Abstract
The streptothricin antibiotics were among the first antibiotics to be discovered from the environment and remain some of the most recovered antimicrobials in natural product screens. Increasing rates of antibiotic resistance and recognition that streptothricin antibiotics may play a role in countering so-called super-bugs has led to the re-evaluation of their clinical potential. Here we will review the current state of knowledge of streptothricins and their resistance in bacteria, with a focus on the potential for new resistance mechanisms and determinants to emerge in the context of potential widespread clinical adoption of this antibiotic class.
Similar content being viewed by others
Introduction
Antimicrobial-resistant infections are estimated to cause 700,000 deaths globally each year. By 2050, it is projected that the annual death toll will increase to 10 million and cost the global healthcare system in excess of $100 trillion1. Meanwhile, there has been a drought in the development of new antimicrobial therapeutics2, especially from major pharmaceutical companies which normally drive antibiotic development3,4. One solution to the dual problems of resistance and an anemic antimicrobial pipeline is to re-examine previously discarded antibiotics. Daptomycin, vancomycin, and colistin were previously considered to be too toxic for clinical use in the context of, at the time, sufficient coverage by other antibiotics such as β-lactams2,5,6,7,8,9,10. Increased resistance to first-choice antibiotics led to renewed interest in these more toxic options, and the side effects that kept daptomycin, vancomycin, and colistin off the market were moderated through changes in dosing, increasing purity, and altering pharmacokinetics9,10,11. These formerly discarded antibiotics are now critical medicines in the treatment of multidrug-resistant infections that would otherwise be fatal12,13. Similarly, many antimicrobials recently approved by the FDA are derivatives of compounds that were discovered decades ago, such as plazomicin (a derivative of the aminoglycoside antibiotic sisomycin), lefamulin (a semi-synthetic derivative of the antibiotic pleuromutilin), and the tetracyclines eravacycline and omadacyline (third generation tetracycline antibiotics)14.
It is in this context that interest in streptothricin antibiotics, one of the first discovered antimicrobial classes but one that has been dogged by toxicity issues, has been renewed. Recent work by Dowgiallo et al. and Morgan et al. has highlighted the clinical potential of streptothricins in combatting multidrug-resistant pathogens by suggesting new routes to diversify streptothricins for increased clinical efficacy and by elucidating their molecular targets15,16. Given this reinvigoration of the field, it is worth re-visiting the current state of knowledge of bacterial resistance to streptothricins in the context of their potential therapeutic future. Here, we review important milestones in streptothricin history, its biochemistry and mechanism of action in susceptible bacteria, and the current state of microbial resistance to streptothricins. Our goal is to highlight the apparent disconnect between the abundance of streptothricins in the soil environment and the paucity of mechanisms for their resistance in the soil resistome (the collection of resistance genes and their precursors in an environment), especially compared to other natural product antibiotics17. We propose that parallel to investigating the medicinal potential of streptothricins, the streptothricin soil resistome must be studied and quantified so that this knowledge can be incorporated into any next-generation streptothricin analogs that may eventually reach the clinic.
Streptothricin history and biochemistry
Discovery of streptothricin
The mass production and clinical deployment of penicillin in the 1940s highlighted the potential for life-saving drugs to come from the soil environment. Selman Waksman and colleagues at Rutgers University sought to make systematic what Alexander Fleming found by serendipity: a procedure for studying the ability of extracts from soil-dwelling bacteria to inhibit pathogenic bacteria, now referred to as the Waksman platform18,19. As early as 1940 this approach identified a soil actinomycete that produced a compound capable of killing E. coli19 and resulted in the purification of another compound, actinomycin, with activity against Gram-positive pathogens20. The Waksman platform fully blossomed by 1944 with the discovery of streptomycin, the first anti-tuberculosis antibiotic, and continued to provide additional discoveries18,21. In 1942, prior to the more famous streptomycin, Waksman and Woodruff discovered an antibiotic they termed streptothricin, produced by a Streptomyces lavendulae isolate (Fig. 1, 1942)22. Streptothricin was met with substantial optimism as it appeared to be the first broad-spectrum antibiotic, meaning it was able to kill both Gram-positive and Gram-negative bacteria. This spectrum of bactericidal activity was much wider than that of penicillin, allowing streptothricin to potentially meet a vital healthcare need22,23. Initial tests appeared to support this optimism as streptothricin treatment cured Brucella abortus model infections of chicken eggs and guinea pigs24 and protected mice against a number of Gram-negative pathogens25.
1942: initial discovery, 1946: detailed report of major toxicity during animal testing, 1952–1961: characterization of the main molecular components, 1972: complete structural description, 1978: mechanism of action described, 1982: first total synthesis completed, 1987: first genetic isolation of a streptothricin acetyltransferase (STAT), 1997: resistance-guided discovery of the biosynthetic gene cluster, 2006: first isolation of a streptothricin hydrolase (SttH), 2021: first genetic characterization of a putative streptothricin resistance rRNA methyltransferase, 2023: discovery of the precise molecular targets of streptothricins.
Structure of streptothricins and streptothricin-like compounds
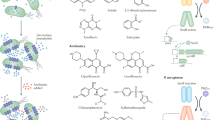
Structurally, the streptothricins are defined by three key molecular features: a streptolidine lactam ring (Fig. 2a, blue component), a gulosamine sugar (Fig. 2a, black component), and a β-lysine homopolymer with lengths of a single β-lysine to seven residues (Fig. 2a, red component)26,27,28. These three core components were identified following chemical degradation of streptothricins29,30,31,32 (Fig. 1, 1952, 1956, 1961) and confirmed the hypothesis that streptothricin antibiotics (also known then as yazumycins and racemomycins) differ primarily by how many β-lysine residues are attached to the gulosamine sugar33,34. The diversity of congeners differing only by β-lysine homopolymer length led to the proliferation of synonyms for streptothricins and streptothricin mixes, including nourseothricin, zhongshengmycin, streptolin, racemomycin, geomycin, pleocidin, yazumycin, phytobacteriomycin, grisein, and polymycin26. By convention, individual streptothricins (A–E, X) are identified by the number of β-lysine residues they contain (Fig. 2a). In contrast, the commercially available mixture of streptothricin congeners termed nourseothricin contains streptothricin D (29.6%), E (trace), and F (65.5%)15,16,35. In 1972 Khokhlov and Shutova declared that the chemical structure of the streptothricins was fully understood, followed a decade later by confirmation through total synthesis of streptothricin F (Fig. 1, 1972, 1982)26,27.
a General structure of the streptothricins (STC) highlighting the streptolidine lactam ring (blue), gulosamine sugar (black), and β-lysine residue(s) (red). Streptothricins are defined by the length of their β-lysine homopolymer chain, from one to seven residues. b Structural elements of the “streptothricin-like” antibiotics, highlighting variation in the streptolidine lactam ring (blue: methylation, hydroxylation, stereochemistry), the sugar (black: carbamoyl location), and the amino acid residue (red: N-methylation, presence of β-lysine or a glycine derivative). R-group features in bold signify the default substituent found in canonical streptothricins.
