Abstract
Water quality testing does not recognise antimicrobial resistance (AMR) and is often limited to indicators of faecal contamination Escherichia coli and Enterococcus species. In Europe, data on AMR in drinking water is scarce. In Ireland, as in many countries, household drinking water is supplied via mains or via private wells or water schemes. Using citizen science, we identified Irish private drinking water supplies as reservoirs of antimicrobial resistant bacteria (ARB). Gram-negative (n = 464) and Gram-positive (n = 72) bacteria were isolated. We identified instances of potentially opportunistic ARB such as Enterobacter cloacae, Acinetobacter baumannii and Enterococcus species. We report reservoirs of multidrug resistance in Enterococcus casseliflavus, E. cloacae, E. coli, Stenotrophomonas maltophilia, and Serratia rubidaea. We also identified linezolid-resistant Enterococcus in Irish drinking water. Linezolid is a last-resort antibiotic used to treat vancomycin-resistant Enterococcus sp. Additionally, we identified mobile AMR in three water samples, two of which were carried on IncF group, one on IncQ and five on Col-like plasmids. Our work suggests that private drinking water is a potential sink and source of AMR pathogens. This highlights a value of drinking water surveillance in a One Health framework as the surveillance would provide information regarding the movement and persistence of ARB and ARGs that are able to survive in drinking water and subsequently have the opportunity to be mobilised through humans; linking the environment to the human and potentially threatening human health.
Similar content being viewed by others
Introduction
The World Health Organisation sets out minimum international guidelines for mitigating faecal contaminants in drinking water1. Drinking water becomes contaminated with faecal coliforms due to anthropogenic activities such as manure spreading and leakage of wastewater treatment systems2,3. While the current water quality guidelines aim to reduce instances of faecal contamination, they fail to consider the potential of water to be a reservoir for antimicrobial resistance genes (ARGs) conferring resistance against clinically relevant antimicrobials. Antimicrobial resistance (AMR) is a global epidemic that is recognised by the World Health Organisation as one of the top ten public health issues facing the human population4 and makes up a key component of the One Health approach to improving public health outcomes5. Horizontal gene transfer (HGT) of mobile resistance elements has been documented as a leading cause for the dissemination and persistence of ARGs6. In recent years, studies have revealed the presence of antimicrobial resistant bacteria (ARB) in drinking water supplies globally7,8,9,10,11,12,13. The emergence of ARB and ARGs in drinking water raises concerns regarding the transmission of AMR from the environment to the human and the potential impact on human health. In the human gut, HGT is a common occurrence14. The potential for bacteria to acquire ARGs via HGT in the gut following consumption of water contaminated with ARB has been explored in mice15 however, the interlink between consumption of ARGs through drinking water and the impact this has on the human gut microbiome is yet to be explored. If this potential is realised in the human gut, this could possibly lead to gastrointestinal illnesses16 and even bloodstream infections that are more challenging to treat17.
The aim of this work was to employ citizen science to isolate and identify bacterial reservoirs of antimicrobial resistance in private sources of drinking water in Ireland and to elucidate the role of mobile AMR in the drinking water resistomes.
Results
Water samples and household background
The water sampling and citizen science project occurred during the COVID pandemic, where tight restrictions on movement were in place across Ireland. We collected data regarding the sampling locations and the use of antimicrobials in each household by means of a written questionnaire (Supplementary Questionnaire 1). Questionnaires from all volunteers (n = 49 households) situated across Ireland were received. Several households (n = 19) relied on privately sourced drinking water. Of those, 18 reported owning a private well, one reported sourcing water from a spring. Additionally, 30 households relied on Group Water Schemes (GWS). The GWS supplied drinking water via springs (n = 16, 53%) or wells (n = 5, 17%). The remainder of households supplied by GWS (n = 9, 30%) did not indicate the source of their drinking water. Of the households that reported wells as their source of drinking water (n = 23), the majority reported having a bored (n = 10, 43%) or drilled well (n = 8, 35%) and, dug wells (n = 3, 13%) were the least common. One household reported that their well was both dug and drilled. Two households did not report how the wells were constructed. A significant number of households did not use filtration (n = 44, 90%) nor treatment systems (n = 43, 88%) to reduce microbial contamination in their drinking water. Only six (12%) households reported the use of chlorination, five of which were supplied water from the same GWS, the other is supplied by a different GWS. Of these households, one also reported the use of ultraviolet (UV) treatment. The GWS and private wells were constructed between 1973 and 2019. The depth of the wells ranged between 1.3 m–160 m.
Overall, the majority (n = 33, 67%) of households reported living on or within 5 km of farmland. Although not asked to specify, one household reported the presence of sheep and cattle on the household farm (Household 6) and one household reported having horses on the farm and the presence of a cattle farm within 1 km of residence (Household 44). Most households (n = 31, 63%) considered their surroundings to be remote rural or rural community. A smaller number (n = 18, 37%) reported proximity to surface waters such as rivers and lakes. Others reported living near quarries (n = 12, 24%), landfills (n = 4, 8%) and/or manufacturing and processing facilities (n = 4, 8%). Households living on farms (n = 12, 24%) did not use antibiotics on their farm animals at any stage since the arrival of the animals to the farms, whilst over 59% (n = 29) of households reported use of antibiotics in the past 10 years to treat human infections. The beta-lactam antibiotics (penicillin, amoxicillin-clavulanate) were the most frequently reported (n = 20, 41%). Cephalosporins and other antibiotics were less common (n = 2, 4%). A small number of households (n = 8, 16%) were either unsure or did not indicate the type of antibiotics they have consumed.
Correlation analysis
We assessed for correlation between the reliance on domestic wastewater systems the detection of ARB in drinking water samples. We identified a moderate positive correlation between the use of septic tanks and the prevalence of antimicrobial resistant bacteria (Spearman’s rS value + 0.649, α = 0.001). Additionally, correlation analysis for households that reported the use of antimicrobials revealed Spearman’s rS value of + 0.716 (α = 0.002), suggesting a positive correlation between the use of antimicrobials and the presence of bacteria of clinical relevance. These households were more likely to harbour potentially pathogenic species (e.g. E. faecium, E. cloacae, S. maltophilia, E. coli).
Isolation and identification of bacterial reservoirs of AMR
We isolated bacteria on selective agars with and without antibiotics and used MALDI-TOF-MS and 16 S rRNA sequencing to speciate the isolates which were subsequently tested against a variety of antimicrobials. Of 49 household samples retrieved, 21 arrived later than 24 h after sampling. We isolated bacteria from all but one sample arriving to our laboratory within 24 h of sample collection (n = 21) (Supplementary Table 1). As this work focuses on AMR bacteria rather than quantification of faecal bacteria, we deviated from ISO quality testing standards and processed all samples, including those arriving beyond the recommended 24 h mark. In some instances, we were able to isolate bacteria from samples arriving beyond the 24 h period (n = 6) but we were only able to isolate bacteria from one household where the water sample arrived 96-h after collection. The longest time between collection and processing was 96 h.
Overall, 536 isolates constituting 464 Gram-negative and 72 Gram-positive bacteria were isolated from 28 of 49 household samples. Bacterial isolates were not detected in samples from 21 households, 20 of which were those that were delayed (>24 h) arriving to the laboratory (Supplementary Table 1) and thus it is possible that a loss of viability occurred in this time period. Bacteria resistant to at least one class of antimicrobial were detected in 22 of 28 (78.6%) households (Fig. 1).
Household and Group Water Scheme numbers are confidential identifiers. The households are classified based on private ownership of the water supply or dependence on Group Water Schemes (S1–S6). The geographical breakdown of households is focused on the East & Midlands, the West or Northern Ireland. For each region, the number of households harbouring viable bacteria is indicated. These bacteria are grouped based on phenotype (AMR or MDR). The households and species which harboured MDR isolates are shown. GWS: Group Water Scheme, AMR: Antimicrobial-Resistant, MDR: Multidrug-Resistant.
In total, 244 of 536 isolates (45.5%) were resistant to one or more of the antimicrobial agents tested. We identified a range of bacteria acting as reservoirs for antimicrobial resistance against a broad-range of antimicrobial classes such as the beta-lactams, cephalosporins and fluoroquinolones amongst others (Table 1). Ampicillin-resistance was found predominantly in species which are known to produce intrinsic AmpC beta-lactamases such as Citrobacter sp. And Buttiauxella sp. AmpC producers made up 68.5% (n = 367) of total isolated bacteria. Acquired ampicillin-resistance was detected in E. faecium and E. coli isolated from Household 36. Separately, five households from the same Group Water Scheme (S6) were also reservoirs for ampicillin-resistant E. faecium.
Carbapenem-resistance was predominantly identified in known carbapenemase producing species S. maltophilia, which made up 8.4% (n = 45) of total isolated bacteria. Only two isolates of H. alvei from the same household were resistant to ertapenem. No other carbapenem-resistance was identified. Cephalosporin-resistance was identified in ten species of bacteria isolated from twelve unique households. Cefotaxime-resistance was more frequently identified compared to ceftazidime.
Enterococcus sp. from two unrelated households were susceptible to vancomycin but were resistant to the last-resort antimicrobial, linezolid. These species were E. durans and E. faecium. Additionally, linezolid-resistance was detected in E. casseliflavus which is known to carry a chromosomal vanC gene allowing the expression of low-level vancomycin-resistance18.
