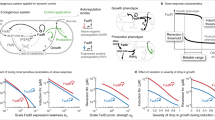
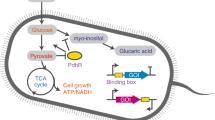
Abstract
Metabolic engineering is a powerful tool to reprogramme cells to produce value-added chemicals. Such engineering strategies require the fine-tuning of a cell’s metabolism to balance competition for resources and prevent negative impacts on growth. Dynamic regulation enables the shifting of resources or metabolic flux toward different pathways based on a received input to increase titres of value-added chemicals in microbial production strains. In this Review, we discuss autonomous dynamic regulation, that is, responses triggered directly by a stimulus without the need for human intervention, and its application to metabolic engineering. We highlight strategies to control the transcription of genes using metabolite-specific regulation, including by transcription factors and through biosensing, and non-specific regulation, in particular, environmental regulation, growth-phase responses and quorum sensing, examining the application of these regulation strategies to the bioproduction of different chemicals.
Key points
-
Autonomous dynamic regulation can be used to shift metabolic flux in microorganisms without external intervention to control the accumulation of biomass or metabolites.
-
Metabolite-specific transcription factors and biosensors provide fine-tuned control of the expression of target genes requiring specific substrates for induction.
-
Globally regulated promoters can regulate genes using native machinery in response to common metabolites but are often subject to carbon catabolite repression and require complex networks of regulation.
-
Environmentally responsive regulators that respond to external stimuli, such as metals, pH, light and temperature, are useful tools for bioremediation and specialized bioproduction.
-
Quorum sensing, which provides consistent regulation based on the population density of the culture, is a versatile regulation strategy but requires cellular resources to produce and detect signalling molecules.
-
Autonomous dynamic regulation may improve the efficiency of bioproduction and bioremediation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Ni, C., Dinh, C. V. & Prather, K. L. J. Dynamic control of metabolism. Annu. Rev. Chem. Biomol. Eng. 12, 519–541 (2021).
Tan, S. Z. & Prather, K. L. Dynamic pathway regulation: recent advances and methods of construction. Curr. Opin. Chem. Biol. 41, 28–35 (2017).
Cui, S. et al. Multilayer genetic circuits for dynamic regulation of metabolic pathways. ACS Synth. Biol. 10, 1587–1597 (2021).
Xu, P. Production of chemicals using dynamic control of metabolic fluxes. Curr. Opin. Biotechnol. 53, 12–19 (2018).
Shen, X., Wang, J., Li, C., Yuan, Q. & Yan, Y. Dynamic gene expression engineering as a tool in pathway engineering. Curr. Opin. Biotechnol. 59, 122–129 (2019).
Lv, Y. et al. Coupling feedback genetic circuits with growth phenotype for dynamic population control and intelligent bioproduction. Metab. Eng. 54, 109–116 (2019).
Zhang, Q. et al. Production of proteins and commodity chemicals using engineered Bacillus subtilis platform strain. Essays Biochem. 65, 173–185 (2021).
Venayak, N., Anesiadis, N., Cluett, W. R. & Mahadevan, R. Engineering metabolism through dynamic control. Curr. Opin. Biotechnol. 34, 142–152 (2015).
Zhang, X., Lin, Y., Wu, Q., Wang, Y. & Chen, G. Q. Synthetic biology and genome-editing tools for improving PHA metabolic engineering. Trends Biotechnol. 38, 689–700 (2020).
Li, C., Zhang, R., Wang, J., Wilson, L. M. & Yan, Y. Protein engineering for improving and diversifying natural product biosynthesis. Trends Biotechnol. 38, 729–744 (2020).
Zubi, Y. S. et al. Metal-responsive regulation of enzyme catalysis using genetically encoded chemical switches. Nat. Commun. 13, 1864 (2022).
Grohmann, C. et al. Development of NanoLuc-targeting protein degraders and a universal reporter system to benchmark tag-targeted degradation platforms. Nat. Commun. 13, 2073 (2022).
Zaslaver, A. et al. Just-in-time transcription program in metabolic pathways. Nat. Genet. 36, 486–491 (2004).
Gao, C., Xu, P., Ye, C., Chen, X. & Liu, L. Genetic circuit-assisted smart microbial engineering. Trends Microbiol. 27, 1011–1024 (2019).