Following the initial description by Waksman and Woodruff22, several additional streptothricin-like analogs were identified that strayed from the three central components outlined above. These compounds have historically been termed “streptothricin-like” (Fig. 2b). Streptothricin-like antibiotics with variation in the streptolidine ring include compounds with N-methylation and loss of the C-4 hydroxyl group in albothricin (with compounds A37812 and N-methyl-Streptothricin-D sharing N-methylation without hydroxyl group loss)36,37,38, lactam ring opening in the streptothricin acids39,40, or stereocenter inversions in the cis-fused streptothricins41,42. A number of streptothricin-like compounds also vary at the gulosamine sugar position, most commonly in changes to the location of the carbamoyl moiety from C-10 in the classic streptothricins to C-12 in a variety of analogs41,42,43. Finally, a number of streptothricin-like molecules have been identified that contain non-lysine amino acid residues linked to the gulosamine sugar, including formiminoglycine and N-methylglycine (sarcosine) (e.g., compounds LL-AB664, LL-BL136, LL-AC541, sclerothricin, BD-12, BY-81, citromycin, E-749-C, SF-701, A-269A, and A-269A’)44,45,46,47,48,49,50,51,52. It has been estimated that across all these combinations, there are at least 45 streptothricin or streptothricin-like metabolites recorded in the literature53,54.
Streptothricin target and mechanism of action
The aqueous solubility of streptothricins and the presence of an amino sugar in their structure has sometimes led to them being categorized with the aminoglycoside antibiotics. While not accurate, fortuitously the streptothricins do to share a similar mechanism of action as the aminoglycosides. Early studies of 14C-leucine and 32P-phosphate incorporation in bacterial cells suggested that streptothricins target the bacterial translation apparatus while not affecting DNA synthesis or general cellular integrity55. Important details on the molecular mechanism of action of the streptothricins were reported by Haupt et al. beginning in 1978 with the observation that, in addition to generally inhibiting protein synthesis, streptothricin F leads to misreading of the mRNA message, making the streptothricins miscoding antibiotics similar to kanamycin and several other aminoglycosides56 (Fig. 1, 1978). A series of in vitro translation assay systems demonstrated that streptothricin F moderately inhibited binding of charged tRNAs to the ribosome but did not inhibit ribosome peptidyltransferase activity. Instead, the greatest effect of streptothricin F on translation appeared to come via blocking of the translocation reaction57. More recently, Morgan et al. used cryogenic electron microscopy (cryo-EM) to solve the structure of streptothricins F and D bound to their molecular target on the Acinetobacter baumannii ribosome (Fig. 1, 2023). They found that the antibiotics principally bind at helix 34 of the 16 S rRNA, specifically interacting with bases A1196, C1054, and U1052 (E. coli numbering conventions). The authors hypothesized that these interactions may stabilize noncognate tRNAs in the A-site, leading to streptothricin’s antimicrobial effects16. The cryo-EM model also revealed additional binding sites on the ribosome, unique to either streptothricin F or streptothricin D. The authors hypothesized that these could reflect nonspecific binding due to the high concentration of the antibiotics used in the study, but noted that the streptothricins have been proposed to have multiple binding sites on the bacterial ribosome, similar to some aminoglycosides16,56,58.
Streptothricin use in industry, agriculture, and medicine
The broad-spectrum activity and unique molecular mechanism of action suggest streptothricins should be useful tools in the clinical antibiotic armamentarium. However, soon after wider laboratory testing was undertaken it became evident that the streptothricins have appreciable toxicity in mammals. In an early study in rabbits, intravenous, intradermal, oral, and topical applications resulted in organ failure and death (Fig. 1, 1946)59. Replicable delayed toxicity of streptothricin and streptothricin-like antibiotics in mammalian test organisms (principally mice) led to delayed toxicity becoming an identifying feature of the whole antimicrobial class33,34,36,46,49,52,60. In an effort to characterize this delayed toxicity, Inamori et al. carried out extensive toxicity tests of streptothricins in mice and rats60,61,62,63. They found that following intravenous administration in mice, streptothricin F distributes largely to the kidneys with very little active antibiotic recoverable from the urine60. Histological examination of kidneys from treated mice and rats confirmed streptothricin-induced nephrotoxicity, most notably in the renal cortex, which developed ~48 h after administration61,62. Before this delayed toxicity was recognized, the pharmaceutical company Merck attempted a clinical trial of streptothricin in humans which resulted in all four volunteers losing their ability to urinate. The apparent kidney failure was reversible and all four eventually recovered64, though this toxicity and the discovery of less toxic broad-spectrum antibiotics resulted in a loss of interest in clinical applications for the streptothricins.
Strategies to mitigate streptothricin toxicity have been partially successful. Treatment of streptothricin D with a streptothricin hydrolase (see section below) was found to result in a 32-fold decrease in activity against E. coli but also a larger 128- to 256-fold decrease in toxicity in eukaryotic cells, suggesting a potential medicinal chemistry approach to combating the drug’s side effects40. Furthermore, with the discovery that streptothricins vary at the β-lysine position34 (Fig. 2a), it was observed that streptothricin toxicity in mice increases with β-lysine chain length. Streptothricin F (one β-lysine residue) was shown to be significantly less toxic than streptothricin D (three β-lysine residues), with reported LD50 values of 300 mg/kg vs <10 mg/kg respectively34. It has been suggested that the lower cytotoxicity of streptothricin F compared to other streptothricins could reflect decreased cellular internalization by host cells. Takuechi et al. noted that bacterial polycationic isopeptides can directly penetrate mammalian membranes to reach the cytosol. They proposed that the higher toxicity of streptothricins A–E and X compared to streptothricin F may be due to greater host cellular uptake as a result of their longer β-lysine homopolymer tails (Fig. 2a)65. A result of this difference in cytotoxicity is that pure streptothricin F, as opposed to a mix of streptothricins, may have a wide enough therapeutic window to justify its use against multidrug-resistant pathogens. Pure streptothricin F has been found to have high activity against pathogens identified by the Center for Disease Control and Prevention to be threats, including vancomycin-resistant Staphylococcus aureus, multidrug-resistant A. baumanni, and β-lactam resistant Enterobacteriaceae (including the pan drug-resistant Klebsiella pneumoniae strain AR-0636), justifying its potential clinical utility12,15. This proposition was tested recently by Morgan et al. in a neutropenic mouse model of A. baumanni infection where a single dose streptothricin F treatment was associated with no or minimal toxicity and a ~10,000-fold decrease in pathogen titer16. This development suggests that streptothricins may follow a similar clinical course to vancomycin where the obstacle of high toxicity was surmounted by increasing antibiotic purity. A recently reported total synthesis of streptothricin F may also lower barriers to the preparation of synthetic streptothricin analogs and allow medicinal chemists to directly tackle toxicity at a molecular level15.
Due to their toxicity, streptothricins have not been used in clinical settings, with the possible exception of a 1945 correspondence reporting their use in a balm for treating athlete’s foot66. Instead, streptothricins have been predominantly adopted as tools for biotechnology, in particular the congener mix nourseothricin. Because of its broad biological activity and lack of cross-resistance from other selectable markers, nourseothricin and nourseothricin-resistance cassettes are available for a wide spectrum of experimental organisms, both prokaryotic and eukaryotic, including bacteria, fungi, protozoa, plants, animals, and mammalian cell lines67,68,69,70,71,72.