As our methods did not follow the ISO standards for faecal contamination detection using E. coli and Enterococcus sp. the presence of E. coli and Enterococcus sp. may be below the limit of detection of the ISO tests due to the variation in our methods. Therefore, we cannot state that faecal contamination has occurred. Escherichia coli and Enterococcus sp were amongst the bacteria identified in 10% (n = 5) and 20% (n = 10) of households, respectively. 60% of all E. coli (n = 24 of 40) and 73% of all Enterococcus (53 of 72) isolated were resistant to at least one class of antimicrobials.
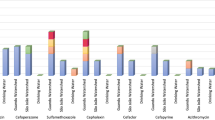
We also identified AMR species of clinical relevance (Fig. 2) including potentially opportunistic pathogens. Antimicrobial-resistant Enterobacter sp. made up 28% (14 of 50) of isolated Enterobacter sp., thirteen of which were E. cloacae and one E. hormaechei. Antimicrobial resistant S. maltophilia made up 27.5% (19 of 69) of isolated S. maltophilia and AMR Acinetobacter sp. made up 9.5% (4 of 42) of isolated Acinetobacter sp. Three were A. baumannii and one A. schindleri. Whilst a large number of Citrobacter sp. were isolated (n = 66) including twenty C. freundii (30%) isolates, AMR was only identified in C. gillenii (n = 3, 4.5%). No AMR Pseudomonas sp. were identified.
The bar chart shows the different species of clinical relevance and the numbers which were identified as being antimicrobial resistant. The blue denotes the total number of isolates from each species of relevance, red signifies the number of isolates identified as resistant to at least one class of antimicrobial and yellow indicates resistance to at minimum one class of antimicrobials. MDR: Multidrug-Resistant.
Three households from the East & Midlands (Household 8, 10 and 36) were reservoirs for MDR bacteria. The MDR bacteria isolated included two S. maltophilia (Household 8) isolates which were resistant to ceftazidime, chloramphenicol, and levofloxacin. One isolate of E. cloacae (Household 11) was resistant to cefotaxime, tetracycline, and ciprofloxacin. From Household 36, sixteen isolates of E. coli and five isolates of S. rubidaea were resistant to multiple combinations of antimicrobial classes (Fig. 3) including the tetracyclines and phenicols. One isolate of E. casseliflavus (Household 36) was resistant to linezolid, tetracycline, and chloramphenicol. These households have a few things in common: they all report living in a remote rural area, their water supply is a well that is 20+ years old and between 15–30 m in depth, all three depend on a domestic wastewater treatment system and none of them report treating their water against microbial contamination. While these characteristics were not unique to these three households, the aforementioned are a few risk factors that may have contributed to the presence of MDR bacteria.
The resistance patterns for multidrug-resistant (a) Escherichia coli and (b) antimicrobial resistant Serratia rubideae shows similarity in resistance patterns against phenicols, aminoglycosides and tetracycline. Blue represents susceptibility, yellow signifies intermediate resistance and red denotes resistance of the isolate to the antimicrobial. The numbers assigned are arbitrary identifiers. AK amikacin, AMP ampicillin, C chloramphenicol, CN gentamicin, COL colistin, CTX cefotaxime, K kanamycin, TET tetracycline, CAZ ceftazidime, CIP ciprofloxacin, IMP imipenem, W trimethoprim.
Overall, the identification of bacteria of clinical relevance (E. coli, Enterococcus sp., Enterobacter sp., S. rubidaea, S. maltophilia) in drinking water as reservoirs for MDR is of most concern.
Mechanisms of antimicrobial resistance
To identify the mechanism of reduced susceptibility towards the fluoroquinolone ciprofloxacin in 20 isolates, we screened for PMQR genes and mutations associated with the QRDR regions. We did not identify any PMQR genes. Mutations in QRDR were mainly identified in isolates exhibiting intermediate susceptibilities towards ciprofloxacin with two exceptions observed in ciprofloxacin-resistant S. rubidaea 512 and 513 (Table 2). In E. coli 505 the GyrA contained a mutation at amino acid position 87 (Asp→His). A mutation in the ParC of H. alvei 207 was identified at amino acid position 60 (Asn→Ser) and position 59 in S. rubidaea 507-513 (Ser→Thr). Mutations were identified in the GyrB of S. rubidaea 507-513 at amino acid position 439 (Arg→Lys) and C. gillenii 87 at amino acid positions 239 (Asn→Thr), 240 (Ile→Val), 287 (Ala→Ser) and 298 (Asp→Glu).
To identify carbapenemase activity, carbapenem-resistant H. alvei were exposed to EDTA in the presence of ertapenem to inhibit potential MBL activity. No MBL production in ertapenem-resistant H. alvei was detected. We also screened these isolates against a cohort of carbapenemase genes, none of which were detected by PCR. Additionally, we screened LRE isolates for mobile resistance genes optrA, poxtA and cfr but we did not detect any of these genes. Screening for mutations in the 23 S rRNA, L3, L4 and L22 ribosomal regions revealed a mutation in E. durans 62 in the 23 S rRNA sequence (A2227G) and in the L3-region at position 45 (Ser→Gly). No mutations were detected in the remaining LRE (Supplementary Table 5).
Antimicrobial resistance plasmid isolation and characterisation
Exogenous plasmids were successfully transferred from three of the 28 investigated household drinking water samples to E. coli CV601, the recipient strain. No transconjugant growth was observed for the remaining household samples (n = 25). Antimicrobial susceptibility tests of transconjugants showed that transconjugants had acquired multiple antibiotic resistances (Fig. 4). Resistances against ampicillin, tetracycline, ciprofloxacin, and chloramphenicol were identified. Two instances of MDR transconjugants were observed (Transconjugants t17 and c40). Transconjugant t17 was selected in the presence of tetracycline and was resistant to ampicillin, tetracycline, and chloramphenicol. Transconjugant c40 was selected in the presence of ciprofloxacin and was resistant to ampicillin, ciprofloxacin, and chloramphenicol. These MDR transconjugants were obtained from households 14 and 36.
The resistance patterns of E. coli transconjugants from Households 36, 14 and 43 are shown where blue represents susceptibility, yellow signifies intermediate resistance and red denotes resistance of the transconjugant to the antimicrobial. The numbers assigned to each transconjugant are arbitrary identifiers. Letters next to the numbers represent the antimicrobial used to select the transconjugants: tetracycline (t), ciprofloxacin (c), amoxicillin (a). AMP ampicillin, TET tetracycline, CIP ciprofloxacin, C chloramphenicol, AK amikacin.
For household 36, the phenotype of Transconjugant t17 (ampicillin, tetracycline, chloramphenicol resistant) mirrored the phenotypes observed for the MDR E. coli and S. rubidaea (Fig. 3a, b) isolated from the same household which may be indicative of the host bacteria. However, the phenotypes of transconjugants c36-c45 from household 14 and transconjugants a58 and a60 from household 42 did not match to resistance phenotypes of isolates from the corresponding household.
We reduced the percentage identity to 50% (from the default 80%) when screening our plasmids. Doing so revealed one potential homologue of the cfrA gene in plasmid c36 which had 58.67% coverage and 99.52% identity match. All remaining ARGs and virulence factors identified were over 80% identical to those already deposited in the databases. Screening of plasmids identified conjugative machinery, replicon types, virulence factors and ARGs (Table 3). Plasmid sizes ranged between 33.6–118.8 kb. Transconjugant t17 carried multi-replicon plasmid with identity match 98.24% and 95.59% to IncFIB (Accession: JN233704) and IncFIC (Accession: AP001918) plasmids, respectively. The t17 plasmid was also 100% identical to IncQ (Accession: M28829), although the coverage was 66.46%. The t17 plasmid contained the beta-lactam resistance blaTEM-1 gene which confers the ampicillin-resistant phenotype observed. The tetB gene gave rise to a tetracycline-resistant phenotype. In addition, aminoglycoside-resistance genes of the aphA-variants were present. However, these genes did not confer resistance to gentamicin or amikacin. The aphA(3’)-Ia gene is known to confer resistance to kanamycin19. As the plasmids were expressed in an E. coli that was chromosomally resistant to kanamycin, we could not determine through disk testing if the aphA-variants were conferring resistance to kanamycin. The phenicol-resistance genes cfrA, catI and floR were present, which are responsible for the chloramphenicol-resistant phenotype. Sulfonamide-resistance gene, sul2 was also present. Sequence analysis also identified virulence-associated iroBCDEN gene cluster, iucABC and iutA.
Transconjugant a60 carried multi-replicon plasmid similar in identity to IncFII (Accession: CP000670, 97.83%) and IncFIB (Accession: JN233704, 97.51%), with a coverage of 98.26 and 99.29%, respectively. No ARGs were identified using CARD or resfinder, although an ampicillin-resistant phenotype was observed. Both t17 and a60 carried conjugation machinery, MOBF and MOBP suggesting that the plasmids are self-mobilizable.
