tumours from 597 patients with CRC from The Human Protein Atlas (THPA) [25]. RNA-seq data was reported as median number of fragments per kilobase of exon per million reads (FPKM) [26]. Samples were classified as high expression using the thresholds recommended by THPA (for CDH1 FPKM was >137; https://www.proteinatlas.org/ENSG00000039068-CDH1/pathology/colorectal+cancer). A log-rank P-value was obtained for a difference in survival between patients with CRCs with high and low expression levels. We also performed survival analysis using a linear Cox-proportional hazards model.
Results
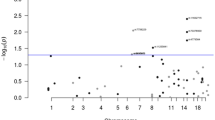
In total, we had survival, clinicopathological and germline genotyping data on 1926 patients with advanced CRC (Table 1). We found that eight CRC-risk SNPs (rs13086367 at 3q13.2, rs280097 at 4q22.2, rs16892766 at 8q23.3, rs117079142 at 8q24.11, rs11255841 at 10p14, rs4444073 at 11p15.4, rs1497077 at 14q22.1 and rs9924886 at 16q22.1) and three CRC-risk genes (EPB41L2, ADAMTS15 and F2), were nominally associated with survival under an additive model (Table 2, Supplementary Table 1).
Only rs117079142 (MAF = 0.06, HR = 2.79, 95% CI = 1.70–4.58, P = 4.7 × 10−5) and rs9924886 (MAF = 0.25, HR = 1.24, 95% CI = 1.12–1.38, P = 5.2 × 10−5) passed the threshold for multiple testing when considered under a recessive model (Table 2). Patients homozygous for the rs117079142 minor allele (n = 4) had a median survival of 198 days compared to 420 days for heterozygotes (n = 204) and 497 days for patients homozygous for the major allele (n = 1724) (Fig. 1). Patients homozygous for the rs9924886 minor allele (n = 113) had a median survival of 385 days compared to 487 days for heterozygotes (n = 715) and 507 days for patients homozygous for the major allele (n = 1026) (Fig. 1).
Kaplan–Meier Plots for a rs117079142 and b rs9924886. P-values are for multivariate recessive Cox-regression models and patients are grouped by number of copies of the minor allele. The relationship between genotype and overall survival was adjusted for eleven prognostic covariates: WHO performance status, resection status of the primary tumour, white blood cell count, platelet count, alkaline phosphatase levels, number of metastatic sites, liver metastases, site of primary tumour, surface area of primary tumour, time from diagnosis to metastases and metachronous versus synchronous metastases.
rs117079142 was an eQTL for UTP23 (Supplementary Table 2) and rs9924886 was an eQTL for CDH1, CDH3 and ZFP90 (Supplementary Table 2) in multiple tissues, but neither were significant in the sigmoid or transverse colon. Low CDH1 expression in CRCs was associated with worse survival in patients from THPA (5-year survival: low CDH1 expression = 58%, high CDH1 expression = 71%, HR = 2.18, 95% CI = 1.3–3.5, P = 1.8 × 10−3; linear Cox-proportional hazards model P = 2.8 × 10−2). UTP23, EIF3H and CDH3 expression levels were not associated with survival.
Discussion
In this study, we investigated the relationship between CRC-risk variants and patient outcome. We identified two SNPs associated with survival under a recessive model that were significant beyond the threshold for multiple testing. Interestingly, both SNPs were only nominally significant under additive analyses and others have previously reported on the value of considering recessive models to uncover associations potentially missed [22]. rs117079142 had a modest effect size (HR = 2.79), but relatively low frequency in our cohort; furthermore, in the 1000 genomes dataset the MAF ranges from 0.0076 in the African population to 0.073 in South Asians. In contrast, rs9924886 was more commonly observed in our cohort (and was 0.178 in the African population and 0.3095 in East Asians), but the effect size was lower. These data suggest that neither SNPs are likely to have a direct clinical impact although their identification helps inform potential therapeutic targets.