Outside the laboratory, streptothricin use was largely limited until a period between 1981 and 1989 during which nourseothricin was used as an ergotropic compound in pork husbandry in the German Democratic Republic (GDR). Within one year of this practice beginning, it became possible to isolate streptothricin-resistant E. coli from pig rectal swabs, agricultural sewage, and from fecal samples from agricultural workers at those farms. These resistant strains were found to contain plasmids encoding multidrug-resistant transposon cassettes with genes for a streptothricin acetyltransferase (see section below) and an aminoglycoside adenyltransferase73. Within two years of agricultural nourseothricin use, E. coli carrying plasmid-encoded streptothricin resistance could be found in fecal samples from non-farm associated individuals living in the same village as farms employing the antibiotic as a growth-enhancer. Farm workers and individuals from villages where nourseothricin was not used in this way yielded no E. coli strains with this resistance phenotype74. These represented the first discovered examples of transferable streptothricin resistance genes. The rapid emergence and spread of these resistance determinants as the result of the solely agricultural use of nourseothricin highlights the connection between agricultural antibiotic use and antibiotic resistance in potential human pathogens. Even after discontinuation of this practice, identical streptothricin resistance cassettes appeared to continue to spread into other pathogenic taxa and on other transposons, including ESKAPE pathogens Enterococcus faecium and A. baumannii, potentially due to selective pressure on the multidrug resistance cassettes from non-streptothricin antibiotic use3,75,76. Within the last few decades, a mix of streptothricins termed zhongshengmycin has entered use in Chinese agriculture as a microbial control agent for crops77,78. Based on prior experience with nourseothricin in the GDR and, more recently, with colistin79, it is likely that extensive agricultural use will result in the appearance of new mobilizable streptothricin resistance determinants.
Streptothricin biosynthesis and ecology
An interesting aspect of the ecology of streptothricin, and perhaps and explanation for the rapid development of streptothricin resistance in the GDR73,74, is the widespread potential for its biosynthesis among soil bacteria. Streptothricin production among actinomycetes bacteria, the taxa most often mined for potential natural product antibiotics80, is apparently common. Between 10% and 42% of members of actinomycetes strain libraries have been identified as streptothricin producers across studies (Fig. 3)18,80,81,82. This regularity is supported by the historical record: streptothricin was discovered before any other broad-spectrum antibiotics and was only preceded by the narrow-spectrum compounds penicillin and actinomycin D22. The streptothricin biosynthetic gene cluster was discovered in 1997, based on its synteny with a self-protection streptothricin acetyltransferase (Fig. 1, 1997), and this gene cluster has been found across the globe in phylogenetically diverse bacteria81. When researchers looked at all publicly available genomes from the Streptomyces genus specifically, they found that nearly 4% of genomes encoded a recognizable streptothricin biosynthetic gene cluster83. Streptothricins are discovered often enough in natural product screens that methods for streptothricin dereplication have been developed to lower their signal and allow for the discovery of novel compounds81,84.
Pie graph charting the prevalence of streptothricin (red), streptomycin (light green), macrolide or tetracycline (dark green), or other natural product (purple) production in an actinomycetes culture collection. Roughly 42% of actinomycetes were found to produce streptothricin. Figure based on data from ref. 81.
Bacterial streptothricin resistance mechanisms and determinants
Overview of the streptothricin resistance landscape
The widespread biosynthesis of streptothricins (Fig. 3) suggests them to be ecologically important compounds in the soil microbiome. One current understanding of antibiotic ecology is that resistance genes are common in the soil microbiome (the resistome) due to the biosynthesis of many natural product antibiotics in that environment17,79,85 and that many resistance genes originate from producers of antimicrobials as self-protection genes85,86,87. This understanding of the interactions between antimicrobial production and resistance suggests that a commonly produced compound like streptothricin (Fig. 3) should be associated with widespread resistance in the soil microbiome. However, as described below, streptothricin resistance genes are limited to a few streptothricin acetyltransferases families88, two hydrolases for which resistance may be a moonlighting activity40,89, and a single putative methyltransferase90 (Fig. 4a). This paucity of mechanisms (two to three) and biochemically validated enzymes (single digits across all mechanisms) is in sharp contrast with the resistomes of other antimicrobials. A striking example of this inequality can be found by comparison to the aminoglycoside family of antibiotics. The aminoglycosides were discovered soon after streptothricin and are also characterized by high aqueous solubility, targeting of the 30 S ribosome, and biosynthesis by soil-derived actinomycetes91. Taking kanamycin B as a model aminoglycoside, there are at least eight atomic targets for resistance found on the molecule itself, with three different classes of transferases acting at these sites92,93. Resistance to kanamycin B and other aminoglycosides is also conferred through active efflux and protection/alteration of the ribosome target88,94 (Fig. 4b). While not as extensively modified as aminoglycosides, other ribosome-targeting antibiotics are resisted through significantly more mechanisms as well. For example, chloramphenicol is modified by acetyltransferases (including type A and type B classes encompassing dozens of gene families), hydrolases, oxidases, and nitroreductases and resisted via efflux pumps and ribosome methylation95,96,97,98,99,100, tetracyclines are modified by multiple families of tetracycline destructases and are resisted through efflux, ribosome methylation, and ribosome protection101,102,103, and, finally, macrolides are modified both by esterases and phosphotransferases and are subject to efflux, ribosome methylation, and ribosome protection as well104.
a Biochemically confirmed streptothricin resistance mechanisms are limited to β-lysine acetylation and (potentially nonspecific) hydrolysis of the streptolidine lactam ring. b Kanamycin B resistance mechanisms include acetyl, phosphoryl, and nucleotidyltransferases targeting multiple sites, drug efflux, and 16 S ribosome subunit modification through methylation.
This inequality in resistance determinants between streptothricins and comparable antibiotics contrasts with what ecological theory would predict given the apparent abundance of streptothricin producers in the soil microbiome. We pose that it is an interesting question whether this imbalance reflects a true paradox or not. One resolution to this paradox is that streptothricins are somehow more immune to bacterial resistance than other antibiotics and are therefore excellent candidates for the clinic. A second resolution is that the lack of known resistance reflects the lack of widespread use (and selection for resistance) and in-depth study, suggesting that if clinical use is pursued it should be coupled with scrupulous monitoring for the transfer of as-yet unknown environmental streptothricin resistance genes into human pathogens. The resolution of this paradox has important implications as streptothricin is re-examined for clinical utility and suggests it would be appropriate to review the current state of knowledge of streptothricin resistance.
