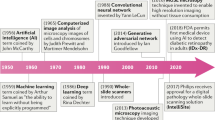
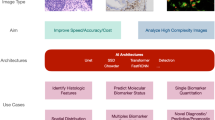
Abstract
Advances in digitizing tissue slides and the fast-paced progress in artificial intelligence, including deep learning, have boosted the field of computational pathology. This field holds tremendous potential to automate clinical diagnosis, predict patient prognosis and response to therapy, and discover new morphological biomarkers from tissue images. Some of these artificial intelligence-based systems are now getting approved to assist clinical diagnosis; however, technical barriers remain for their widespread clinical adoption and integration as a research tool. This Review consolidates recent methodological advances in computational pathology for predicting clinical end points in whole-slide images and highlights how these developments enable the automation of clinical practice and the discovery of new biomarkers. We then provide future perspectives as the field expands into a broader range of clinical and research tasks with increasingly diverse modalities of clinical data.
Key points
-
Supported by advances in artificial intelligence, curation of multi-institutional cohorts and the development of high-performance computing, computational pathology is now reaching clinical-grade performance for certain tasks.
-
Artificial intelligence-based methods in computational pathology can be distinguished into methods for predicting clinical end points from tissue specimens and assistive tools for clinical or research tasks.
-
Multiple instance learning is a rapidly growing paradigm for predicting clinical end points, such as disease diagnosis and molecular alterations, from whole-slide images.
-
Computational pathology can be used for automating tasks that pathologists already perform in daily practice and for discovering morphological biomarkers for clinical outcomes of interest.
-
Initiatives for collecting larger, well-curated and multimodal datasets, together with advances in artificial intelligence frameworks, are required for the clinical adoption of computational pathology tools.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Abels, E. et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the digital pathology association. J. Pathol. 249, 286–294 (2019).
Lu, M. Y. et al. AI-based pathology predicts origins for cancers of unknown primary. Nature 594, 106–110 (2021). This article presents an attention-based MIL approach for the prediction of the primary cancer site, one of the most challenging tasks in oncology.
Bulten, W. et al. Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat. Med. 28, 154–163 (2022). This article presents the results of the PANDA challenge, which aimed to automate Gleason grading of prostate biopsies using over 10,000 digitized samples.
Skrede, O.-J. et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet 395, 350–360 (2020).
Ehteshami Bejnordi, B. et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017). This article presents the outcome of the CAMELYON16 challenge for the detection of breast cancer lymph node metastases.
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015). This article provides an overview of deep learning, highlighting its superior performance to traditional machine learning approaches.
Bostrom, R., Sawyer, H. & Tolles, W. Instrumentation for automatically prescreening cytological smears. Proc. IRE 47, 1895–1900 (1959).
Prewitt, J. M. S. & Mendelsohn, M. L. The analysis of cell images. Ann. N. Y. Acad. Sci. 128, 1035–1053 (1966).
Fuchs, T. J., Wild, P. J., Moch, H. & Buhmann, J. M. Computational pathology analysis of tissue microarrays predicts survival of renal clear cell carcinoma patients. Int. Med. Image Comput. Comput. Assist. Interv. 11, 1–8 (2008).
Beck, A. H. et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci. Transl. Med. 3, 108ra113 (2011).
Madabhushi, A. & Lee, G. Image analysis and machine learning in digital pathology: challenges and opportunities. Med. Image Anal. 33, 170–175 (2016).
Madabhushi, A., Agner, S., Basavanhally, A., Doyle, S. & Lee, G. Computer-aided prognosis: predicting patient and disease outcome via quantitative fusion of multi-scale, multi-modal data. Comput. Med. Imaging Graph. 35, 506–514 (2011).
Tarantino, P., Mazzarella, L., Marra, A., Trapani, D. & Curigliano, G. The evolving paradigm of biomarker actionability: histology-agnosticism as a spectrum, rather than a binary quality. Cancer Treat. Rev. 94, 102169 (2021).
Lee, Y. et al. Derivation of prognostic contextual histopathological features from whole-slide images of tumours via graph deep learning. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-022-00923-0 (2022).
Saltz, J. et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 23, 181–193.e7 (2018).