The ColRNAI (Accession: DQ298019) was identified in transconjugants c36 (98.46% coverage, 85.50% identity), c43 (99.23% coverage, 86.26% identity), c44 (99.23% coverage, 86.26% identity), a58 (99.23% coverage, 83.21% identity) and a60 (99.23% coverage, 83.21% identity). The ARGs cfrA and blaTEM-116 were present in c36, c43 and c44. None of these plasmids conferred resistance to chloramphenicol, despite the presence of the phenicol-resistance gene cfrA. Only c44 was ampicillin-resistant and contained the blaTEM-116 gene, c36 had reduced susceptibility to ampicillin, whilst c42 was susceptible. For c37 and c45, we identified linear contigs while circularisation of plasmids was possible for c40, c41 and c42. However, we could not match any of the linear or circular contigs to known plasmids in the database therefore these potential plasmids require further study to identify and characterise them.
Discussion
The presence of ARB and ARGs in drinking water is well documented across the globe. However, this has not been incorporated in surveillance systems for drinking water to-date, which leads to the lack of standards on the acceptable levels for consuming AMR bacteria or genes. In Ireland, very few studies have analysed bacteria in drinking water as a reservoir of antimicrobial resistance. Up until recently, studies that have assessed AMR in Irish supplies focused on E. coli and P. aeruginosa20. A more recent publication has evaluated coliforms collected by Public Health Laboratories during routine water quality testing and found a variety of bacterial species with AMR phenotypes12. However, the study focused solely on Enterobacterales, and mechanisms associated with ESBL and MBL phenotypes.
Our study incorporated citizen science to identify a comprehensive range of bacterial reservoirs of antimicrobial resistance in drinking water supplied privately to Irish households. We identified a broad range of AMR bacteria, including those usually assessed as indicators of faecal contamination (Escherichia coli and Enterococcus sp.), as well as potentially pathogenic bacteria that have yet to be recommended in water quality assessments1,21,22.
Most of our sampling cohort obtained their water through GWS (n = 30, 61%). The remaining households depended on personal private wells (n = 19, 39%). Contamination of privately sourced drinking water has been identified globally23,24,25. Our metadata suggests that many of the households in this study do not take action to mitigate potential microbial contamination. In 2020, the Irish Environmental Protection Agency reported 5% non-compliance rate for private supplies of drinking water. With use of membrane filtration technique for water quality testing (ISO 9308-1:2014 and ISO 7899-2:2000), it is possible that low level faecal contamination for many households is below the detection limit. Due to delays, the water samples received were at times processed beyond the 24 h timeline recommended. As a result, the loss of viable bacteria was inevitable. However, we focused on AMR analysis as faecal contamination has been previously positively correlated with ARGs26,27. Therefore we enriched our filtered water samples to increase the abundance of bacteria for AMR analysis.
In Ireland, septic tank leakages are often cited as the most common route of contamination of privately supplied drinking water28,29. We performed correlation analysis to determine associations between the reliance on septic tanks and the occurrence of antimicrobial resistant bacteria. We identified a moderate positive correlation (Spearman’s rS value + 0.649, α = 0.001) suggesting that perhaps the septic tanks may have a role in introducing ARB to the drinking water source. We also wanted to assess whether the use of antimicrobials in the household would coincide with the detection of ARB in drinking water, as it could suggest that the household residents have (A) underwent treatment for a pathogen present in the water or (B) the use of antimicrobials have selected for these pathogens. We found a stronger correlation (Spearman’s rS value of + 0.716, α = 0.002) in this instance. Since antimicrobials are not fully metabolised30, and a positive link exists between the reliance on septic tanks and the presence of ARB, it is possible that households that underwent antimicrobial treatments have selected for ARB through excrements which may have contaminated the drinking water source via septic tanks.
Our work corroborates a previous report of high prevalence of amoxicillin and amoxicillin-clavulanate resistance in Enterobacterales of Irish drinking water12 as most of our isolates were known natural producers of AmpC (e.g. Enterobacter sp., Klebsiella sp., Citrobacter sp.). In contrast, Daly et al., identified acquired AmpC production in E. coli which we did not identify in this work. While E. coli were isolated from four households (n = 40 E. coli), AMR E. coli (n = 25) were identified in two of the households. A total of 16 AMR E. coli exhibited a MDR phenotype. All of which were isolated from Household 36. The AMR phenotypes corroborated findings in another study, which identified high prevalence of ampicillin and tetracycline resistance in E. coli from private drinking water in Ireland20. In addition to this, E. coli isolates in our work also showed high prevalence of chloramphenicol resistance (n = 16, 40%) and aminoglycoside-resistance (n = 24, 60%). Phenicols and aminoglycosides make up a small percentage of veterinary sales in Ireland (3.3 and 7.3%, respectively)31. However, amongst the antimicrobials prescribed to humans, phenicols and aminoglycosides are grouped alongside the least prescribed antimicrobials, which collectively make up <10% of consumption32. Non-MDR E. coli were isolated from the two households that reported the presence of horses, sheep and cattle on or near their residence (Household 6 and Household 44), although AMR E. coli was only identified in one of these households (Household 6, cattle & sheep on farm) were an E. coli strain exhibited resistance to kanamycin. The antimicrobial resistance in our isolates echoes patterns found in clinical isolates. Escherichia coli is the primary cause of urinary tract (UTI) and bloodstream infection (BSI) across the EU/EEA. The majority of UTI E. coli isolates have been reported as resistant to at least one antimicrobial class (54.0%, 53.1%)33,34. High levels of AMR E. coli have been reported in isolates from human blood samples between 2017-2021, especially against aminopenicillins (53.1–58.7%)34. Cyprus, Ireland, and Bulgaria are amongst the most recurrent reporters of high levels of resistance to aminopenicillins (>60%) (https://atlas.ecdc.europa.eu/). On a global level multidrug-resistant E. coli have been reported in drinking water sources in Northern Tanzania and Peru35,36. Similar to our work, both studies report the co-existence of ampicillin and tetracycline resistance in MDR E. coli. While these three countries differ in terms of climate, socio-economic status, culture, antimicrobial prescribing regulations and agriculture, a commonality of the presence of these MDR E. coli in drinking water exists. This suggests that something else is common to all three countries. However, what this common factor is still needs to be determined. The household from which we isolated MDR E. coli with resistances against ampicillin and tetracycline have indicated living within 1 km from farmland. This may explain the occurrence of ampicillin and tetracycline-resistance in the isolates as penicillins and tetracycline antimicrobials made up the majority of antimicrobial sales in the veterinary industry in Ireland, making up 26.3% and 35.8% of sales in 202231.
The presence of extended-spectrum β-lactamases (ESBLs) in Enterobacterales has been well documented in a variety of settings, including the natural environment and hospitals37,38. ESBL-producing Enterobacterales have been previously reported in drinking water in Ethiopia (K. pneumonia), Bangladesh (E. coli), and the United States (E. coli, Klebsiella sp., Citrobacter sp.)39,40,41. We did not identify ESBL phenotypes in any of our isolates. As the only species demonstrating cephalosporin-resistance in our study were known AmpC producers, AmpC over-production may have been responsible for the cephalosporin-resistant phenotypes observed42. In addition, the only previous study to test for ESBL-production in Irish drinking water isolates also failed to detect any ESBL phenotypes, corroborating our finding12.
The emergence of carbapenem-resistance amongst Gram-negative pathogens in clinical isolates has seen a rise on a global-scale43. Carbapenemase-producing species such as those that produce metallo-β-lactamases (MBLs) are of most concern due to the potential for horizontal gene transfer of resistance genes to pathogenic species44. This is significant as carbapenem antimicrobials are often used as a last-resort treatment option for MDR infections45. We identified carbapenem-resistance in Stenotrophomonas maltophilia, an emerging pathogen of concern due to its intrinsic resistance to a wide range of antimicrobial classes, including carbapenems through a chromosomally encoded MBL called L146. Although considered an environmental microorganism that thrives in aquatic settings47, S. maltophilia has been identified in hospital water sources such as handwash sinks and showers48,49,50. This is likely due to both their intrinsic resistance to a broad-range of antimicrobials as well as their ability to form biofilms allowing for long term colonisation of pipes and drains51. This may also explain the identification of S. maltophilia in our household samples, as they may have colonised the water piping systems for these households. Few reports of S. maltophilia in drinking water are available52,53. The Irish EPA reports that wells are generally 60–120 m deep (https://www.epa.ie/), however five of the six households in which S. maltophilia was isolated in our study reported wells of depths 30 m or less. The sixth household relied on spring water. The shallow depth of wells and the exposure of springs to the natural environment may have provided an opportunity for S. maltophilia to enter into drinking water supplies as this species are also inhabitants of soils and rivers54,55.
We identified Hafnia alvei as a reservoir for acquired carbapenem-resistance demonstrating ertapenem-resistant phenotype. In Europe, carbapenem-resistance has mostly been associated with clinical isolates of K. pneumonia, P. aeruginosa and Acinetobacter sp.56 We were unable to elucidate the mechanism of carbapenem-resistance in our H. alvei isolate. No previous reports of aquatic carbapenem-resistant H. alvei exist. H. alvei is generally isolated from faeces of humans, animals and birds57 therefore its presence in drinking water could be a result of contamination by septic tank leakages or agricultural practices such as farming or manure spreading. However, the ertapenem-resistant phenotype is unusual, as the carbapenem of choice in Irish hospitals is meropenem (https://www.hpsc.ie/) which is rarely used. However, the ertapenem-resistant H. alvei was identified from a household that reported living in close proximity to a quarry which perhaps may be leaching heavy metals which may have co-selected for this phenotype. An OXA-48 producing H. alvei isolate has been reported in a clinical isolate exhibiting an ertapenem-resistance phenotype58. Our results suggest that the patient with the carbapenem resistant H. alvei may have been exposed to it from a drinking water source. This is an important finding as in recent years, infections due to this microorganism have increased predominantly in intra-abdominal focus and in immunosuppressed patients59.