Resistance through drug modification: streptothricin acetyltransferases
By far the best-studied streptothricin resistance mechanism is acetylation targeting the β-lysine amino group (Fig. 5a). Acetylation was first identified as a mechanism of resistance in a streptothricin producer in 1983105. In the earliest characterization of this mechanism, it was noted that ribosomes from a streptothricin-producing Streptomyces strain were susceptible to inhibition by the antibiotic in vitro, suggesting that a self-protection mechanism must exist. In analogy to self-protection transferases in aminoglycoside-producing organisms, the authors screened protein fractions from the producer for streptothricin-inactivation activity. A fraction with this activity was identified that functioned in the presence of acetyl coenzyme A (acetyl-CoA) but not when ATP was substituted, suggesting an acetyltransferase. Incubation of this fraction with acetyl-CoA and kanamycin, neomycin, or chloramphenicol did not result in attenuation of the antibacterial activity of those compounds, demonstrating specificity for streptothricin105. Similar experiments on protein fractions from two other producers, Streptomyces noursei and S. lavendulae, confirmed streptothricin acetyltransferase activity in those organisms as well106,107. The gene responsible for this activity, stat, was soon identified and sequenced, followed by purification of the active enzyme itself (Fig. 1, 1987)107,108 (notably, streptothricin acetyltransferases have been given the following names: STAT, Nat, Sta, and Sat. We propose that Sat become standard moving forward).
a Streptothricin acetyltransferase (Sat, STAT, or NAT) mechanism of action. Enzymatic transfer of an acetyl group from acetyl-CoA to the streptothricin amino group of β-lysine results in loss of antibiotic activity. b Phylogenetic tree of three novel predicted streptothricin acetyltransferases (“Soil NTC SatA/sta”, blue dots) captured by functional metagenomic selection in the context of CARD validated STAT, SatA, Sat-2, Sat-3, and Sat-4 enzymes, predicted streptothricin acetyltransferases, and other related acetyltransferases. b is modified from ref. 90 (CC BY 4.0).
Initial in vitro characterization of the STAT enzyme with streptothricin F demonstrated use of acetyl-CoA as a cofactor and the product was confirmed by 13C and 1H nuclear magnetic resonance (NMR) spectroscopy to be streptothricin F mono-acetylated at the β-amino group of the β-lysine moiety. Reaction kinetics were found to be consistent with Michaelis-Menten models of enzymatic activity and low KM values for acetyl-CoA and streptothricin of 69 μM and 2.3 μM, respectively, were physiologically relevant109. An additional study found similar results for a Sat-type enzyme from E. coli110. More recently, detailed in vitro studies performed on the SatA enzyme from Bacillus sp. recorded streptothricin acetyltransferase catalytic efficiencies (kcat/KM) of 5 × 106 M−1 s−1 (B. subtilis SatA) and 8.9 × 108 M−1 s−1 (B. anthracis SatA), in line with many other bona fide antibiotic resistance enzymes111,112. Following random and site-directed mutagenesis of the B. anthracis SatA, Burckhardt and Escalante-Semerena suggested that a conserved glutamate at position 137 may catalyze the nucleophilic attack on the acetyl-CoA carbonyl and that a cluster of aromatic (Y149, F154, and Y164) and hydrophobic (L136 and A145) residues are important for binding of streptothricin112.
The initially discovered streptothricin acetyltransferases from producers of the antibiotic, such as S. noursei113 and Streptomyces rochei114, were joined by genes termed “sat’” from non-producers such as Campylobacter coli115, E. coli73,116,117,118, and B. anthracis and B. subtilis111. Following the advent of metagenomic sequencing, homologs of these acetyltransferases have been predicted across many bacterial taxa. However, as of July 2023, the UniProt database119 contains just three reviewed streptothricin acetyltransferase proteins while the Comprehensive Antibiotic Resistance Database (CARD)120 contains just five canonical streptothricin acetyltransferase protein families (STAT, SAT-2, SAT-3, SAT-4, and SatA). In contrast, CARD describes fourteen unique kanamycin AAC(6’) acetyltransferase families alone, suggesting that streptothricin acetyltransferase diversity is under-sampled. Supporting this, in a 1992 survey of streptothricin-resistant bacteria, Smalla et al. found that 77.5% of environmental isolates and 100% of resistant isolates from the soil lacked known acetyltransferase genes as measured by DNA hybridization assays. At most this indicates the presence of other novel streptothricin resistance mechanisms, and at the very least indicates substantial undiscovered acetyltransferase diversity121.
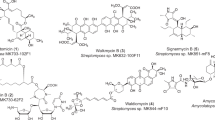
In line with this prediction, we recently proposed a significant increase in Sat enzyme diversity following the use of a functional metagenomic selection for nourseothricin resistance90. Briefly, a 162 Gb functional metagenomic library was prepared from soil metagenomic DNA by METa assembly90 and housed in E. coli. The library was selected by plating cells on agar containing 64 μg/ml nourseothricin and resistance-conferring gene fragments were recovered from colonies arising and were sequenced. Our analyses suggested the screen captured streptothricin acetyltransferases with low to modest amino acid identity to their closest CARD counterparts (26% to STAT, 50% to SatA, and 48% to Sat-4). The phylogenetic distribution relative to canonical streptothricin acetyltransferases from CARD and putative streptothricin acetyltransferase hits strongly suggests that most streptothricin acetyltransferase diversity remains uncharacterized, particularly for the acetyltransferase with 26% identity to STAT (Fig. 5b). If verified by biochemical characterization, this diversity would suggest that streptothricin acetyltransferases are abundant in the soil resistome. The development of next-generation streptothricins for clinical use would benefit from resistance-proofing against these acetyltransferases, much in the same way that the natural products sisomycin and chloramphenicol have been used as templates to prepare semi-synthetic derivatives that are immune to their respective common antibiotic-modifying enzymes. In the case of sisomycin, hydroxyaminobutyryl and hydroxyethyl moieties have been added to primary amines to protect them from modification by a variety of transferases122 (Fig. 6a). Chloramphenicol has seen conversion of a hydroxy group into a fluorine to prevent inactivation by acetyltransferases, with the resulting veterinary antibiotic being termed florfenicol123 (Fig. 6b). Similarly, modification of the β-lysine target of streptothricin antibiotics could potentially render them immune to modification. The naturally occurring streptothricin analog BD-12 contains a formimidoylglycine group in place of β-lysine(s), notably lacking the β-amino group targeted by streptothricin acetyltransferases48. Neither in vitro reactions between BD-12 and a streptothricin acetyltransferase nor the aminoglycoside modifying enzyme AAC(6’)-Ie-APH(6”)-Ia resulted in an acetylated product, suggesting BD-12 may avoid inactivation by acetylating resistance enzymes124. Additional streptothricin derivatives with unnatural β-lysine modifications have been prepared through enzymatic modification in vitro, including replacement of β-lysine by 3-aminoproprionyl, 4-aminobutyl, or β-homolysine groups124,125. Many of these streptothricin analogs have lower antimicrobial activity compared to the canonical streptothricins, suggesting that incorporating residues more similar to β-lysine, such as N-methylated ones, could potentially retain amine-ribosome electrostatic interactions necessary for high levels of antimicrobial activity (with ribose O2’ groups at positions U1052 and C105416) while avoiding disruptive acetylation by resistance enzymes (Fig. 6c).
a, b are natural product (left) and semi-synthetic (right) antibiotic pairs where modifications (red) have been introduced to combat antibiotic-modifying enzymes. a Sisomycin and plazomicin. b Chloramphenicol and florfenicol (replacement of the nitro group with a methyl-sulfonyl group, blue, is to combat toxicity, not resistance). c BD-12, a formimidoylglycyl-streptothricin and a hypothetical N-methylated streptothricin derivative could potentially avoid streptothricin acetyltransferase inactivation while maintaining activity.