Marusyk, A., Janiszewska, M. & Polyak, K. Intratumor heterogeneity: the Rosetta Stone of therapy resistance. Cancer Cell 37, 471–484 (2020).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Dosovitskiy, A. et al. An image is worth 16×16 words: transformers for image recognition at scale. In International Conference on Learning Representations (ICLR, 2021). This article explores the application of the transformer, initially developed for natural language processing, to the field of computer vision.
Krishnan, R., Rajpurkar, P. & Topol, E. J. Self-supervised learning in medicine and healthcare. Nat. Biomed. Eng. 6, 1346–1352 (2022).
Jiang, P. et al. Big data in basic and translational cancer research. Nat. Rev. Cancer 22, 625–639 (2022).
Andreou, C., Weissleder, R. & Kircher, M. F. Multiplexed imaging in oncology. Nat. Biomed. Eng. 6, 527–540 (2022).
Marx, V. Method of the Year: spatially resolved transcriptomics. Nat. Methods 18, 9–14 (2021).
Liu, J. T. et al. Harnessing non-destructive 3D pathology. Nat. Biomed. Eng. 5, 203–218 (2021).
Stockman, G. & Shapiro, L. G. Computer Vision (Prentice Hall PTR, 2001).
Bándi, P. et al. Resolution-agnostic tissue segmentation in whole-slide histopathology images with convolutional neural networks. PeerJ 7, e8242 (2019).
Deng, J. et al. in IEEE Conference on Computer Vision and Pattern Recognition. 248–255 (IEEE, 2009).
Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
Vitale, I., Shema, E., Loi, S. & Galluzzi, L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 27, 212–224 (2021).
Kather, J. N. et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 1, 789–799 (2020).
Ilse, M., Tomczak, J. & Welling, M. in Proc. 35th International Conference on Machine Learning (eds Dy, J. & Krause, A.) 2127–2136 (PMLR, 2018). This article introduces attention-based MIL, which uses a neural network to assign importance scores to instances.
Dietterich, T. G., Lathrop, R. H. & Lozano-Pérez, T. Solving the multiple instance problem with axis-parallel rectangles. Artif. Intell. 89, 31–71 (1997).
Dundar, M. M. et al. in 20th International Conference on Pattern Recognition 2732–2735 (IEEE, 2010).
Bahdanau, D., Cho, K. & Bengio, Y. Neural machine translation by jointly learning to align and translate. In International Conference on Learning Representations (ICLR, 2015).
Lu, M. Y. et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5, 555–570 (2021).
He, K., Zhang, X., Ren, S. & Sun, J. in Proc. IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016). This article introduces a deep CNN with residual connections, enabling better performance on computer vision tasks such as classification.
Campanella, G. et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301–1309 (2019). This study applies MIL to a cohort of 15,000 patients, demonstrating the power of weakly supervised learning at scale.
Shaban, M. et al. Context-aware convolutional neural network for grading of colorectal cancer histology images. IEEE Trans. Med. Imaging 39, 2395–2405 (2020).
Tellez, D. et al. Neural image compression for gigapixel histopathology image analysis. IEEE Trans. Pattern Anal. Mach. Intell. 43, 567–578 (2019).
Wang, X. et al. Transformer-based unsupervised contrastive learning for histopathological image classification. Med. Image Anal. 81, 102559 (2022).
Chen, R. J. et al. in Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 16144–16155 (IEEE, 2022).
Ciga, O., Xu, T. & Martel, A. L. Self supervised contrastive learning for digital histopathology. Mach. Learn. Appl. 7, 100198 (2022).
Chen, C.-L. et al. An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning. Nat. Commun. 12, 1193 (2021).
Pinckaers, H., Van Ginneken, B. & Litjens, G. Streaming convolutional neural networks for end-to-end learning with multi-megapixel images. IEEE Trans. Pattern Anal. and Mach. Intell. 44 1581–1590 (2022).
Huang, S.-C. et al. Deep neural network trained on gigapixel images improves lymph node metastasis detection in clinical settings. Nat. Commun. 13, 3347 (2022).
Wulczyn, E. et al. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS ONE 15, e0233678 (2020).
Wulczyn, E. et al. Interpretable survival prediction for colorectal cancer using deep learning. NPJ Digit. Med. 4, 71 (2021).