The prescription of quinolone antimicrobials to treat human infections has reduced in the European Union as a result of a 2019 review which identified rare but serious side effects60. However, findings of bacteria demonstrating reduced susceptibility to the quinolones have already been established in the aquatic environments and clinical settings61,62. Plasmid-mediated quinolone resistance (PMQR) has been reported as early as the 1990s associated with the emergence of qnr, qep and aac(6’)-Ib genes63,64,65. Alternatively, mutations in the quinolone-resistance determining-regions (QRDRs) such as GyrA, GyrB and ParC can result in reduced susceptibility or resistance to the quinolones66. Plasmid-associated resistances against quinolone antibiotics have previously been reported in drinking water in Portugal67 and in other aquatic environments68,69,70. We did not identify any PMQR genes in the tested isolates. We could not elucidate the mechanism of reduced susceptibility to ciprofloxacin for some isolates of Enterobacter sp., C. gillenii, S. fonticola and H. alvei. However, we identified mutations mainly in the QRDRs of isolates exhibiting intermediate susceptibilities towards ciprofloxacin. The most common GyrA mutations in E. coli are known to occur in positions Ser-83 and Asn-8771,72. We identified a mutation at Asn-87 in an E. coli isolate. Mutations in the gyrB gene have mostly been reported in Mycobacterium tuberculosis isolated from hospital patients73,74 but have also been identified in Salmonella sp. from stool samples75. We identified mutations in the GyrB region of C. gillenii isolate 87. All mutations (positions 239, 240, 287 and 298) identified are previously unreported GyrB mutations. We also identified mutations in the GyrB region of S. rubidaea which may explain the reduced susceptibility in our isolates, although there are currently no other reports of GyrB mutations in Serratia sp. Mutations in positions 59 and 60 of the ParC region have previously been reported in clinical isolates of Serratia marcescens76. A mutation in position 57 from a clinical E. coli isolate (Accession: ABLAPL010000001) is reported in the NCBI Pathogen Detection Reference Gene Catalogue. Our S. rubidaea isolates had mutations in position 59. No previous reports of ParC mutations in S. rubidaea nor H. alvei were available. This may be because S. rubidaea and H. alvei are not considered common causes of infections and therefore have not been previously screened for QRDR mutations.
Occurrence of resistance against last-resort oxazolidinone antimicrobial linezolid has been reported in species of Staphylococcus and Enterococcus77,78. Linezolid is often used as a last-line defence against methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus species79. The EARS-Net reported vancomycin-resistance in 15-18.3% of E. faecium between 2017 to 2021. Ireland has one of the highest prevalence of vancomycin-resistant E. faecium in Western Europe since 2008 (https://atlas.ecdc.europa.eu/). The only instance of VRE identified in our study was that in E. casseliflavus which exhibit low-level resistance against vancomycin due to a chromosomal vanC80. Linezolid-resistance in enterococci is associated with mutations in the 23 S rRNA or ribosomal L-proteins81, or by acquisition of ARGs optrA, poxtA or cfr via mobile genetic elements or plasmids82,83,84. Our study presents LRE species that are deficient in known mobile resistance genes (optrA, poxtA, cfr) but some of which contained chromosomal target site mutations. Linezolid-resistant Enterococci were identified across three household samples, in E. durans, E. faecium and E. casseliflavus. Linezolid-resistant Enterococcus sp. were resistant to at least one other antimicrobial, namely tetracycline or ampicillin. Mutation G2576T in the 23 S rRNA are frequently associated with linezolid-resistance in Enterococcus sp.81, however the mutation we identified in E. durans was at a previously unreported position of the 23 S rRNA, position A2227G. Reports of mutations in the L-proteins in Enterococcus sp. are rare but mutation of the L3 region at codon V149I has been reported in E. faecalis isolated from swine85. Our study identified a mutation at codon S45G of the L3 protein in E. durans. However, contribution of this mutation to linezolid resistance requires further studies. We could not elucidate potential mechanisms of linezolid-resistance in E. casseliflavus and E. faecium as no mutations were identified in their 23 S rRNA or the ribosomal L-proteins. Linezolid-resistant Enterococcus sp. have been reported in clinical settings and in surface water77,86,87. There is currently no data available on the use of linezolid in Ireland, however, considering the high prevalence of VRE in Irish hospitals relative to other European countries, it is plausible that linezolid is frequently used for treating VRE infections. In fact, the earliest report of LRE outbreaks in an Irish hospitals began in 201488. This means that the potential for selection of linezolid resistant Enterococcus species in Ireland in general could be higher than other countries but also the potential to select for novel linezolid resistance mechanisms is also higher.
Enrichment of our water samples for AMR analysis may have been biased towards certain species. Culture-based analysis is limited to bacteria that can withstand or thrive under the specific laboratory conditions provided. This excludes a potentially large number of species that were not culturable under these conditions and therefore the results presented may not be representative of the true composition of the drinking water samples as non-cultured bacteria may have acquired ARGs or mutations. We were unable to isolate bacteria from 21 of 49 water samples due to complete loss of viability therefore alternative approaches may have been useful such as the inclusion of molecular-based methods to overcome these caveats. For example, analysis on each drinking water sample such as metagenomics89 would have detected the species present, irrespective of their viability or growth requirements. Metagenomics would also allow for screening of ARGs that may have been overlooked by analysing only culturable and viable bacteria. However, studies using metagenomics at times report processing excessive volumes (20 L–2000 L) of drinking water in order to extract DNA89,90,91 making it difficult to do so without the necessary resources in place.
Our culture-dependent screening failed to identify the mechanism of resistance for a number of bacterial isolates that demonstrated an AMR phenotype. These included cephalosporin-resistant, quinolone-resistant and carbapenem-resistant Enterobacterales and linezolid-resistant Enterococci. In all of these cases, the screening involved isolating bacteria and amplifying known genes of interest via conventional polymerase chain reaction. The potential for identifying novel genes or mutations is overlooked using PCR. An alternative approach would be to perform genome sequencing of the individual isolates and analyse the entire genome for potential mutations or genes that could have contributed to the phenotypes observed92.
As many bacteria in drinking water tend to be viable but nonculturable93, we used the exogenous isolation method to capture potential mobile resistance elements. We captured plasmids within the frequently identified replicon type IncF group94. IncF plasmids have played a pivotal role in the successes of extraintestinal pathogenic E. coli (ExPEC) such as ST131 and ST41095. The IncF group of plasmids have previously been reported in drinking water in France and Tanzania35,96. They generally carried tetA, blaTEM-1 and blaCTX-M genes. Our IncF-type multi-replicon plasmid t17 also contained the blaTEM-1 but carried the tetracycline-resistance tetB rather than tetA gene alongside other ARGs associated with phenicol, aminoglycoside and sulfonamide resistance. Plasmid t17 also carried virulence genes associated with iron acquisition: iroBCDEN, iucABC and, iutA. The iroBCDEN cluster originated in the chromosome of Salmonella enterica but has later been found on transmissible plasmids in uropathogenic E. coli97. A comprehensive study of extraintestinal pathogenic E. coli from veal calves found correlation between the IncFIB replicon and the presence of iucABC-iutA98. This supports our plasmid analysis of the multi-replicon t17 which had the IncFIB plasmid. However, this was not the case for plasmid a60 which did not carry any virulence factor genes but contained the IncFIB replicon.
The most common replicon identified in our work were the Col-type plasmids. Col-plasmids encode bacteriocin proteins called colicins, which target and kill related strains of E. coli99. This provides a colonisation advantage for the plasmid-carrying E. coli over related E. coli. The ColRNAI plasmid has been previously reported in water environments100,101,102. Other Col-plasmids identified included Col(Ye4449) and Col(MGD2). Col(Ye4449) is rarely reported but has been identified in hospital wastewater103 and associated with animals intended for human consumption104,105. Col(MGD2) has been associated with clinical and environmental settings106,107,108.
None of the ColRNAI plasmids c36, c43 and c44 conferred resistance to chloramphenicol, despite the presence of the phenicol-resistance gene cfrA. However, chloramphenicol-resistance in E. coli has frequently been associated with cmlA, floR and catA genes109,110 rather than cfr. This may explain the resistance phenotype observed in t17 which carried both floR and catA genes. As the cfr gene was identified on plasmids exogenously introduced into E. coli, the gene may have conferred AMR in its original host. The cfr genes are associated with phenicol and oxazolidinone-resistance amongst others in Gram-positive bacteria such as Staphylococcus and Enterococcus111,112. Instances of gene presence in the absence of resistance phenotypes in our transconjugants highlights the necessity of combining both phenotypic and molecular analysis for a more representative overview of the composition of AMR in drinking water. In addition, the purpose of using the exogenous method was to identify mobile resistance in a sample without relying on the cultivability of the host strain, yet mobile resistance was only found in three household samples despite the abundance of AMR bacteria in a number of household samples, some of which were multidrug-resistant. It is likely that the use of only one recipient, E. coli for capturing plasmids may have led to under-representation of mobile resistance as some plasmids have a narrow-host range113. It would have been beneficial to include an additional Gram-negative recipient such as Klebsiella pneumonia and Gram-positive recipients such as Enterococcus faecium and Staphylococcus aureus to reduce bias towards E. coli-compatible plasmids. This bias is evident in our work, as multiple types of colicin-plasmids (MGD2, Ye4449, RNAI) usually associated with E. coli99 were identified.