rs117079142 lies 4 kb downstream of UTP23. UTP23 codes for part of the 90S pre-ribosome and is required for 18S rRNA early processing. Reduced UTP23 expression has been associated with poor prognosis in patients with ovarian cancer possibly by affecting sensitivity to paclitaxel-based chemotherapy [27]. rs117079142 also lies 23 kb downstream of EIF3H, which regulates translation through its interaction with the 40S ribosome and other initiation factors. EIF3 subunits are thought to have oncogenic potential [28] through increased protein synthesis of oncoproteins such as cyclinD1, c-Myc, FGF2 and ornithine decarboxylase [29].
rs9924886 in CDH3 is a strong proxy for rs9929218 (D’ = 0.95 and r2 = 0.80) and rs9939049 (D’ = 0.96 and r2 = 0.80) in CDH1 (encoding E-cadherin) that we previously identified as a prognostic biomarker in CRC [12, 30]. Others have also demonstrated a relationship between rs9929218 and survival in CRC patients from Korea [31] and Spain [5]. rs9924886, rs9929218 and rs9939049 are in strong LD with rs16260 [32] in the CDH1 promoter, which down-regulates CDH1 expression [33]. Patients homozygous for the minor alleles of these variants would be expected to have reduced E-cadherin expression. Mechanistically, our data are consistent with the downregulation of CDH1 affecting survival. First, we found that patients homozygous for the rs9924886 minor allele had worse survival and second, we observed that patients with low CDH1 expression in their colorectal tumours had worse outcome. E-cadherin functions as a transmembrane glycoprotein involved in intercellular adhesion, cell polarity and tissue morphology and regeneration [34], and its loss is a key feature of epithelial to mesenchymal transition during metastasis. Together, these data support a prognostic role for CDH1 in colorectal tumourigenesis.
rs10161980 has been previously associated with survival from CRC under a recessive model [22]. However, we failed to replicate this SNP in COIN and COIN-B despite having over 98% power. rs10161980 may therefore represent a false-positive or a prognostic biomarker that is specific to patients with earlier stages of disease (we only considered patients with advanced disease in our analyses).
In conclusion, our work provides support for the importance of germline variation as a determinant of patient outcome. Understanding the biological basis of these relationships provides a focus for future work with the goal of identifying novel therapeutic targets for the treatment of CRC.
Data availability
Additional data are available in the Supplementary Information. Scripts are available at https://github.com/Chris-Wills/Wills_et_al_2023_Survival_SNPs.
Change history
05 April 2024
A Correction to this paper has been published: https://doi.org/10.1038/s44276-024-00040-2
References
Law PJ, Timofeeva M, Fernandez-Rozadilla C, Broderick P, Studd J, Fernandez-Tajes J, et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat Commun. 2019;10:2154 https://doi.org/10.1038/s41467-019-09775-w.
Phipps AI, Newcomb PA, Garcia-Albeniz X, Hutter CM, White E, Fuchs CS, et al. Association between colorectal cancer susceptibility loci and survival time after diagnosis with colorectal cancer. Gastroenterology. 2012;143:51–U555. https://doi.org/10.1053/j.gastro.2012.04.052.
Dai JY, Gu J, Huang MS, Eng C, Kopetz ES, Ellis LM, et al. GWAS-identified colorectal cancer susceptibility loci associated with clinical outcomes. Carcinogenesis. 2012;33:1327–31. https://doi.org/10.1093/carcin/bgs147.
Garcia-Albeniz X, Nan H, Valeri L, Morikawa T, Kuchiba A, Phipps AI, et al. Phenotypic and tumor molecular characterization of colorectal cancer in relation to a susceptibility SMAD7 variant associated with survival. Carcinogenesis. 2013;34:292–8. https://doi.org/10.1093/carcin/bgs335.
Abuli A, Lozano JJ, Rodriguez-Soler M, Jover R, Bessa X, Munoz J, et al. Genetic susceptibility variants associated with colorectal cancer prognosis. Carcinogenesis. 2013;34:2286–91. https://doi.org/10.1093/carcin/bgt179.