Laleh, N. G. et al. Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology. Med. Image Anal. 79, 102474 (2022).
Jaume, G., Song, A. & Mahmood, F. Integrating context for superior cancer prognosis. Nat. Biomed. Eng. 6, 1323–1325 (2022).
Taube, J. M. et al. Implications of the tumor immune microenvironment for staging and therapeutics. Mod. Pathol. 31, 214–234 (2018).
Pati, P. et al. Hierarchical graph representations in digital pathology. Med. Image Anal. 75, 102264 (2022).
Zhao, Y. et al. in IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 4836–4845 (IEEE, 2020).
Adnan, M., Kalra, S. & Tizhoosh H. R. in IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPR) 4254–4261 (IEEE, 2020).
Scarselli, F., Gori, M., Tsoi, A. C., Hagenbuchner, M. & Monfardini, G. The graph neural network model. IEEE Trans. Neural Netw. 20, 61–80 (2008).
Gunduz, C., Yener, B. & Gultekin, S. H. The cell graphs of cancer. Bioinformatics 20, i145–i151 (2004).
Chen, R. J. et al. Pathomic fusion: an integrated framework for fusing histopathology and genomic features for cancer diagnosis and prognosis. IEEE Trans. Med. Imaging 41, 757–770 (2022).
Zhou, Y. et al. in IEEE/CVF International Conference on Computer Vision Workshop (ICCVW) 388–398 (IEEE, 2019).
Ahmedt, D. et al. A survey on graph-based deep learning for computational histopathology. Comput. Med. Imaging Graph. 95, 102027 (2022).
Vaswani, A. et al. in Advances in Neural Information Processing Systems (eds Guyon, I. et al.) (Curran Associates, Inc., 2017).
Shao, Z. et al. TransMIL: transformer based correlated multiple instance learning for whole slide image classification. In Advances in Neural Information Processing Systems (eds Beygelzimer, A. et al.) (Curran Associates, Inc., 2021).
Wu, H., Wu, J., Xu, J., Wang, J. & Long, M. in Proc. 39th International Conference on Machine Learning (eds Chaudhuri, K. et al.) 24226–24242 (PMLR, 2022).
Iizuka, O. et al. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci. Rep. 10, 1504 (2020).
Kalra, S., Adnan, M., Taylor, G. & Tizhoosh, H. R. in European Conference on Computer Vision (eds Vedaldi, A. et al.) 677–693 (Springer, 2020).
Lipkova, J. et al. Deep learning-enabled assessment of cardiac allograft rejection from endomyocardial biopsies. Nat. Med. 28, 575–582 (2022).
Sirinukunwattana, K., Alhan, N. K., Verril, C. & Rittscher, J. in International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Frangi, A. F. et al.) 192–200 (Springer, 2018).
Thandiackal, K. et al. in European Conference on Computer Vision (eds Avidan, S. et al.) 699–715 (Springer, 2022).
Katharopoulos, A. & Fleuret, F. in International Conference on Machine Learning (eds Chaudhuri, K. & Salakhutdinov R.) 3282–3291 (PMLR, 2019).
Kong, S. & Henao, R. in IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2374–2384 (IEEE/CVF, 2021).
Malon, C., Miller, M., Burger, H. C., Cosatto, E. & Graf, H. P. in Proc. 5th International Conference on Soft Computing as Transdisciplinary Science and Technology 450–456 (ACM, 2008).
Bulten, W. et al. Epithelium segmentation using deep learning in H&E-stained prostate specimens with immunohistochemistry as reference standard. Sci. Rep. 9, 864 (2019).
Anklin, V. et al. in International Conference on Medical Image Computing and Computer Assisted Intervention (eds de Bruijne, M. et al.) 636–646 (Springer, 2021).
Chan, L., Hosseini, M. S., Rowsell, C., Plataniotis, K. N. & Damaskinos, S. in Proc. IEEE/CVF International Conference on Computer Vision 10661–10670 (IEEE/CVF, 2019).
Graham, S. et al. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 58, 101563 (2019). This article proposes a deep learning framework for simultaneous nuclear segmentation and classification from tissue images.
Sirinukunwattana, K. et al. Gland segmentation in colon histology images: the GLAS challenge contest. Med. Image Anal. 35, 489–502 (2017).