Annual reports on the quality of privately supplied drinking water show that Irish private supplies generally have a high rate of compliance (>95%) in respect to the absence of E. coli28,29,114. Our identification of clinically relevant and ARB in drinking water highlights the need for more robust water quality testing and surveillance. This is to minimise the risk of infection and disease in the consumers and to prolong the use of currently available antimicrobials. The results presented suggest that private Irish drinking water is a route of spread and persistence of AMR and ARG.
Overall, we demonstrate that AMR persistence and spread extends beyond the clinical setting. We have identified private household drinking water in Ireland as reservoirs for clinically relevant, antimicrobial resistant and potentially pathogenic bacteria. The detection of MDR bacteria and bacteria resistant to last-resort antibiotics was of particular concern. The transferability of AMR and virulence genes should be considered in relation to surveillance and quality testing as current water quality guidelines do not recognise ARGs as contaminants, whilst surveillance data is scarce. Surveillance of AMR may inform the transmission of mobile AMR to improve public health measures in cases of outbreaks and to subsequently preserve antimicrobials. Further studies are required to assess the pathogenicity and risk factors of the ARB identified, as drinking water is a direct link between humans and the environment in the One Health framework. This is especially important for MDR bacteria which likely carry conjugative plasmids that may support the survival of pathogens against antimicrobials.
Methods
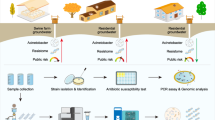
Citizen science: water sampling
Households supplied with drinking water through personal private wells (n = 19, 39%) or Group Water Schemes (GWS) (n = 30, 61%) were invited to participate in this study. Participants were recruited on a voluntary basis, some of which were enroled through the National Federation of Group Water Schemes. The participating households were located in the East and Midlands of Ireland (n = 33, 67%) or in the West of Ireland (n = 16, 33%). A Group Water Scheme (GWS) is defined as any private, community-based scheme that manages and distributes drinking water to private households.
The citizens were provided with nitrile gloves and a sterile 50 mL tube. As the focus of this study was on consumed AMR, the volunteers were asked to collect water only from the tap they use for drinking water. The participants were requested to use any household disinfectant spray or wipes to sanitise the handles on their drinking water tap and countertops. The volunteers were required to fully open the tap and allow to run for 2–3 min before reducing the flow of the water to fill the tube provided. The volunteers were instructed to avoid touching the tube against the nozzle. The samples were then transported by courier to our laboratory. The participants were invited to complete a questionnaire (Supplementary Questionnaire 1) regarding the location of residence and antibiotic use in the household. They were given the option to opt-out of answering all or any of the questions.
Isolation and identification of antimicrobial resistant bacteria from private household drinking water
Water samples (50 mL) were processed using the membrane filtration method, which is an ISO method115. Bacteria were subsequently enriched from membranes in Brain-Heart Infusion (Oxoid) broth at 37 °C with shaking at 225 rpm for 18 to 48 h. To select for resistant bacteria, enrichment broths were cultured on Eosin Methylene Blue (Sigma) and Slanetz & Bartley Medium agars (Oxoid) in the presence of antibiotics at breakpoint concentrations: amoxicillin (32 µg/mL) (Sigma), tetracycline (16 µg/mL) (Sigma) and ciprofloxacin (1 µg/mL) (Fluka). Bacteria were also selected in the absence of antibiotics.
Bacteria were speciated using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) as previously described116,117 using a microflex LT MALDI-TOF mass spectrometer (Bruker Daltonics) and the associated flexControl software (ver. 3.4). Spectra were classified using the Bruker Taxonomy main spectra database (MBT Compass ver. 4.1, 9607 spectra). Bacterial identification was reported to the species level if the score value was ≥ 2.00.
Any bacteria with a score between 1.70–1.99 can only be reliably identified at the genus level118. For each isolate with a MALDI-ToF MS score of 1.70–1.99 a single colony was suspended in 50 µL sterile deionised water and boiled in the thermocycler at 95° C for 10 min to release DNA. A PCR reaction was performed in 50 µL volumes consisting of 2.5 µL of boiled bacterial suspension, 25 µL Readymix RedTaq X2 (Sigma-Aldrich), 1 µL each of forward primer (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′)119 and reverse primers (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3’)119 at 0.2 µM concentrations. The remainder volume was made up with sterilised deionised water. Thermocycling conditions used were as follows: initial denaturation (95 °C, 5 min), 2 cycles of denaturation (95 °C, 40 s), annealing (55 °C, 2 min), and extension (72 °C, 1 min). And a final extension (72 °C, 7 min) step. The resulting PCR products were cleaned up using AxyPrep Mag PCR Clean-up beads (Axygen) and were sent to Eurofins Genomics for Sanger sequencing. The 16 S rRNA PCR product sequences were analysed using NCBI BLASTN.
Antimicrobial susceptibility testing using disk diffusion and broth microdilution
Antimicrobial susceptibility testing (AST) was determined by the Kirby-Bauer disk diffusion test or broth dilution method using the Clinical and Laboratory Standards Institute (CLSI) guidelines120.
Correlation analysis
To assess potential correlation between reliance on domestic wastewater systems and the presence of ARB, Spearman’s rank correlation was used to analyse the relationship for households from which bacteria were isolated (n = 28). For this, the absence of septic tanks (n = 2) was denoted “1” and the use of septic tanks (n = 26) was denoted “2”. Similarly, the absence of viable ARB (n = 5) was denoted “1” while the presence of ARB (n = 23 households) was denoted “2”.
Further analysis was performed to investigate if there were correlations between the use of antimicrobials to treat bacterial infections, and the isolation of species of clinical relevance in household water supplies. The species of clinical relevance included E. coli, Pseudomonas aeruginosa, Enterobacter sp., Enterococcus sp., S. maltophilia and Acinetobacter sp. Households that did not indicate whether or not they have used antimicrobials in the past 10 years have been excluded from analysis (n = 6). Households that did not consume antibiotics (n = 5 households) were denoted “1”, while those who did (n = 17 households) were denoted “2”. The absence of species of clinical relevance was denoted “1”, while the presence was denoted “2”. The Spearman’s correlation coefficients were then compared against Spearman’s rank correlation table of critical values121 to identify the level of significance (α), where n is the number of household samples and rS is the absolute value of the test statistic.
Phenotypic identification of the mechanism of antimicrobial resistant Enterobacterales
Screening for AmpC, Extended Spectrum β-Lactamase (ESBL) and Metallo-β-Lactamase (MBL) production in the Enterobacterales was performed using inhibitor-based tests as previously described122,123,124. Known AmpC-producers Citrobacter sp., Enterobacter sp., Hafnia alvei, Raoultella sp., and Serratia sp. were excluded from AmpC testing.
Genotype screening for carbapenemase resistance genes
Carbapenem resistant bacteria were screened for carbapenemase resistance genes using multiplex PCR. A single colony was suspended in 50 µL sterile deionised water and boiled in the thermocycler at 95 °C for 10 min to release DNA. The primers used included blaVIM, blaKPC, blaOXA-40, blaNDM, blaOXA-48, blaOXA-23, blaIMI, blaOXA-58, blaGES, blaGIM, blaIMP, and blaOXA-51 and thermocycling conditions were followed as described125.
Screening for plasmid-mediated quinolone resistance and mutations in the quinolone resistance-determining regions
Isolates of potential clinical relevance showing reduced susceptibilities to ciprofloxacin were screened for plasmid-mediated quinolone resistance (PMQR) genes qnrA, qnrB, qnrS, qepA and aac(6’)-Ib-cr and mutations in their quinolone-resistance determining regions (QRDR) gyrA, gyrB and parC genes. A single colony was suspended in 50 µL sterile deionised water and boiled in the thermocycler at 95 °C for 10 min to release DNA. Genes were amplified via PCR (primers listed in Supplementary Table 2 and Table 3) and sequenced by Sanger Sequencing. BLASTX and ClustalW against reference were used to identify mutations in amino acid sequences of the QRDR regions.
Analysis of the molecular mechanism of resistance in linezolid-resistant Enterococcus sp
Linezolid-resistant Enterococcus sp. (LRE) were screened for mobile resistance genes optrA, poxtA and cfr using PCR. Furthermore, LRE were screened for chromosomal mutations in the 23 S rRNA, L3, L4 and L22 regions. DNA was extracted using the NucleoSpin Microbial DNA kit as per manufacturer’s protocols. Primers used to amplify all genes are listed in Supplementary Table 4. The PCR products were sequenced by Sanger sequencing and analysed by BLASTN for 23 S rRNA products and BLASTX for the L-regions against reference genomes (Supplementary Table 5).