Moser, F. et al. Dynamic control of endogenous metabolism with combinatorial logic circuits. Mol. Syst. Biol. 14, e8605 (2018).
Liang, C. et al. Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit. Metab. Eng. 57, 239–246 (2020).
Zhang, D. et al. Global and gene-specific translational regulation in Escherichia coli across different conditions. PLoS Comput. Biol. 18, e1010641 (2022).
Balleza, E. et al. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol. Rev. 33, 133–151 (2009).
Engstrom, M. D. & Pfleger, B. F. Transcription control engineering and applications in synthetic biology. Synth. Syst. Biotechnol. 2, 176–191 (2017).
Mitchler, M. M., Garcia, J. M., Montero, N. E. & Williams, G. J. Transcription factor-based biosensors: a molecular-guided approach for natural product engineering. Curr. Opin. Biotechnol. 69, 172–181 (2021).
Zhou, S., Alper, H. S., Zhou, J. & Deng, Y. Intracellular biosensor-based dynamic regulation to manipulate gene expression at the spatiotemporal level. Crit. Rev. Biotechnol. 43, 646–663 (2023).
Lyu, M. et al. AccR, a TetR family transcriptional repressor, coordinates short-chain acyl coenzyme A homeostasis in Streptomyces avermitilis. Appl Environ Microbiol. 86, e00508–e00520 (2020).
Chen, Y. et al. Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 9, 64 (2018).
d’Oelsnitz, S. et al. Using fungible biosensors to evolve improved alkaloid biosyntheses. Nat. Chem. Biol. 18, 981–989 (2022).
Xiong, D. et al. Improving key enzyme activity in phenylpropanoid pathway with a designed biosensor. Metab. Eng. 40, 115–123 (2017).
Zhou, S. et al. Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli. Metab. Eng. 67, 41–52 (2021).
Wu, Y. et al. CRISPR–dCas12a-mediated genetic circuit cascades for multiplexed pathway optimization. Nat. Chem. Biol. 19, 367–377 (2023).
Brückner, R. & Titgemeyer, F. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209, 141–148 (2002).
Kunitake, E. et al. cAMP signaling factors regulate carbon catabolite repression of hemicellulase genes in Aspergillus nidulans. AMB Express 12, 126 (2022).
Galinier, A. et al. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc. Natl Acad. Sci. USA 95, 1823–1828 (1998).
Weinhandl, K., Winkler, M., Glieder, A. & Camattari, A. Carbon source dependent promoters in yeasts. Microb. Cell Fact. 13, 5 (2014).
Ferreira, R. et al. Metabolic engineering of Saccharomyces cerevisiae for overproduction of triacylglycerols. Metab. Eng. Commun. 6, 22–27 (2018).
Görke, B. & Stülke, J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624 (2008).
Yu, T. et al. Metabolic reconfiguration enables synthetic reductive metabolism in yeast. Nat. Metab. 4, 1551–1559 (2022).
Kong, W., Qian, Y., Stewart, P. S. & Lu, T. De novo engineering of a bacterial lifestyle program. Nat. Chem. Biol. 19, 488–497 (2022).
Deng, J. et al. A synthetic promoter system for well-controlled protein expression with different carbon sources in Saccharomyces cerevisiae. Microb. Cell Fact. 20, 202 (2021).
Blazeck, J., Garg, R., Reed, B. & Alper, H. S. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol. Bioeng. 109, 2884–2895 (2012).
Callaghan, J. D. et al. Xylose-inducible promoter tools for Pseudomonas species and their use in implicating a role for the type II secretion system protein XcpQ in the inhibition of corneal epithelial wound closure. Appl. Environ. Microbiol. 86, e00250-20 (2020).
Wei, W. et al. Engineering prokaryotic transcriptional activator xylr as a xylose-inducible biosensor for transcription activation in yeast. ACS Synth. Biol. 9, 1022–1029 (2020).
Kim, S., Lee, K., Bae, S. J. & Hahn, J. S. Promoters inducible by aromatic amino acids and γ-aminobutyrate (GABA) for metabolic engineering applications in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 99, 2705–2714 (2015).
den Haan, R. et al. Heterologous production of cellulose- and starch-degrading hydrolases to expand Saccharomyces cerevisiae substrate utilization: lessons learnt. Biotechnol. Adv. 53, 107859 (2021).