Resistance through drug modification: streptothricin hydrolases
Aside from the streptothricin acetyltransferases, the only other biochemically validated streptothricin resistance enzymes in the literature come from the streptothricin hydrolase family. This enzyme family was first reported in 2006 when Hamano et al. observed that a strain of Streptomyces albulus showed greater streptothricin resistance than the streptothricin-producing strain S. lavendulae (Fig. 1, 2006). Attempted PCR amplification of nat genes from S. albulus genomic DNA did not result in any amplicons, suggesting involvement of a novel resistance gene. The authors transformed a streptothricin-sensitive Streptomyces lividans strain with a genomic library prepared from S. albulus genomic DNA and selected for streptothricin-resistant colonies. Selection of this library resulted in the capture of a 2.9 kb DNA fragment which sequencing revealed to contain three candidate resistance genes. None of the predicted genes showed significant identity to known streptothricin acetyltransferase genes. Genetic dissection of the resistance-conferring fragment revealed a predicted isochorismatase-like hydrolase to be responsible for the observed phenotype40. The responsible gene was termed sttH and its role in resistance was confirmed by the streptothricin-susceptible phenotype of a S. albulus sttH knockout strain and streptothricin-resistant phenotype of E. coli expressing sttH from a plasmid40,126. Using a similar strategy, Maruyama and Hamano captured another resistance-conferring gene from a streptothricin nonproducing S. noursei strain. The gene, referred to as sttH-sn, came from a homologous region of the S. noursei genome and the predicted protein sequence showed 74% identity to S. albus SttH89.
Recombinant SttH and SttH-sn were prepared from E. coli and modification of streptothricins F and D was confirmed using liquid chromatography. Mass spectrometry and 1H NMR determined that the resulting products were streptothricin acids in which the amide of the streptolidine lactam was hydrolyzed, producing a new primary amine and carboxylic acid (Fig. 7). Streptothricin acids were found by the authors of the study and another group to have significantly lower antimicrobial activity than their corresponding streptothricins, supporting the feasibility of streptolidine hydrolysis as a resistance mechanism40,43. Measurement of hydrolysis kinetics demonstrated the two enzymes to be broadly similar in activity, with each showing an apparent slight preference for streptothricin F over D. Interestingly, the KM values for streptothricin F and D with both enzymes were measured to be around 1 mM and 3 mM to 6 mM40,89, respectively. These KM values are orders of magnitude greater than the measured streptothricin F and D minimal inhibitory concentrations of 30 μM and 8 μM for E. coli40. This KM, high for an enzymatic reaction where microbial survival is on the line and high compared to SatA (1 μM) and STAT (2.8 μM) enzymes109,111,112, suggests that streptothricins are not the native substrates for the SttH enzymes. Supporting this, the authors noted that the genomic context of the two investigated sttH genes include open reading frames predicted to function in molybdopterin metabolism, suggesting that the true biological function of the SttH enzymes remains to be determined89. The genomic context of sttH and high KM of the enzyme for its substrate suggest streptothricin hydrolysis is a side activity, making SttH a potential example of a housekeeping enzyme with the ability to evolve into a resistance enzyme86.
Resistance through target modification
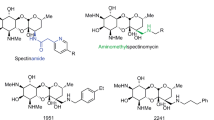
Antibiotics that target the ribosome can also be resisted by protection or modification of the ribosome itself. At its simplest, ribosomal point mutations can decrease antibiotic binding while retaining translational activity. Many aminoglycoside antibiotics can be partially or fully resisted by bacteria with mutations in the helix 44 region of their 16 S rRNA, often in bases 1400–1410 and 1490–1500 (Fig. 8a). In contrast, Morgan et al. found that streptothricin resistance is conferred by mutations (C1054 and A1196) mapping to helix 34 of the 16 S rRNA (Fig. 1, 2023) (Fig. 8a)16. Methyltransferase enzymes that methylate specific ribosome bases provide another route to resistance and over a dozen 16 S rRNA methyltransferases are known to confer kanamycin resistance (Fig. 8b, i.e., 16 S rRNA methyltransferases)88. In contrast, no biochemically validated methyltransferase enzymes are known to confer streptothricin resistance. Our functional metagenomic selection for nourseothricin resistance, in addition to identifying novel streptothricin acetyltransferases (Fig. 5b), captured a predicted S-adenosylmethionine-dependent methyltransferase gene. The predicted novel methyltransferase did not form an outgroup on a phylogenetic tree of rRNA methyltransferase enzymes, but it also did not cluster with the aminoglycoside resistance 16 S rRNA methyltransferases (Fig. 8b). Expression of the methyltransferase-containing DNA fragment in E. coli resulted in high-level nourseothricin resistance, shifting the minimal inhibitory concentration for this mix of streptothricins from 4 μg/ml to 1024 μg/ml90. Since neither the expression of an Erm 23 S (ermC) nor a 16 S (rmtB) rRNA methyltransferase conferred streptothricin resistance, it is likely that the novel methyltransferase represents a new class of ribosome methyltransferase. We are currently characterizing the soil_nt_13615 methyltransferase and if biochemical characterization demonstrates its ability to methylate rRNA it would fill one of the missing resistance mechanisms suggested by the comparison of streptothricin to kanamycin (Fig. 4).
a E. coli 16 S rRNA representation showing approximate regions where mutations confer resistance to kanamycin (space filling, blue) or streptothricin (space filling, red). b Phylogenetic tree of ribosome methyltransferases from the comprehensive antibiotic resistance database (CARD) alongside a putative methyltransferase that confers resistance to streptothricin (soil_nt_13615, red). b is adapted from ref. 90 (CC BY 4.0).
Ecologically predicted resistance mechanisms and future steps
To the best of our knowledge, acetyltransferases targeting the β-amino group of β-lysine (Fig. 5a), streptolidine hydrolases (Fig. 7), ribosome mutation (Fig. 8a), and a putative rRNA methyltransferase (Fig. 8b) are the only documented bacterial resistance mechanisms against streptothricins, despite the apparent abundance of this antibiotic class in the soil microbiome (Fig. 3). To continue the comparison used above, kanamycin resistance mechanisms dwarf that of streptothricin and include acetyltransferases, phosphotransferases, nucleotidyltransferases, and rRNA methyltransferases (each targeting multiple sites), as well as efflux pumps (Fig. 4b). We therefore predict that some, or all, of these mechanisms exist in the soil resistome awaiting discovery. Genome mining for self-protection genes in streptothricin producers, further streptothricin functional metagenomic selections, and comparative genome analyses between resistant and susceptible bacterial taxa offer paths forward to uncovering these predicted mechanisms.
If streptothricins are developed for clinical use in humans, it would behoove medicinal chemists to consider the myriad ways bacteria may develop resistance to the antibiotic and attempt to future-proof second-generation streptothricins against this possibility (Fig. 6c). A thorough cataloging of resistance mechanisms in the soil microbiome that could mobilize into pathogens is the first step of this process, and the divergence between the expected prevalence of streptothricin in the environment and the paucity of resistance mechanisms suggests that significant work remains before this is accomplished.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
References
O’Neill, J. Tackling drug-resistant infections globally: final report and recommendations. https://wellcomecollection.org/works/thvwsuba (2016).
Wright, G. D. Opportunities for natural products in 21st century antibiotic discovery. Nat. Prod. Rep. 34, 694–701 (2017).
Tommasi, R., Brown, D. G., Walkup, G. K., Manchester, J. I. & Miller, A. A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 14, 529–542 (2015).
Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 (2007).