Exogenous plasmid isolation, extraction, and sequencing
Plasmids were exogenously isolated using biparental mating as previously described using kanamycin/rifampicin-resistant E. coli CV601 as recipient126. The transconjugants were selected on LB agar supplemented with rifampicin (100 µg/mL) (Duchefa) and amoxicillin (32 µg/mL) or tetracycline (16 µg/mL) or ciprofloxacin (0.5 µg/mL). Transconjugants were screened on CHROMagar Orientation supplemented with kanamycin (64 µg/mL) (Sigma) to further confirm selection of the recipient strain. Transconjugants were subjected to AST using the CLSI guidelines for Enterobacterales120. Plasmids were extracted from all transconjugants using the Machery-Nagel NucleoSpin plasmid isolation kit and the DNA concentration quantified using Qubit-Fluorometric Quantitation.
Plasmid sequencing was performed using Oxford Nanopore Technology (ONT) MinION with the Rapid Barcoding Sequencing Kit (SQK-RBK004). Briefly, library preparation involved attaching unique barcodes and adaptors to each sample. The flow cells were primed, and the DNA library was loaded onto the flow cell for long read sequencing. The raw fast5 files were base called and demultiplexed using ONT Guppy v6.1.2 (https://github.com/nanoporetech/rerio) with GPU acceleration. Quality control (QC) was performed using Guppy during base calling. An additional QC filter step using Filtlong v0.2.1 (https://github.com/rrwick/Filtlong) was performed on https://usegalaxy.eu/. To remove short read sequences, the default settings were changed to exclude contigs <1000 bp.
https://usegalaxy.eu/ was used as follows: Sequences were assembled using Unicycler v0.5.0 (https://github.com/rrwick/Unicycler) with long reads only. Reads were visualised using Bandage v0.8.1 (https://github.com/rrwick/Bandage). ABRicate v1.0.1 (https://github.com/tseemann/abricate) identified ARGs, virulence factors and plasmid replicon types. For all databases, the minimum DNA identity and coverage were initially set to 80% to screen for ARGs and then a second screen was performed at 50% to identify potentially more distant homologues and account for potential database bias towards clinical samples. MOB-typer v3.0.3 (https://github.com/phac-nml/mob-suite) at default settings screened for conjugative machinery.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
GenBank Accession numbers for plasmid sequences are included in Table 3.
References
World Health Organization. Guidelines for Drinking-Water Quality: Fourth Edition Incorporating the First Addendum. Guidelines for Drinking-Water Quality: Fourth Edition Incorporating the First Addendum (WHO, 2017).
Unc, A. & Goss, M. J. Transport of bacteria from manure and protection of water resources. Appl. Soil Ecol. 25, 1–18 (2004).
Ahmed, W., Neller, R. & Katouli, M. Evidence of septic system failure determined by a bacterial biochemical fingerprinting method. J. Appl. Microbiol. 98, 910–920 (2005).
WHO. Ten Threats to Global Health. https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (2019).
WHO. One Health. https://www.who.int/europe/initiatives/one-health (2023).
Lin, Z. et al. Impact factors of the accumulation, migration and spread of antibiotic resistance in the environment. Environ. Geochem. Health 43, 1741–1758 (2021).
Alpay-Karaoglu, S. et al. Investigation of antibiotic resistance profile and TEM-type β-lactamase gene carriage of ampicillin-resistant Escherichia coli strains isolated from drinking water. Ann. Microbiol. 57, 281–288 (2007).
Narciso-da-Rocha, C., Vaz-Moreira, I., Svensson-Stadler, L., Moore, E. R. B. & Manaia, C. M. Diversity and antibiotic resistance of Acinetobacter spp. in water from the source to the tap. Appl. Microbiol. Biotechnol. 97, 329–340 (2013).
Abdel Rahim, K. A. A., Hassanein, A. M. & Abd El Azeiz, H. A. E. H. Prevalence, plasmids and antibiotic resistance correlation of enteric bacteria in different drinking water resources in sohag, Egypt. Jundishapur J. Microbiol. 8, e18648 (2015).
Odonkor, S. T. & Addo, K. K. Prevalence of multidrug-resistant escherichia coli isolated from drinking water sources. Int. J. Microbiol. 2018, 7204013 (2018).
Mahmoud, N. E., Altayb, H. N. & Gurashi, R. M. Detection of carbapenem-resistant genes in escherichia coli isolated from drinking water in khartoum, Sudan. J. Environ. Public Health 2020, 2571293 (2020).
Daly, M., Powell, J., O’Connell, N. H., Murphy, L. & Dunne, C. P. Antimicrobial resistance Is prevalent in e.coli and other enterobacterales isolated from public and private drinking water supplies in the Republic of Ireland. Microorganisms 11, 1224 (2023).
Li, S., Ondon, B. S., Ho, S.-H., Zhou, Q. & Li, F. Drinking water sources as hotspots of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs): Occurrence, spread, and mitigation strategies. J. Water Process Eng. 53, 103907 (2023).
McInnes, R. S., McCallum, G. E., Lamberte, L. E. & van Schaik, W. Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 53, 35–43 (2020).
Khan, H., Miao, X., Liu, M., Ahmad, S. & Bai, X. Behavior of last resort antibiotic resistance genes (mcr-1 and blaNDM-1) in a drinking water supply system and their possible acquisition by the mouse gut flora. Environ. Pollut. 259, 113818 (2020).
Anthony, W. E., Burnham, C.-A. D., Dantas, G. & Kwon, J. H. The Gut microbiome as a reservoir for antimicrobial resistance. J. Infect. Dis. 223, S209–S213 (2021).
Iacob, S. & Iacob, D. G. Infectious threats, the intestinal barrier, and its trojan horse: dysbiosis. Front. Microbiol. 10, 1676 (2019).
Navarro, F. & Courvalin, P. Analysis of genes encoding D-alanine-D-alanine ligase-related enzymes in Enterococcus casseliflavus and Enterococcus flavescens. Antimicrob. Agents Chemother. 38, 1788–1793 (1994).
Oka, A., Sugisaki, H. & Takanami, M. Nucleotide sequence of the kanamycin resistance transposon Tn9 03. J. Mol. Biol. 147, 217–226 (1981).
Andrade, L., Chique, C., Hynds, P., Weatherill, J. & O’Dwyer, J. The antimicrobial resistance profiles of Escherichia coli and Pseudomonas aeruginosa isolated from private groundwater wells in the Republic of Ireland. Environ. Pollut. 317, 120817 (2023).
The Council of the European Union. European Union Drinking Water Regulations. https://www.irishstatutebook.ie/eli/2023/si/99/made/en/pdf (2014).
Water, N. & Management, Q. Australian Drinking Water Guidelines 6 (Commonwealth of Australia, 2017).
Rutter, M., Nichols, G. L., Swan, A. & De Louvois, J. A survey of the microbiological quality of private water supplies in England. Epidemiol. Infect. 124, 417–425 (2000).
Schets, F. M. et al. Escherichia coli O157:H7 in drinking water from private water supplies in the Netherlands. Water Res. 39, 4485–4493 (2005).
Murray, R. T. et al. Prevalence of microbiological and chemical contaminants in private drinking water wells in maryland, usa. Int. J. Environ. Res. Public. Health 15, 1686 (2018).
Reynolds, L. J. et al. Correlation between antimicrobial resistance and faecal contamination in small urban streams and bathing waters. Sci. Total Environ. 739, 140242 (2020).
Karkman, A., Pärnänen, K. & Larsson, D. G. J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 10, 80 (2019).
EPA. Focus on private water supplies 2018. Focus on Private Water Supplies 2018. (Environmental Protection Agency, Ireland, 2020).
EPA. Focus on private water supplies 2019. Focus on Private Water Supplies 2019. (Environmental Protection Agency, Ireland, 2021).
Du, L. & Liu, W. Occurrence, fate, and ecotoxicity of antibiotics in agro-ecosystems. a review. Agron. Sustain. Dev. 32, 309–327 (2012).
HPRA. Sales of Veterinary Antibiotics in Ireland During 2022 (HPRA, 2022).
ECDC. Antimicrobial Consumption (ECDC, 2018).
EARS-Net. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report for 2020. https://www.ecdc.europa.eu/sites/default/files/documents/AER-EARS-Net-2021_2022-final.pdf (2021).
EARS-Net. Antimicrobial resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report for 2021. https://www.ecdc.europa.eu/sites/default/files/documents/AER-EARS-Net-2021_2022-final.pdf (2022).
Lyimo, B. et al. IncF plasmids are commonly carried by antibiotic resistant Escherichia coli isolated from drinking water sources in northern Tanzania. Int. J. Microbiol. 2016, 3103672 (2016).
Larson, A. et al. Antibiotic-resistant Escherichia coli in drinking water samples from rural Andean households in Cajamarca, Peru. Am. J. Trop. Med. Hyg. 100, 1363–1368 (2019).
Hooban, B., Joyce, A., Fitzhenry, K., Chique, C. & Morris, D. The role of the natural aquatic environment in the dissemination of extended spectrum beta-lactamase and carbapenemase encoding genes: a scoping review. Water Res. 180, 115880 (2020).