Zhang, M. et al. MULTiPly: a novel multi-layer predictor for discovering general and specific types of promoters. Bioinformatics 35, 2957–2965 (2019).
Li, F. et al. Computational prediction and interpretation of both general and specific types of promoters in Escherichia coli by exploiting a stacked ensemble-learning framework. Brief. Bioinform. 22, 2126–2140 (2021).
Tang, H. et al. Promoter architecture and promoter engineering in Saccharomyces cerevisiae. Metabolites 10, 320 (2020).
Yang, Y. et al. Structural visualization of transcription activated by a multidrug-sensing MerR family regulator. Nat. Commun. 12, 2702 (2021).
Su, Y., Liu, C., Jiang, X. & Wei, W. Different bacterial host-based lux reporter array for fast identification and toxicity indication of multiple metal ions. Anal. Bioanal. Chem. 412, 8127–8134 (2020).
Travis, B. A. et al. Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria. Nat. Commun. 13, 3793 (2022).
Stirling, F. et al. Synthetic cassettes for pH-mediated sensing, counting, and containment. Cell Rep. 30, 3139–3148.e4 (2020).
Chin, M. Y. et al. Genetically encoded, pH-sensitive mTFP1 biosensor for probing lysosomal pH. ACS Sens. 6, 2168–2180 (2021).
Chien, T. et al. Enhancing the tropism of bacteria via genetically programmed biosensors. Nat. Biomed. Eng. 6, 94–104 (2022).
Ji, H., Lu, X., Zong, H. & Zhuge, B. A synthetic hybrid promoter for D-xylonate production at low pH in the tolerant yeast Candida glycerinogenes. Bioengineered 8, 700–706 (2017).
Yin, X. et al. P gas, a low-pH-induced promoter, as a tool for dynamic control of gene expression for metabolic engineering of Aspergillus niger. Appl. Environ. Microbiol. 83, e03222-16 (2017).
Healey, E. M. et al. Effects of nitrate and ammonium on assimilation of nitric oxide by Heterosigma akashiwo. Sci. Rep. 13, 621 (2023).
Rohac, R. et al. Structural determinants of DNA recognition by the NO sensor NsrR and related Rrf2-type [FeS]-transcription factors. Commun. Biol. 5, 769 (2022).
Huang, M. et al. Efficient production of succinic acid in engineered Escherichia coli strains controlled by anaerobically-induced nirB promoter using sweet potato waste hydrolysate. J. Environ. Manag. 237, 147–154 (2019).
Magerand, R., Rey, P., Blanchard, L. & de Groot, A. Redox signaling through zinc activates the radiation response in Deinococcus bacteria. Sci. Rep. 11, 4528 (2021).
Xia, P. et al. Zinc is an important inter-kingdom signal between the host and microbe. Vet. Res. 52, 39 (2021).
Harms, A., Brodersen, D. E., Mitarai, N. & Gerdes, K. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell 70, 768–784 (2018).
Zhen, X. et al. Molecular mechanism of toxin neutralization in the HipBST toxin-antitoxin system of Legionella pneumophila. Nat. Commun. 13, 4333 (2022).
Mobed, A. & Hasanzadeh, M. Sensitive recognition of Shiga toxin using biosensor technology: an efficient platform towards bioanalysis of pathogenic bacterial. Microchem. J. 172, 106900 (2022).
Bertani, P. & Lu, W. Cyanobacterial toxin biosensors for environmental monitoring and protection. Med. Nov. Technol. Devices 10, 100059 (2021).
Selim, A. S., Perry, J. M., Nasr, M. A., Pimprikar, J. M. & Shih, S. C. C. A synthetic biosensor for detecting putrescine in beef samples. ACS Appl. Bio Mater. 5, 5487–5496 (2022).
Schmauder, L., Sima, S., Hadj, A. B., Cesar, R. & Richter, K. Binding of the HSF-1 DNA-binding domain to multimeric C. elegans consensus HSEs is guided by cooperative interactions. Sci Rep 12, 8984 (2022).
Almblad, H. et al. Bacterial cyclic diguanylate signaling networks sense temperature. Nat. Commun. 12, 1986 (2021).