Talkington, K., Shore, C. & Kothari, P. A Scientific Roadmap for Antibiotic Discovery. https://www.pewtrusts.org/-/media/assets/2016/05/ascientificroadmapforantibioticdiscovery.pdf (2016).
Čivljak, R., Giannella, M., Di Bella, S. & Petrosillo, N. Could chloramphenicol be used against ESKAPE pathogens? A review of in vitro data in the literature from the 21st century. Expert Rev. Anti Infect. Ther. 12, 249–264 (2014).
Li, J. et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601 (2006).
Carpenter, C. F. & Chambers, H. F. Daptomycin: another novel agent for treating infections due to drug‐resistant Gram‐positive pathogens. Clin. Infect. Dis. 38, 994–1000 (2004).
Falagas, M. E., Grammatikos, A. P. & Michalopoulos, A. Potential of old-generation antibiotics to address current need for new antibiotics. Expert Rev. Anti Infect. Ther. 6, 593–600 (2008).
Levine, D. P. Vancomycin: a history. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 42(Suppl 1), S5–S12 (2006).
Eisenstein, B. I., Oleson, F. B. Jr. & Baltz, R. H. Daptomycin: from the mountain to the clinic, with essential help from Francis Tally, MD. Clin. Infect. Dis. 50, S10–S15 (2010).
Center for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013. https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (2013).
WHO model list of essential medicines: 20th list. http://www.who.int/medicines/publications/essentialmedicines/20th_EML2017_FINAL_amendedAug2017.pdf?ua=1 (2017).
Chahine, E. B., Dougherty, J. A., Thornby, K.-A. & Guirguis, E. H. Antibiotic approvals in the last decade: are we keeping up with resistance? Ann. Pharmacother. 56, 441–462 (2022).
Dowgiallo, M. G. et al. The convergent total synthesis and antibacterial profile of the natural product streptothricin F. Chem. Sci. 13, 3447–3453 (2022).
Morgan, C. E. et al. Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant Gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome. PLoS Biol. 21, e3002091 (2023).
Wright, G. D. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186 (2007).
Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 12, 371–387 (2013).
Waksman, S. A. & Woodruff, H. B. The soil as a source of microorganisms antagonistic to disease-producing bacteria. J. Bacteriol. 40, 581–600 (1940).
Waksman, S. A. & Woodruff, H. B. Bacteriostatic and bactericidal substances produced by a soil actinomyces. Exp. Biol. Med. 45, 609–614 (1940).
Schatz, A., Bugle, E. & Waksman, S. A. Streptomycin, a substance exhibiting antibiotic activity against Gram-positive and Gram-negative bacteria. Exp. Biol. Med. 55, 66–69 (1944).
Waksman, S. A. & Woodruff, H. B. Streptothricin, a new selective bacteriostatic and bactericidal agent, particularly active against Gram-negative bacteria. Proc. Soc. Exp. Biol. Med. 49, 207–210 (1942).
Waksman, S. A. Production and activity of streptothricin. J. Bacteriol. 46, 299–310 (1943).
Metzger, H. J., Waksman, S. A. & Pugh, L. H. In vivo activity of streptothricin against Brucella abortus. Exp. Biol. Med. 51, 251–252 (1942).
Robinson, H. J., Graessle, O. E. & Smith, D. G. Studies on the toxicity and activity of streptothricin. Science 99, 540–542 (1944).
Khokhlov, A. S. & Shutova, K. I. Chemical structure of streptothricins. J. Antibiot. 25, 501–508 (1972).
Kusumoto, S., Imaoka, S., Kambayashi, Y. & Shiba, T. Total synthesis of antibiotic streptothricin F. Tetrahedron Lett. 23, 2961–2964 (1982).
Kusumoto, S., Kambayashi, Y., Imaoka, S., Shima, K. & Shiba, T. Total chemical structure of streptothricin. J. Antibiot. 35, 925–927 (1982).
Carter, H. E. et al. Structure of the diaminohexanoic acid from streptothricin. J. Am. Chem. Soc. 74, 3704–3704 (1952).
Van Tamelen, E. E., Dyer, J. R., Carter, H. E., Pierce, J. V. & Daniels, E. E. Structure of the aminosugar derived from streptothricin and streptolin B. J. Am. Chem. Soc. 78, 4817–4818 (1956).
Carter, H. E. et al. Streptothricin and streptolin: the structure of streptolidine (roseonine). J. Am. Chem. Soc. 83, 4296–4297 (1961).
Van Tamelen, E. E., Dyer, J. R., Whaley, H. A., Carter, H. E. & Whitfield, G. B. Constitution of the streptolin-streptothricin group of Streptomyces antibiotics. J. Am. Chem. Soc. 83, 4295–4296 (1961).
Taniyama, H., Sawada, Y. & Kitagawa, T. The identity of yazumycins A and C with Racemomycins A and C. J. Antibiot. 24, 390–392 (1971).
Taniyama, H., Sawada, Y. & Kitagawa, T. Characterization of Racemomycins. Chem. Pharm. Bull. 19, 1627–1634 (1971).
Bradler, G. & Thrum, H. Nourseothricin A und B, zwei neue antibakterielle Antibiotica einer Streptomyces-noursei- variante. Z. Für Allg. Mikrobiol. 3, 105–112 (1963).
Ohba, K. et al. Albothricin, a new streptothricin antibiotic. J. Antibiot. 39, 872–875 (1986).
Hunt, A. H., Hamill, R. L., Deboer, J. R. & Presti, E. A. A37812: N-methylstreptothricin F. J. Antibiot. 38, 987–992 (1985).
Kim, B. T. et al. N-Methylstreptothricin D-A new streptothricin-group antibiotic from a Streptomyces spp. J. Antibiot. 47, 1333–1336 (1994).
Ji, Z., Wang, M., Wei, S., Zhang, J. & Wu, W. Isolation, structure elucidation and antibacterial activities of streptothricin acids. J. Antibiot. 62, 233–237 (2009).
Hamano, Y., Matsuura, N., Kitamura, M. & Takagi, H. A novel enzyme conferring streptothricin resistance alters the toxicity of streptothricin D from broad-spectrum to bacteria-specific. J. Biol. Chem. 281, 16842–16848 (2006).
Gan, M. et al. Two streptothricins with a cis-streptolidine lactam moiety from Streptomyces sp. I08A 1776. J. Antibiot. 65, 513–516 (2012).
Gan, M. et al. Streptothricin derivatives from Streptomyces sp. I08A 1776. J. Nat. Prod. 74, 1142–1147 (2011).
Ji, Z., Wang, M., Zhang, J., Wei, S. & Wu, W. Two new members of streptothricin class antibiotics from Streptomyces qinlingensis sp. nov. J. Antibiot. 60, 739–744 (2007).
Borders, D. B. et al. Structures of LL-AC541 and LL-AB664. Tetrahedron 26, 3123–3133 (1970).
Taniyama, H. & Sawada, Y. The identity of citromycin with LL-AG541, E-749-C, and BY-81. J. Antibiot. 24, 708–709 (1971).
Kusakabe, Y. et al. Citromycin, a new antibiotic. I Isolation and characterization. J. Antibiot. 22, 112–118 (1969).
Sawada, Y., Kawakami, S. & Taniyama, H. Glycinothricin, a new streptothricin-class antibiotic from Streptomyces griseus. J. Antibiot. 30, 460–467 (1977).