Castanheira, M., Simner, P. J. & Bradford, P. A. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC-Antimicrob. Resist 3, dlab092 (2021).
Abera, B., Kibret, M. & Mulu, W. Extended-spectrum beta (β)-lactamases and antibiogram in Enterobacteriaceae from clinical and drinking water sources from bahir dar city, Ethiopia. PLOS ONE. 11, e0166519 (2016).
Mahmud, Z. H. et al. Extended-spectrum beta-lactamase-producing Escherichia coli in drinking water samples from a forcibly displaced, densely populated community setting in Bangladesh. Front. Public Health 8, 100526 (2020).
Tanner, W. D. et al. Multi-state study of Enterobacteriaceae harboring extended-spectrum beta-lactamase and carbapenemase genes in U.S. drinking water. Sci. Rep. 9, 3938 (2019).
Mizrahi, A. et al. Infections caused by naturally AmpC-producing Enterobacteriaceae: can we use third-generation cephalosporins a narrative review. Int. J. Antimicrob. Agents 55, 105834 (2020).
Nordmann, P. & Poirel, L. Epidemiology and diagnostics of carbapenem resistance in Gram-negative bacteria. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 69, S521–S528 (2019).
van Duin, D. & Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469 (2017).
Meletis, G. Carbapenem resistance: overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 3, 15 (2016).
Gil-Gil, T., Martínez, J. L. & Blanco, P. Mechanisms of antimicrobial resistance in Stenotrophomonas maltophilia: a review of current knowledge. Expert Rev. Anti Infect. Ther. 18, 335–347 (2020).
Looney, W. J., Narita, M. & Mühlemann, K. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect. Dis. 9, 312–323 (2009).
Cervia, J. S., Ortolano, G. A. & Canonica, F. P. Hospital tap water as a source of Stenotrophomonas maltophilia infection. Clin. Infect. Dis. 46, 1485–1486 (2008).
Verweij, P. E. et al. Nosocomial outbreak of colonization and infection with Stenotrophomonas maltophilia in preterm infants associated with contaminated tap water. Epidemiol. Infect. 120, 251–256 (1998).
Guyot, A., Turton, J. F. & Garner, D. Outbreak of Stenotrophomonas maltophilia on an intensive care unit. J. Hosp. Infect. 85, 303–307 (2013).
Brooke, J. S. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 25, 2–41 (2012).
Silbaq, F. S. Viable ultramicrocells in drinking water. J. Appl. Microbiol. 106, 106–117 (2009).
Hoefel, D., Monis, P. T., Grooby, W. L., Andrews, S. & Saint, C. P. Profiling bacterial survival through a water treatment process and subsequent distribution system. J. Appl. Microbiol. 99, 175–186 (2005).
Deredjian, A. et al. Occurrence of Stenotrophomonas maltophilia in agricultural soils and antibiotic resistance properties. Res. Microbiol. 167, 313–324 (2016).
Tacão, M., Correia, A. & Henriques, I. S. Low prevalence of carbapenem-resistant bacteria in river water: resistance Is mostly related to intrinsic mechanisms. Microb. Drug Resist. Larchmt. N. 21, 497–506 (2015).
European Centre for Disease Prevention and Control & World Health Organization. Antimicrobial Resistance Surveillance in Europe 2023: 2021 Data (Publications Office, 2023).
Padilla, D. et al. The pathogen Hafnia alvei in veterinary medicine: a review. J. Appl. Anim. Res. 43, 231–235 (2015).
Sevillano, L. et al. First report of a carbapenemase OXA-48-producing Hafnia alvei clinical isolate. Access Microbiol. 5, acmi000498.v3 (2023).
Cardile, A. P. et al. Hafnia alvei pyelonephritis in a renal transplant recipient: case report and review of an under-recognized nosocomial pathogen. Transpl. Infect. Dis 13, 407–410 (2011).
European Medicines Agency. Fluoroquinolone Antibiotics: Reminder of Measures to Reduce the Risk Long-Lasting, Disabling and Potentially Irreversible Side Effects. https://www.ema.europa.eu/en/news/fluoroquinolone-antibiotics-reminder-measures-reduce-risk-long-lasting-disabling-potentially (2023).
Castrignanò, E. et al. (Fluoro)quinolones and quinolone resistance genes in the aquatic environment: a river catchment perspective. Water Res. 182, 116015 (2020).
Akova, M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence 7, 252–266 (2016).
Martínez-Martínez, L., Pascual, A. & Jacoby, G. A. Quinolone resistance from a transferable plasmid. Lancet. 351, 797–799 (1998).
Yamane, K. et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51, 3354–3360 (2007).
Shen, P. et al. Complete nucleotide sequence of pKP96, a 67 850 bp multiresistance plasmid encoding qnrA1, aac(6’)-Ib-cr and blaCTX-M-24 from Klebsiella pneumoniae. J. Antimicrob. Chemother 62, 1252–1256 (2008).
Jacoby, G. A., Strahilevitz, J. & Hooper, D. C. Plasmid-mediated quinolone resistance. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.PLAS-0006-2013 (2014).
Mendonça, N., Ramalho, J., Vieira, P. & Da Silva, G. J. Association of plasmid-mediated quinolone resistance and virulence markers in Escherichia coli isolated from water. J. Water Health 10, 288–294 (2012).
Wen, Y., Pu, X., Zheng, W. & Hu, G. High Prevalence of plasmid-mediated quinolone resistance and IncQ plasmids carrying qnrS2 gene in bacteria from rivers near hospitals and aquaculture in China. PLoS One 11, e0159418 (2016).
Tomova, A., Ivanova, L., Buschmann, A. H., Godfrey, H. P. & Cabello, F. C. Plasmid-mediated quinolone resistance (PMQR) genes and class 1 integrons in quinolone-resistant marine bacteria and clinical isolates of Escherichia coli from an aquacultural area. Microb. Ecol. 75, 104–112 (2018).
Amin, M. B. et al. High prevalence of plasmid-mediated quinolone resistance (PMQR) among E. coli from aquatic environments in Bangladesh. PLoS One 16, e0261970 (2021).
Jaktaji, R. P. & Mohiti, E. Study of mutations in the DNA gyrase gyrA gene of Escherichia coli. Iran. J. Pharm. Res. 9, 43–48 (2010).
Johnning, A., Kristiansson, E., Fick, J., Weijdegård, B. & Larsson, D. G. J. Resistance mutations in gyrA and parC are common in Escherichia communities of both fluoroquinolone-polluted and uncontaminated aquatic environments. Front. Microbiol. 6, 1355 (2015).
Chien, J.-Y. et al. Mutations in gyrA and gyrB among fluoroquinolone- and multidrug-resistant mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 60, 2090–2096 (2016).
Disratthakit, A. et al. Role of gyrB mutations in pre-extensively and extensively drug-resistant tuberculosis in Thai clinical isolates. Antimicrob. Agents Chemother. 60, 5189–5197 (2016).
Qian, H. et al. Discovery of seven novel mutations of gyrB, parC and parE in salmonella typhi and paratyphi strains from jiangsu province of China. Sci. Rep. 10, 7359 (2020).
Yang, H. F., Cheng, J., Hu, L. F., Ye, Y. & Li, J. B. Identification of a Serratia marcescens clinical isolate with multiple quinolone resistance mechanisms from China. Antimicrob. Agents Chemother. 56, 5426–5427 (2012).
Bi, R., Qin, T., Fan, W., Ma, P. & Gu, B. The emerging problem of linezolid-resistant enterococci. J. Glob. Antimicrob. Resist. 13, 11–19 (2018).
Stefani, S., Bongiorno, D., Mongelli, G. & Campanile, F. Linezolid resistance in staphylococci. Pharmaceuticals 3, 1988–2006 (2010).
Zurenko, G. E. et al. In vitro activities of U-100592 and U-100766, novel oxazolidinone antibacterial agents. Antimicrob. Agents Chemother. 40, 839–845 (1996).
Koganemaru, H. & Hitomi, S. Bacteremia caused by VanC-type enterococci in a university hospital in Japan: a 6-year survey. J. Infect. Chemother. 14, 413–417 (2008).
Mendes, R. E., Deshpande, L. M. & Jones, R. N. Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist. Updat. 17, 1–12 (2014).
Bourgeois-Nicolaos, N. et al. Dose dependence of emergence of resistance to linezolid in Enterococcus faecalis in vivo. J. Infect. Dis. 195, 1480–1488 (2007).
Diaz, L. et al. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob. Agents Chemother. 56, 3917–3922 (2012).
Papagiannitsis, C. C., Tsilipounidaki, K., Malli, E. & Petinaki, E. Detection in greece of a clinical enterococcus faecium isolate carrying the novel oxazolidinone resistance gene poxtA. J. Antimicrob. Chemother 74, 2461–2462 (2019).
Filsner, P. H. L. N. et al. Identification of the cfr methyltransferase gene in Enterococcus faecalis isolated from swine: first report in Brazil. J. Glob. Antimicrob. Resist. 8, 192–193 (2017).
Nüesch-Inderbinen, M., Raschle, S., Stevens, M. J. A., Schmitt, K. & Stephan, R. Linezolid-resistant Enterococcus faecalis ST16 harbouring optrA on a Tn6674-like element isolated from surface water. J. Glob. Antimicrob. Resist. 25, 89–92 (2021).