Cireşan, D. C., Giusti, A., Gambardella, L. M. & Schmidhuber, J. in International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Mori, K. et al.) 411–418 (Springer, 2013).
Li, C., Wang, X., Liu, W. & Latecki, L. J. DeepMitosis: mitosis detection via deep detection, verification and segmentation networks. Med. Image Anal. 45, 121–133 (2018).
Long, J., Shelhamer, E. & Darrell, T. in Proc. IEEE Conference on Computer Vision and Pattern Recognition 3431–3440 (IEEE, 2015).
Ronneberger, O., Fischer, P. & Brox, T. in International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Navab, N. et al.) 234–241 (Springer, 2015).
He, K., Gkioxari, G., Dollár, P. & Girshick, R. in Proc. IEEE International Conference on Computer Vision 2980–2988 (IEEE, 2017).
Alemi Koohbanani, N., Jahanifar, M., Zamani Tajadin, N. & Rajpoot, N. NuClick: a deep learning framework for interactive segmentation of microscopic images. Med. Image Anal. 65, 101771 (2020).
Kumar, N. et al. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans. Med. Imaging 36, 1550–1560 (2017).
Greenwald, N. F. et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40, 555–565 (2022).
Han, W., Cheung, A. M., Yaffe, M. J. & Martel, A. L. Cell segmentation for immunofluorescence multiplexed images using two-stage domain adaptation and weakly labeled data for pre-training. Sci. Rep. 12, 4399 (2022).
Martinelli, A. L. & Rapsomaniki, M. A. ATHENA: analysis of tumor heterogeneity from spatial omics measurements. Bioinformatics 38, 3151–3153 (2022).
Tellez, D. et al. H and E stain augmentation improves generalization of convolutional networks for histopathological mitosis detection. In Medical Imaging 2018: Digital Pathology Vol. 10581 (eds Tomaszewski, J. E. & Gurcan, M. N.) (SPIE, 2018).
Zanjani, F. G., Zinger, S., Bejnordi, B. E., van der Laak, J. A. W. M. & de With, P. H. N. in IEEE 15th International Symposium on Biomedical Imaging 573–577 (IEEE, 2018).
Macenko, M. et al. in IEEE International Symposium on Biomedical Imaging: From Nano to Macro 1107–1110 (IEEE, 2009).
Vahadane, A. et al. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans. Med. Imaging 35, 1962–1971 (2016).
Cho, H., Lim, S., Choi, G. & Min, H. Neural stain-style transfer learning using GAN for histopathological images. Preprint at arXiv https://doi.org/10.48550/arXiv.1710.08543 (2017).
Zhou, N., Cai, D., Han, X. & Yao, J. in International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Shen, D. et al.) 694–702 (Springer, 2019).
Kang, H. et al. StainNet: a fast and robust stain normalization network. Front. Med. 8, 746307 (2021).
Ozyoruk, K. B. et al. A deep-learning model for transforming the style of tissue images from cryosectioned to formalin-fixed and paraffin-embedded. Nat. Biomed. Eng. 6, 1407–1419 (2022).
He, B. et al. AI-enabled in silico immunohistochemical characterization for Alzheimer’s disease. Cell Rep. Methods 2, 100191 (2022).
Ghahremani, P. et al. Deep learning-inferred multiplex immunofluorescence for immunohistochemical image quantification. Nat. Mach. Intell. 4, 401–412 (2022).
Cao, R. et al. Label-free intraoperative histology of bone tissue via deep-learning-assisted ultraviolet photoacoustic microscopy. Nat. Biomed. Eng. 7, 124–134 (2023).
Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. in IEEE International Conference on Computer Vision (ICCV) 2242–2251 (IEEE, 2017).
Park, T., Efros, A. A., Zhang, R. & Zhu, J.-Y. in European Conference on Computer Vision (eds Vedaldi, A. et al.) 319–345 (Springer, 2020).
Vasiljević, J., Nisar, Z., Feuerhake, F., Wemmert, C. & Lampert, T. CycleGAN for virtual stain transfer: is seeing really believing? Artif. Intell. Med. 133, 102420 (2022).
Holzinger, A. et al. Towards the augmented pathologist: challenges of explainable-AI in digital pathology. Preprint at arXiv https://doi.org/10.48550/arXiv.1712.06657 (2017).