Wang, X. et al. Reversible thermal regulation for bifunctional dynamic control of gene expression in Escherichia coli. Nat. Commun. 12, 1411 (2021).
Lovelett, R. J. et al. Dynamical modeling of optogenetic circuits in yeast for metabolic engineering applications. ACS Synth. Biol. 10, 219–227 (2021).
Zhao, E. M. et al. Optogenetic amplification circuits for light-induced metabolic control. ACS Synth. Biol. 10, 1143–1154 (2021).
Zhao, E. M. et al. Design and characterization of rapid optogenetic circuits for dynamic control in yeast metabolic engineering. ACS Synth. Biol. 9, 3254–3266 (2020).
Lalwani, M. A. et al. Optogenetic control of the lac operon for bacterial chemical and protein production. Nat. Chem. Biol. 17, 71–79 (2021).
An-adirekkun, J. et al. A yeast optogenetic toolkit (yOTK) for gene expression control in Saccharomyces cerevisiae. Biotechnol. Bioeng. 117, 886–893 (2020).
Zhang, J. et al. Near-infrared light-activatable spherical nucleic acids for conditional control of protein activity. Angew. Chem. Int. Ed. 61, e202117562 (2022).
Dahl, R. H. et al. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 31, 1039–1046 (2013).
Yang, S., Du, G., Chen, J. & Kang, Z. Characterization and application of endogenous phase-dependent promoters in Bacillus subtilis. Appl. Microbiol. Biotechnol. 101, 4151–4161 (2017).
Boo, A., Amaro, R. L. & Stan, G.-B. Quorum sensing in synthetic biology: a review. Curr. Opin. Syst. Biol. 28, 100378 (2021).
Mukherjee, S. & Bassler, B. L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382 (2019).
Gu, P. et al. Application of quorum sensing system in microbial synthesis of valuable chemicals: a mini-review. World J. Microbiol. Biotechnol. 38, 192 (2022).
Garg, N., Manchanda, G. & Kumar, A. Bacterial quorum sensing: circuits and applications. Antonie van Leeuwenhoek 105, 289–305 (2014).
Choudhary, S. & Schmidt-Dannert, C. Applications of quorum sensing in biotechnology. Appl. Microbiol. Biotechnol. 86, 1267–1279 (2010).
Wu, S., Liu, J., Liu, C., Yang, A. & Qiao, J. Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 77, 1319–1343 (2020).
Kylilis, N., Tuza, Z. A., Stan, G. B. & Polizzi, K. M. Tools for engineering coordinated system behaviour in synthetic microbial consortia. Nat. Commun. 9, 2677 (2018).
Papenfort, K. & Bassler, B. L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588 (2016).
Tekel, S. J. et al. Engineered orthogonal quorum sensing systems for synthetic gene regulation in Escherichia coli. Front. Bioeng. Biotechnol. 7, 80 (2019).
Jiang, W. et al. Two completely orthogonal quorum sensing systems with self-produced autoinducers enable automatic delayed cascade control. ACS Synth. Biol. 9, 2588–2599 (2020).
Grant, P. K. et al. Orthogonal intercellular signaling for programmed spatial behavior. Mol. Syst. Biol. 12, 849 (2016).
Scott, S. R. et al. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat. Microbiol. 2, 17083 (2017).
Soma, Y. & Hanai, T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab. Eng. 30, 7–15 (2015).
Gupta, A., Reizman, I. M. B., Reisch, C. R. & Prather, K. L. J. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 35, 273–279 (2017).
Soma, Y. et al. Design of synthetic quorum sensing achieving induction timing-independent signal stabilization for dynamic metabolic engineering of E. coli. ACS Synth. Biol. 10, 1384–1393 (2021).
Qin, H.-B., Zhou, J.-P., Zhang, B., Liu, Z.-Q. & Zheng, Y.-G. Combing with redox regulation via quorum-sensing system and fermentation strategies for improving D-pantothenic acid production. Process Biochem. 121, 681–688 (2022).
Ge, C. et al. Redesigning regulatory components of quorum-sensing system for diverse metabolic control. Nat. Commun. 13, 2182 (2022).