Biggel, M., Nüesch-Inderbinen, M., Jans, C., Stevens, M. J. A. & Stephan, R. Genetic context of optrA and poxtA in florfenicol-resistant enterococci isolated from flowing surface water in Switzerland. Antimicrob. Agents Chemother. 65, e0108321 (2021).
O’Driscoll, C. et al. First outbreak of linezolid-resistant vancomycin-resistant Enterococcus faecium in an Irish hospital, February to September 2014. J. Hosp. Infect. 91, 367–370 (2015).
Dang, C. et al. Metagenomic insights into the profile of antibiotic resistomes in a large drinking water reservoir. Environ. Int. 136, 105449 (2020).
Ma, L., Li, B. & Zhang, T. New insights into antibiotic resistome in drinking water and management perspectives: a metagenomic based study of small-sized microbes. Water Res. 152, 191–201 (2019).
Zhao, Q., He, H., Gao, K., Li, T. & Dong, B. Fate, mobility, and pathogenicity of drinking water treatment plant resistomes deciphered by metagenomic assembly and network analyses. Sci. Total Environ. 804, 150095 (2022).
NIHR Global Health Research Unit on Genomic Surveillance of AMR. Whole-genome sequencing as part of national and international surveillance programmes for antimicrobial resistance: a roadmap. BMJ Glob. Health 5, e002244 (2020).
Byrd, J. J., Xu, H. S. & Colwell, R. R. Viable but nonculturable bacteria in drinking water. Appl. Environ. Microbiol. 57, 875–878 (1991).
Carattoli, A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238 (2009).
Pitout, J. D. D. & Chen, L. The significance of epidemic plasmids in the success of multidrug-resistant drug pandemic extraintestinal pathogenic Escherichia coli. Infect. Dis. Ther. 12, 1029 (2023).
Laroche-Ajzenberg, E. et al. Conjugative multiple-antibiotic resistance plasmids in Escherichia coli isolated from environmental waters contaminated by human faecal wastes. J. Appl. Microbiol. 118, 399–411 (2015).
Sorsa, L. J., Dufke, S., Heesemann, J. & Schubert, S. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of Uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect. Immun. 71, 3285–3293 (2003).
Haley, B. J., Kim, S. W., Salaheen, S., Hovingh, E. & Kessel, J. A. S. V. Virulome and genome analyses identify associations between antimicrobial resistance genes and virulence factors in highly drug-resistant Escherichia coli isolated from veal calves. PLoS One 17, e0265445 (2022).
Cascales, E. et al. Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229 (2007).
Nascimento, T. et al. International high-risk clones of Klebsiella pneumoniae KPC-2/CC258 and Escherichia coli CTX-M-15/CC10 in urban lake waters. Sci. Total Environ. 598, 910–915 (2017).
Hooban, B. et al. A point prevalence survey of antibiotic resistance in the Irish environment, 2018–2019. Environ. Int. 152, 106466 (2021).
Campana, E. H., Kraychete, G. B., Montezzi, L. F., Xavier, D. E. & Picão, R. C. Description of a new non-Tn4401 element (NTEKPC-IIe) harboured on IncQ plasmid in citrobacter werkmanii from recreational coastal water. J. Glob. Antimicrob. Resist. 29, 207–211 (2022).
Takizawa, S. et al. Genomic landscape of blaGES-5- and blaGES-24-harboring Gram-negative bacteria from hospital wastewater: emergence of class 3 integron-associated blaGES-24 genes. J. Glob. Antimicrob. Resist. 31, 196–206 (2022).
Liu, Y. et al. Prevalence and genomic investigation of Salmonella isolates recovered from animal food-chain in Xinjiang, China. Food Res. Int. 142, 110198 (2021).
Yue, Y. et al. Whole-genome sequencing-based prediction and analysis of antimicrobial resistance in Yersinia Enterocolitica from Ningxia, China. Front. Microbiol. 13, 936425 (2022).
Flament-Simon, S.-C. et al. High diversity and variability of pipolins among a wide range of pathogenic Escherichia coli strains. Sci. Rep. 10, 12452 (2020).
Zhao, Q. et al. Dissemination of blaNDM-5 via IncX3 plasmids in carbapenem-resistant Enterobacteriaceae among humans and in the environment in an intensive vegetable cultivation area in eastern China. Environ. Pollut. 273, 116370 (2021).
Rocha, J., Henriques, I., Gomila, M. & Manaia, C. M. Common and distinctive genomic features of Klebsiella pneumoniae thriving in the natural environment or in clinical settings. Sci. Rep. 12, 10441 (2022).
Travis, R. M. et al. Chloramphenicol and kanamycin resistance among porcine Escherichia coli in Ontario. J. Antimicrob. Chemother. 58, 173–177 (2006).
Williams, C. T., Musicha, P., Feasey, N. A., Adams, E. R. & Edwards, T. ChloS-HRM, a novel assay to identify chloramphenicol-susceptible Escherichia coli and Klebsiella pneumoniae in Malawi. J. Antimicrob. Chemother. 74, 1212–1217 (2019).
Morales, G. et al. Resistance to linezolid Is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant staphylococcus aureus. Clin. Infect. Dis. 50, 821–825 (2010).
Shen, J., Wang, Y. & Schwarz, S. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative. bacteria. J. Antimicrob. Chemother. 68, 1697–1706 (2013).
Weaver, K. E., Kwong, S. M., Firth, N. & Francia, M. V. The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61, 94–109 (2009).
EPA. Drinking Water Quality in Private Group Schemes and Small Private Supplies (EPA, 2022).
Novo, A. & Manaia, C. M. Factors influencing antibiotic resistance burden in municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 87, 1157–1166 (2010).
Mateo-Estrada, V. et al. Acinetobacter baumannii from grass: novel but non-resistant clones. Microb. Genomics 9, 001054 (2023).
Gekenidis, M.-T., Studer, P., Wüthrich, S., Brunisholz, R. & Drissner, D. Beyond the matrix-assisted laser desorption ionization (MALDI) biotyping workflow: in search of microorganism-specific tryptic peptides enabling discrimination of subspecies. Appl. Environ. Microbiol. 80, 4234–4241 (2014).
Normand, A.-C. et al. Decision criteria for MALDI-TOF MS-based identification of filamentous fungi using commercial and in-house reference databases. BMC Microbiol. 17, 25 (2017).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 (2013).
EM100 Connect. CLSI M100 ED31 (2021) http://em100.edaptivedocs.net/GetDoc.aspx?doc=CLSI%20M100%20ED31:2021&scope=user
Ramsey, P. H. Critical Values for Spearman’s Rank Order Correlation. J. Educ. Stat. 14, 245–253, (1989).
Gupta, G., Tak, V. & Mathur, P. Detection of AmpC β lactamases in Gram-negative bacteria. J. Lab. Physicians 6, 001–006 (2014).
Poulou, A. et al. Modified CLSI extended-spectrum β-lactamase (ESBL) confirmatory test for phenotypic detection of ESBLs among Enterobacteriaceae producing various β-lactamases. J. Clin. Microbiol. 52, 1483–1489 (2014).
Sachdeva, R., Sharma, B. & Sharma, R. Evaluation of different phenotypic tests for detection of metallo-β-lactamases in imipenem-resistant Pseudomonas aeruginosa. J. Lab. Physicians 9, 249–253 (2017).
Cerezales, M. et al. Novel multiplex PCRs for detection of the most prevalent carbapenemase genes in Gram-negative bacteria within Germany. J. Med. Microbiol. https://doi.org/10.1099/jmm.0.001310 (2021).
Heuer, H. et al. Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol. Ecol. 42, 289–302 (2006).
Acknowledgements
The authors would like to thank the National Federation of Group Water Schemes and the volunteers who were involved in the citizen science element of this work. We are grateful to Mirjam Zeiher at the Chemical and Veterinary Investigation Office Sigmaringen (CVUA SIG) for providing measuring time at the MALDI Biotyper and to the staff of the Microbiology Department at the CVUA SIG for excellent collaboration. This work was funded by the Irish Environmental Protection Agency and the Kathleen Lonsdale Human Health Institute co-fund scholarship to Marwa Alawi “Antimicrobial resistance in private drinking water supplies”. The citizen science component of this work was funded by a grant from the Irish Research Council awarded to Dr. Thi Thuy Do.
Author information
Authors and Affiliations
Contributions
M. Alawi: Conceptualization, methodology, software, data curation, writing—original draft preparation, visualization, investigation. C. Smyth: Data processing. D. Drissner: Writing—review and editing, methodology, data curation. A. Zimmerer: Data curation. D. Leupold: Data curation. D. Müller: Data curation. T. T. Do: Funding acquisition, conceptualization, methodology, writing—review and editing. T. Velasco-Torrijos: Writing—review and editing, supervision, funding acquisition. F. Walsh: Conceptualization, methodology, writing—review and editing, supervision, funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alawi, M., Smyth, C., Drissner, D. et al. Private and well drinking water are reservoirs for antimicrobial resistant bacteria. npj Antimicrob Resist 2, 7 (2024). https://doi.org/10.1038/s44259-024-00024-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44259-024-00024-9