Takatsuno Y, Mimori K, Yamamoto K, Sato T, Niida A, Inoue H, et al. The rs6983267 SNP is associated with MYC transcription efficiency, which promotes progression and worsens prognosis of colorectal cancer. Ann Surg Oncol. 2013;20:1395–402. https://doi.org/10.1245/s10434-012-2657-z.
Morris EJA, Penegar S, Whiffin N, Broderick P, Bishop DT, Northwood E, et al. A retrospective observational study of the relationship between single nucleotide polymorphisms associated with the risk of developing colorectal cancer and survival. PLoS ONE. 2015;10:11 https://doi.org/10.1371/journal.pone.0117816.
Tenesa A, Theodoratou E, Din FV, Farrington SM, Cetnarskyj R, Barnetson RA, et al. Ten common genetic variants associated with colorectal cancer risk are not associated with survival after diagnosis. Clin Cancer Res. 2010;16:3754–9. https://doi.org/10.1158/1078-0432.Ccr-10-0439.
Hoskins JM, Ong PS, Keku TO, Galanko JA, Martin CF, Coleman CA, et al. Association of eleven common, low-penetrance colorectal cancer susceptibility genetic variants at six risk loci with clinical outcome. PLoS ONE. 2012;7 https://doi.org/10.1371/journal.pone.0041954.
Sanoff HK, Renfro LA, Poonnen P, Ambadwar P, Sargent DJ, Goldberg RM, et al. Germline variation in colorectal risk Loci does not influence treatment effect or survival in metastatic colorectal cancer. PLoS ONE. 2014;9:e94727 https://doi.org/10.1371/journal.pone.0094727.
He Y, Theodoratou E, Li X, Din FVN, Vaughan-Shaw P, Svinti V, et al. Effects of common genetic variants associated with colorectal cancer risk on survival outcomes after diagnosis: a large population-based cohort study. Int J Cancer. 2019;145:2427–32. https://doi.org/10.1002/ijc.32550.
Summers MG, Maughan TS, Kaplan R, Law PJ, Houlston RS, Escott-Price V, et al. Comprehensive analysis of colorectal cancer-risk loci and survival outcome: a prognostic role for CDH1 variants. Eur J Cancer. 2020;124:56–63. https://doi.org/10.1016/j.ejca.2019.09.024.
Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, et al. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103–14. https://doi.org/10.1016/s0140-6736(11)60613-2.
Adams RA, Meade AM, Seymour MT, Wilson RH, Madi A, Fisher D, et al. Intermittent versus continuous oxaliplatin and fluoropyrimidine combination chemotherapy for first-line treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet Oncol. 2011;12:642–53. https://doi.org/10.1016/s1470-2045(11)70102-4.
Fernandez-Rozadilla C, Timofeeva M, Chen Z, Law P, Thomas M, Schmit S, et al. Deciphering colorectal cancer genetics through multi-omic analysis of 100,204 cases and 154,587 controls of European and east Asian ancestries. Nat Genet. 2023;55:89–99. https://doi.org/10.1038/s41588-022-01222-9.
Al-Tassan NA, Whiffin N, Hosking FJ, Palles C, Farrington SM, Dobbins SE, et al. A new GWAS and meta-analysis with 1000 Genomes imputation identifies novel risk variants for colorectal cancer. Sci Rep. 2015;5:10442 https://doi.org/10.1038/srep10442.
Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529 https://doi.org/10.1371/journal.pgen.1000529.
Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3: Genes|Genom|Genet. 2011;1:457 https://doi.org/10.1534/g3.111.001198.
Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, et al. A global reference for human genetic variation. Nature. 2015;526:68 https://doi.org/10.1038/nature15393.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:19 https://doi.org/10.1371/journal.pcbi.1004219.
Wills C, He Y, Summers MG, Lin Y, Phipps AI, Watts K, et al. A genome-wide search for determinants of survival in 1926 patients with advanced colorectal cancer with follow-up in over 22,000 patients. Eur J Cancer. 2021;159:247–58. https://doi.org/10.1016/j.ejca.2021.09.047.
He YZ, Timofeeva M, Zhang XM, Xu W, Li X, Din FVN, et al. Colorectal cancer risk variants rs10161980 and rs7495132 are associated with cancer survival outcome by a recessive mode of inheritance. Int J Cancer. 2021;148:2774–8. https://doi.org/10.1002/ijc.33465.