Styles, M. J. et al. Autoinducer-fluorophore conjugates enable FRET in LuxR proteins in vitro and in cells. Nat. Chem. Biol. 18, 1115–1124 (2022).
Gao, C. et al. Dynamic consolidated bioprocessing for direct production of xylonate and shikimate from xylan by Escherichia coli. Metab. Eng. 60, 128–137 (2020).
Liu, H. & Lu, T. Autonomous production of 1,4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli. Metab. Eng. 29, 135–141 (2015).
Hawver, L. A., Jung, S. A. & Ng, W. L. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 40, 738–752 (2016).
Tian, J. et al. Developing an endogenous quorum-sensing based CRISPRi circuit for autonomous and tunable dynamic regulation of multiple targets in Streptomyces. Nucleic Acids Res. 48, 8188–8202 (2020).
Gamby, S. et al. Altering the communication networks of multispecies microbial systems using a diverse toolbox of AI-2 analogues. ACS Chem. Biol. 7, 1023–1030 (2012).
Hu, F., Liu, Y. & Li, S. Rational strain improvement for surfactin production: enhancing the yield and generating novel structures. Microb. Cell Fact. 18, 42 (2019).
van Gestel, J. et al. Short-range quorum sensing controls horizontal gene transfer at micron scale in bacterial communities. Nat. Commun. 12, 2324 (2021).
Rutherford, S. T. & Bassler, B. L. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2, a012427 (2012).
Lin, J., Cheng, J., Wang, Y. & Shen, X. The Pseudomonas quinolone signal (PQS): not just for quorum sensing anymore. Front. Cell. Infect. Microbiol. 8, 230 (2018).
Hauk, P. et al. Homologous quorum sensing regulatory circuit: a dual-input genetic controller for modulating quorum sensing-mediated protein expression in E. coli. ACS Synth. Biol. 9, 2692–2702 (2020).
Dinh, C. V. & Prather, K. L. Layered and multi-input autonomous dynamic control strategies for metabolic engineering. Curr. Opin. Biotechnol. 65, 156–162 (2020).
Doong, S. J., Gupta, A. & Prather, K. L. J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc. Natl Acad. Sci. USA 115, 2964–2969 (2018).
Biarnes-Carrera, M., Lee, C. K., Nihira, T., Breitling, R. & Takano, E. Orthogonal regulatory circuits for Escherichia coli based on the γ-butyrolactone system of Streptomyces coelicolor. ACS Synth. Biol. 7, 1043–1055 (2018).
Wu, J. et al. Developing a pathway-independent and full-autonomous global resource allocation strategy to dynamically switching phenotypic states. Nat. Commun. 11, 5521 (2020).
Malešević, M. et al. Pseudomonas aeruginosa quorum sensing inhibition by clinical isolate Delftia tsuruhatensis 11304: involvement of N-octadecanoylhomoserine lactones. Sci. Rep. 9, 16465 (2019).
Meschwitz, S. M. et al. Antagonism of quorum sensing phenotypes by analogs of the marine bacterial secondary metabolite 3-methyl-N-(20-phenylethyl)-butyramide. Mar. Drugs 17, 389 (2019).
Swem, L. R. et al. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell 35, 143–153 (2009).
Kato, L. M., Kawamoto, S., Maruya, M. & Fagarasan, S. The role of the adaptive immune system in regulation of gut microbiota. Immunol. Rev. 260, 67–75 (2014).
Yue, B. et al. Regulation of the intestinal microbiota: an emerging therapeutic strategy for inflammatory bowel disease. World J. Gastroenterol. 26, 4378–4393 (2020).
Wen, J., Tian, L., Liu, Q., Zhang, Y. & Cai, M. Engineered dynamic distribution of malonyl-CoA flux for improving polyketide biosynthesis in Komagataella phaffii. J. Biotechnol. 320, 80–85 (2020).
Kunjapur, A. M. & Prather, K. L. J. Development of a vanillate biosensor for the vanillin biosynthesis pathway in E. coli. ACS Synth. Biol. 8, 1958–1967 (2019).
Li, C. et al. Intelligent microbial cell factory with genetic pH shooting (GPS) for cell self-responsive base/acid regulation. Microb. Cell Fact. 19, 202 (2020).