Ito, Y. et al. New basic water-soluble antibiotics BD-12 and BY-81. II Isolation, purification, and properties. J. Antibiot. 21, 307–312 (1968).
Tsuruoka, T., Shoumura, T., Ezaki, N., Niwa, T. & Niida, T. SF-701, a new streptothricin-like antibiotic. J. Antibiot. 21, 237–238 (1968).
Kido, Y. et al. A streptothricin-like antibiotic mixture, A-269A (and A-269A’). J. Antibiot. 40, 1698–1706 (1987).
Borders, D. B., Kirby, J. P., Wetzel, E. R., Davies, M. C. & Hausmann, W. K. Analytical method for streptothricin-type antibiotics: structure of antibiotic LL-BL136. Antimicrob. Agents Chemother. 1, 403–407 (1972).
Kono, Y., Makino, S., Takeuchi, S. & Yonehara, H. Sclerothricin, a new basic antibiotic. J. Antibiot. 22, 583–589 (1969).
Elshahawi, S. I., Shaaban, K. A., Kharel, M. K. & Thorson, J. S. A comprehensive review of glycosylated bacterial natural products. Chem. Soc. Rev. 44, 7591–7697 (2015).
Khokhlov, A. S. Streptothricins and related antibiotics. J. Chromatogr. Library 15, 617–713 (1978).
Misra, T. K. & Sinha, R. K. Mechanism of action of boseimycin. Experientia 27, 642–644 (1971).
Haupt, I., Hübener, R. & Thrum, H. Streptothricin F, an inhibitor of protein synthesis with miscoding activity. J. Antibiot. 31, 1137–1142 (1978).
Haupt, I., Jonák, J., Rychlík, I. & Thrum, H. Action of streptothricin F on ribosomal functions. J. Antibiot. 33, 636–641 (1980).
Davies, J. & Davis, B. D. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. J. Biol. Chem. 243, 3312–3316 (1968).
Stanley, A. R. Toxicity of streptothricin. J. Bacteriol. 52, 399 (1946).
Inamori, Y. et al. Toxicological approachs to streptothricin antibiotics. I. Implications of delayed toxicity in mice. Chem. Pharm. Bull. 26, 1147–1152 (1978).
Inamori, Y. et al. Toxicological approaches to streptothricin antibiotics. II. The developmental mechanism of delayed toxicity in mice and rats. Chem. Pharm. Bull. 27, 230–234 (1979).
Inamori, Y. et al. Toxicological approaches to streptothricin antibiotics. III. Biological studies on delayed toxicity of streptothricin antibiotics in rats. Chem. Pharm. Bull. 27, 2570–2576 (1979).
Kato, Y. et al. Toxicological approaches to streptothricin antibiotics. IV. Toxicity of streptothricin antibiotics to the blood. Chem. Pharm. Bull. 29, 580–584 (1981).
Illingworth, S. & Molnar, N. An interview with H. Boyd Woodruff for the Rutgers oral history archives. https://oralhistory.rutgers.edu/interviewees/30-interview-html-text/60-woodruff-h-boyd (2004).
Takeuchi, Y. et al. First direct evidence for direct cell-membrane penetrations of polycationic homopoly(amino acid)s produced by bacteria. Commun. Biol. 5, 1132 (2022).
Greenbaum, S. S. Clavacin. J. Am. Med. Assoc. 129, 1045 (1945).
Farley, K. R. & Metcalf, W. W. The streptothricin acetyltransferase (sat) gene as a positive selectable marker for methanogenic archaea. FEMS Microbiol. Lett. 366, fnz216 (2019).
Joshi, P. B., Webb, J. R., Davies, J. E. & McMaster, W. R. The gene encoding streptothricin acetyltransferase (sat) as a selectable marker for Leishmania expression vectors. Gene 156, 145–149 (1995).
Jelenska, J., Tietze, E., Tempé, J. & Brevet, J. Streptothricin resistance as a novel selectable marker for transgenic plant cells. Plant Cell Rep. 19, 298–303 (2000).
Goldstein, A. L. & McCusker, J. H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553 (1999).
Obinata, H., Sugimoto, A. & Niwa, S. Streptothricin acetyl transferase 2 (Sat2): a dominant selection marker for Caenorhabditis elegans genome editing. PLoS ONE 13, e0197128 (2018).
Kochupurakkal, B. S. & Iglehart, J. D. Nourseothricin N-acetyl transferase: a positive selection marker for mammalian cells. PLoS ONE 8, e68509 (2013).
Tschäpe, H. et al. Plasmid-borne streptothricin resistance in gram-negative bacteria. Plasmid 12, 189–196 (1984).
Hummel, R., Tschäpe, H. & Witte, W. Spread of plasmid-mediated nourseothricin resistance due to antibiotic use in animal husbandry. J. Basic Microbiol. 26, 461–466 (1986).
Webb, H. E., Angulo, F. J., Granier, S. A., Scott, H. M. & Loneragan, G. H. Illustrative examples of probable transfer of resistance determinants from food animals to humans: streptothricins, glycopeptides, and colistin. F1000Research 6, 1805 (2017).
Sundström, L., Roy, P. H. & Sköld, O. Site-specific insertion of three structural gene cassettes in transposon Tn7. J. Bacteriol. 173, 3025–3028 (1991).
Taylor, P. & Reeder, R. Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agric. Biosci. 1, 1 (2020).
Zhang, T. et al. Is business linkage affecting agricultural advisory services. Int. J. Agric. Extens. 5, 59–77 (2017).
Crofts, T. S., Gasparrini, A. J. & Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 15, 422–434 (2017).
Baltz, R. H. Antimicrobials from actinomycetes: back to the future. Microbe 2, 125–131 (2007).
Cox, G. et al. A common platform for antibiotic dereplication and adjuvant discovery. Cell Chem. Biol. 24, 98–109 (2017).
Culp, E. J. et al. Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics. Nat. Biotechnol. 37, 1149–1154 (2019).
Chevrette, M. G. & Handelsman, J. Needles in haystacks: reevaluating old paradigms for the discovery of bacterial secondary metabolites. Nat. Prod. Rep. 38, 2083–2099 (2021).
Ji, Z., Wei, S., Zhang, J., Wu, W. & Wang, M. Identification of streptothricin class antibiotics in the earlystage of antibiotics screening by electrospray ionization mass spectrometry. J. Antibiot. 61, 660–667 (2008).
D’Costa, V. M., McGrann, K. M., Hughes, D. W. & Wright, G. D. Sampling the antibiotic resistome. Science 311, 374–377 (2006).
Wencewicz, T. A. Crossroads of antibiotic resistance and biosynthesis. J. Mol. Biol. 431, 3370–3399 (2019).
Benveniste, R. & Davies, J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 70, 2276–2280 (1973).
Van Hoek, A. H. A. M. et al. Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2, 1–27 (2011).
Maruyama, C. & Hamano, Y. The biological function of the bacterial isochorismatase-like hydrolase SttH. Biosci. Biotechnol. Biochem. 73, 2494–2500 (2009).
Crofts, T. S., McFarland, A. G. & Hartmann, E. M. Mosaic ends tagmentation (METa) assembly for highly efficient construction of functional metagenomic libraries. mSystems 6, e0052421 (2021).