Owzar K, Li Z, Cox N, Jung S-H. Power and sample size calculations for SNP association studies with censored time-to-event outcomes. Genet Epidemiol. 2012;36:538–48.
Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobanking. 2015;13:307–8. https://doi.org/10.1089/bio.2015.29031.hmm.
Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science. 2015;347:10 https://doi.org/10.1126/science.1260419.
Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:660 https://doi.org/10.1126/science.aan2507.
Fu ZQ, Wang CH, Chen YQ, Zhang X, Wang XY, Xie X. Down-regulation of UTP23 promotes paclitaxel resistance and predicts poorer prognosis in ovarian cancer. Pathol Res Pract. 2019;215:6 https://doi.org/10.1016/j.prp.2019.152625.
Zhang LL, Pan XY, Hershey JWB. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282:5790–800. https://doi.org/10.1074/jbc.M606284200.
Zhang LL, Smit-McBride Z, Pan XY, Rheinhardt J, Hershey JWB. An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3. J Biol Chem. 2008;283:24047–60. https://doi.org/10.1074/jbc.M800956200.
Smith CG, Fisher D, Harris R, Maughan TS, Phipps AI, Richman S, et al. Analyses of 7,635 patients with colorectal cancer using independent training and validation cohorts show that rs9929218 in CDH1 is a prognostic marker of survival. Clin Cancer Res. 2015;21:3453–61. https://doi.org/10.1158/1078-0432.ccr-14-3136.
Song N, Kim K, Shin A, Park JW, Chang HJ, Shi JJ, et al. Colorectal cancer susceptibility loci and influence on survival. Genes Chromosom Cancer. 2018;57:630–7. https://doi.org/10.1002/gcc.22674.
Pittman AM, Twiss P, Broderick P, Lubbe S, Chandler I, Penegar S, et al. The CDH1-160C > A polymorphism is a risk factor for colorectal cancer. Int J Cancer. 2009;125:1622–5. https://doi.org/10.1002/ijc.24542.
Li LC, Chui RM, Sasaki M, Nakajima K, Perinchery G, Au HC, et al. A single nucleotide polymorphism in the E-cadherin gene promoter alters transcriptional activities. Cancer Res. 2000;60:873–6.
Takeichi M. Cadherin cell-adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–5. https://doi.org/10.1126/science.2006419.
Acknowledgements
We thank the patients and their families who participated and gave their consent for this research, and the investigators and pathologists throughout the UK who submitted samples for assessment. COIN and COIN-B were conducted with the support of the National Institute of Health Research Cancer Research Network. The study sponsors had no involvement in the study design, collection, analysis and interpretation of the data, the writing of the report, nor the decision to submit this article for publication.
Funding
This work was supported by Tenovus Cancer Care, Cancer Research Wales and Cardiff University School of Medicine. The work of the Houlston laboratory was supported by Cancer Research UK (C1298/A8362). DF was funded by the UK Medical Research Council under award number MC_UU_00004/06. The COIN and COIN-B trials were funded by Cancer Research UK and an unrestricted educational grant from Merck-Serono.
Author information
Authors and Affiliations
Contributions
JPC obtained funding for and directed this study. The study was designed by CW and JPC. TSM was CI of COIN and provided clinical advice and supported the translational research. DF facilitated access to the clinical data and RSH oversaw the genotype imputation and quality control. CW and AH undertook the statistical analyses with supervision from VEP and JPC. CW and JPC interpreted the data with input from AH, KW, HDW and VEP. CW wrote the first draft of the paper with subsequent input from JPC, and all authors provided comments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All patients gave fully informed consent for bowel cancer research (approved by REC [04/MRE06/60]).
Consent for publication
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wills, C., Houseman, A., Watts, K. et al. Relationship between 233 colorectal cancer risk loci and survival in 1926 patients with advanced disease. BJC Rep 1, 2 (2023). https://doi.org/10.1038/s44276-023-00003-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44276-023-00003-z