Bandi, C. K. et al. Engineered regulon to enable autonomous azide ion biosensing, recombinant protein production, and in vivo glycoengineering. ACS Synth. Biol. 10, 682–689 (2021).
Li, B. et al. Structural and mechanistic basis for redox sensing by the cyanobacterial transcription regulator RexT. Commun. Biol. 5, 275 (2022).
Brandenberg, O. F., Schubert, O. T. & Kruglyak, L. Towards synthetic PETtrophy: engineering Pseudomonas putida for concurrent polyethylene terephthalate (PET) monomer metabolism and PET hydrolase expression. Microb. Cell Fact. 21, 119 (2022).
Dinh, C. V., Chen, X. & Prather, K. L. J. Development of a quorum-sensing based circuit for control of coculture population composition in a naringenin production system. ACS Synth. Biol. 9, 590–597 (2020).
Kim, E. M. et al. Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli. Metab. Eng. 44, 325–336 (2017).
Taylor, N. D. et al. Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 13, 177–183 (2016).
Charlier, D., Nguyen Le Minh, P. & Roovers, M. Regulation of carbamoylphosphate synthesis in Escherichia coli: an amazing metabolite at the crossroad of arginine and pyrimidine biosynthesis. Amino Acids 50, 1647–1661 (2018).
Kotoky, R., Ogawa, N. & Pandey, P. The structure-function relationship of bacterial transcriptional regulators as a target for enhanced biodegradation of aromatic hydrocarbons. Microbiol. Res. 262, 127087 (2022).
Huang, X., Song, Q., Guo, S. & Fei, Q. Transcription regulation strategies in methylotrophs: progress and challenges. Bioresour. Bioprocess. 9, 126 (2022).
Dong, X. et al. Genetic manipulation of the human gut bacterium Eggerthella lenta reveals a widespread family of transcriptional regulators. Nat. Commun. 13, 7624 (2022).
Karr, J. P., Ferrie, J. J., Tjian, R. & Darzacq, X. The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer–promoter communication. Genes Dev. 36, 7–16, https://doi.org/10.1101/GAD.349160.121 (2022).
Shahein, A. et al. Systematic analysis of low-affinity transcription factor binding site clusters in vitro and in vivo establishes their functional relevance. Nat. Commun. 13, 5273 (2022).
Ali, F. & Seshasayee, A. S. N. Dynamics of genetic variation in transcription factors and its implications for the evolution of regulatory networks in Bacteria. Nucleic Acids Res. 48, 4100–4114 (2021).
Li, H. et al. Inferring transcription factor regulatory networks from single-cell ATAC-seq data based on graph neural networks. Nat. Mach. Intell. 4, 389–400 (2022).
Jayaram, N., Usvyat, D. & Martin, A. C. R. Evaluating tools for transcription factor binding site prediction. BMC Bioinformatics 17, 547 (2016).
Park, P. J. ChIP–seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 10, 669–680 (2009).
Gomes, A. L. C. & Wang, H. H. The role of genome accessibility in transcription factor binding in bacteria. PLoS Comput. Biol. 12, e1004891 (2016).
Furey, T. S. ChIP–seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 13, 840–852 (2012).
Hua, C. et al. Bacterial transcription factors bind to coding regions and regulate internal cryptic promoters. mBio 13, e0164322 (2022).
Tu, X. et al. Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors. Nat. Commun. 11, 5089 (2020).
Park, J. Y., Rimal, H., Bang, I., Nong, L. K. & Kim, D. Genome-wide identification of DNA-protein interaction to reconstruct bacterial transcription regulatory network. Biotechnol. Bioprocess Eng. 25, 944–954 (2020).
Dos Santos, A. L. S. et al. What are the advantages of living in a community? A microbial biofilm perspective! Mem. Inst. Oswaldo Cruz. 113, e180212 (2018).
Acknowledgements
This work was supported by an MIT Bose Research Grant (award number 2116642).
Author information
Authors and Affiliations
Contributions
M.R. and K.L.J.P conceptualized the manuscript. M.R. researched and wrote the manuscript. K.L.J.P. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Bioengineering thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ream, M., Prather, K.L.J. Engineered autonomous dynamic regulation of metabolic flux. Nat Rev Bioeng 2, 233–243 (2024). https://doi.org/10.1038/s44222-023-00140-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44222-023-00140-7