Deng, Z., Yu, Y. & Zhang, Q. Parallel pathways in the biosynthesis of aminoglycoside antibiotics. F1000Research 6, 1–9 (2017).
Ramirez, M. S. & Tolmasky, M. E. Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171 (2010).
Labby, K. J. & Garneau-Tsodikova, S. Strategies to overcome the action of aminoglycoside-modifying enzymes for treating resistant bacterial infections. Future Med. Chem. 5, 1285–1309 (2013).
Doi, Y., Wachino, J. I. & Arakawa, Y. Nomenclature of plasmid-mediated 16S rRNA methylases responsible for panaminoglycoside resistance. Antimicrob. Agents Chemother. 52, 2287–2288 (2008).
Crofts, T. S. et al. Discovery and characterization of a nitroreductase capable of conferring bacterial resistance to chloramphenicol. Cell Chem. Biol. 26, 559–570.e6 (2019).
Green, K. D., Fosso, M. Y., Mayhoub, A. S. & Garneau-Tsodikova, S. Investigating the promiscuity of the chloramphenicol nitroreductase from Haemophilus influenzae towards the reduction of 4-nitrobenzene derivatives. Bioorg. Med. Chem. Lett. 29, 1127–1132 (2019).
Tao, W. et al. Inactivation of chloramphenicol and florfenicol by a novel chloramphenicol hydrolase. Appl. Environ. Microbiol. 78, 6295–6301 (2012).
Schwarz, S., Kehrenberg, C., Doublet, B. & Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542 (2004).
Kehrenberg, C., Schwarz, S., Jacobsen, L., Hansen, L. H. & Vester, B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57, 1064–1073 (2005).
Zhang, L. et al. Bacterial dehydrogenases facilitate oxidative inactivation and bioremediation of chloramphenicol. ChemBioChem https://doi.org/10.1002/cbic.202200632 (2022).
Gasparrini, A. J. et al. Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun. Biol. 3, 241 (2020).
Moore, I. F., Hughes, D. W. & Wright, G. D. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry 44, 11829–11835 (2005).
Grossman, T. H. Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 6, a025387 (2016).
Fyfe, C., Grossman, T. H., Kerstein, K. & Sutcliffe, J. Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb. Perspect. Med. 6, a025395 (2016).
Keeratipibul, S., Sugiyama, M. & Nomi, R. Mechanism of resistance to streptothricin of a producing microorganism. Biotechnol. Lett. 5, 441–446 (1983).
Haupt, I. & Thrum, H. Bacterial resistance to streptothricins. J. Basic Microbiol. 25, 335–339 (1985).
Kobayashi, T., Uozumi, T. & Beppu, T. Cloning and characterization of the streptothricin-resistance gene which encodes streptothricin acetyltransferase from Streptomyces lavendulae. J. Antibiot. 39, 688–693 (1986).
Horinouchi, S., Furuya, K., Nishiyama, M., Suzuki, H. & Beppu, T. Nucleotide sequence of the streptothricin acetyltransferase gene from Streptomyces lavendulae and its expression in heterologous hosts. J. Bacteriol. 169, 1929–1937 (1987).
Kobayashi, T., Horinouchi, S., Uozumi, T. & Beppu, T. Purification and biochemical characterization of streptothricin acetyltransferase coded by the cloned streptothricin-resistance gene of Streptomyces lavendulae. J. Antibiot. 40, 1016–1022 (1987).
Zähringer, U., Voigt, W. & Seltmann, G. Noureseothricin (streptothricin) inactivated by a plasmid pIE636 encoded acetyl transferase of Escherichia coli: location of the acetyl group. FEMS Microbiol. Lett. 110, 331–334 (1993).
Burckhardt, R. M. & Escalante-Semerena, J. C. In Bacillus subtilis, the SatA (formerly YyaR) acetyltransferase detoxifies streptothricin via lysine acetylation. Appl. Environ. Microbiol. 83, 1–11 (2017).
Burckhardt, R. M. & Escalante-Semerena, J. C. Insights into the function of the N-acetyltransferase SatA that detoxifies streptothricin in Bacillus subtilis and Bacillus anthracis. Appl. Environ. Microbiol. 85, e03029–18 (2019).
Krügel, H., Fiedler, G., Smith, C. & Baumberg, S. Sequence and transcriptional analysis of the nourseothricin acetyltransferase-encoding gene nat1 from Streptomyces noursei. Gene 127, 127–131 (1993).
Fernández-Moreno, M. A., Vallín, C. & Malpartida, F. Streptothricin biosynthesis is catalyzed by enzymes related to nonribosomal peptide bond formation. J. Bacteriol. 179, 6929–6936 (1997).
Jacob, J., Evers, S., Bischoff, K., Carlier, C. & Courvalin, P. Characterization of the sat 4 gene encoding a streptothricin acetyltransferase in Campylobacter coli BE/G4. FEMS Microbiol. Lett. 120, 13–17 (1994).
Heim, U., Tietze, E., Weschke, W., Tschäpe, H. & Wobus, U. Nucleotide sequence of a plasmid born streptothricin-acetyl-transferase gene (sat-1). Nucleic Acids Res. 17, 7103 (1989).
Tietze, E. & Brevet, J. Nucleotide sequence of the bacterial streptothricin resistance gene sat3. Biochim. Biophys. Acta BBA Gene Struct. Expr 1263, 176–178 (1995).
Tietze, E. & Brevet, J. Nucleotide sequence of the streptothricin-acetyl-transferase gene sat-2. Nucleic Acids Res. 18, 1283–1283 (1990).
UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015).
Alcock, B. P. et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525 (2020).
Smalla, K. et al. Distribution of streptothricin acetyltransferase encoding determinants among environmental bacteria. Mol. Ecol. 2, 27–33 (1993).
Cox, G. et al. Plazomicin retains antibiotic activity against most aminoglycoside modifying enzymes. ACS Infect. Dis. 4, 980–987 (2018).
Cannon, M., Harford, S. & Davies, J. A comparative study on the inhibitory actions of chloramphenicol, thiamphenicol and some fluorinated derivatives. J. Antimicrob. Chemother. 26, 307–317 (1990).
Wang, Y.-L. et al. N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry. Nat. Commun. 14, 2528 (2023).
Maruyama, C. et al. A stand-alone adenylation domain forms amide bonds in streptothricin biosynthesis. Nat. Chem. Biol. 8, 791–797 (2012).
Hamano, Y., Maruyama, C. & Kimoto, H. Construction of a knockout mutant of the streptothricin-resistance gene in Streptomyces albulus by electroporation. Actinomycetologica 20, 35–41 (2006).
Acknowledgements
We would like to thank Dr. Tomko for the critical reading of the manuscript. This work was supported by a grant from the Florida State University Council for Research and Creativity and by start-up funds from the Florida State University College of Medicine.
Author information
Authors and Affiliations
Contributions
CRediT taxonomy of authorship: Conceptualization, T.S.C.; writing—original draft, E.F. and T.S.C.; writing—review and editing, E.F. and T.S.C.; visualization, T.S.C.; supervision, T.S.C.; funding acquisition, T.S.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Franck, E., Crofts, T.S. History of the streptothricin antibiotics and evidence for the neglect of the streptothricin resistome. npj Antimicrob Resist 2, 3 (2024). https://doi.org/10.1038/s44259-023-00020-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44259-023-00020-5