Selvaraju, R. R. et al. in IEEE International Conference on Computer Vision 618–626 (IEEE, 2017).
Sundararajan, M., Taly, A. & Yan, Q. in Proc. 34th International Conference on Machine Learning (eds Precup, D. & Teh, Y. W.) 3319–3328 (PMLR, 2017).
Barredo Arrieta, A. et al. Explainable artificial intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 58, 82–115 (2020).
Chen, R. J. et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell 40, 865–878 (2022).
Javed, S. A. et al. in Advances in Neural Information Processing Systems (eds Koyejo, S. et al.) vol. 35, 20689–20702 (Curran Associates, Inc., 2022).
Diao, J. A. et al. Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat. Commun. 12, 1613 (2021).
Jaume, G. et al. Quantifying explainers of graph neural networks in computational pathology. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 8102–8112 (IEEE, 2021).
Tellez, D. et al. Whole-slide mitosis detection in H&E breast histology using PHH3 as a reference to train distilled stain-invariant convolutional networks. IEEE Trans. Med. Imaging 37, 2126–2136 (2018).
Kapil, A. et al. Deep semi supervised generative learning for automated tumor proportion scoring on NSCLC tissue needle biopsies. Sci. Rep. 8, 17343 (2018).
Swiderska-Chadaj, Z. et al. Learning to detect lymphocytes in immunohistochemistry with deep learning. Med. Image Anal. 58, 101547 (2019).
Fassler, D. J. et al. Deep learning-based image analysis methods for brightfield-acquired multiplex immunohistochemistry images. Diagn. Pathol. 15, 100 (2020).
Naylor, P. et al. Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE Trans. Med. Imaging 38, 448–459 (2019).
Widmaier, M. et al. Comparison of continuous measures across diagnostic PD-L1 assays in non-small cell lung cancer using automated image analysis. Mod. Pathol. 33, 380–390 (2020).
Matek, C., Schwarz, S., Spiekermann, K. & Marr, C. Human-level recognition of blast cells in acute myeloid leukaemia with convolutional neural networks. Nat. Mach. Intell. 1, 538–544 (2019).
Binder, T. et al. Multi-organ gland segmentation using deep learning. Front. Med. 6, 173 (2019).
Fraz, M. M. et al. FABnet: feature attention-based network for simultaneous segmentation of microvessels and nerves in routine histology images of oral cancer. Neural Comput. Appl. 32, 9915–9928 (2020).
Ing, N. et al. Semantic segmentation for prostate cancer grading by convolutional neural networks. In Medical Imaging 2018: Digital Pathology Vol. 10581 (eds Tomaszewski, J. E. & Gurcan, M. N.) (SPIE, 2018).
Geessink, O. G. F. et al. Computer aided quantification of intratumoral stroma yields an independent prognosticator in rectal cancer. Cell. Oncol. 42, 331–341 (2019).
Amgad, M. et al. Report on computational assessment of tumor infiltrating lymphocytes from the international immuno-oncology biomarker working group. NPJ Breast Cancer 6, 16 (2020).
Taylor-Weiner, A. et al. A machine learning approach enables quantitative measurement of liver histology and disease monitoring in NASH. Hepatology 74, 133–147 (2021).
Heinemann, F. et al. Deep learning-based quantification of NAFLD/NASH progression in human liver biopsies. Sci. Rep. 12, 19236 (2022).
Jaume, G., Pati, P., Anklin, V., Foncubierta, A. & Gabrani, M. in Proc. MICCAI Workshop on Computational Pathology 117–128 (PMLR, 2021).
Ström, P. et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 21, 222–232 (2020).
Bulten, W. et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 21, 233–241 (2020).
Ertosun, M. G. & Rubin, D. L. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. AMIA Annu. Symp. Proc. 2015, 1899–1908 (2015).
Couture, H. D. et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 4, 30 (2018).
Kers, J. et al. Deep learning-based classification of kidney transplant pathology: a retrospective, multicentre, proof-of-concept study. Lancet Digit. Health 4, 18–26 (2022).
Korbar, B. et al. Deep learning for classification of colorectal polyps on whole-slide images. J. Pathol. Inform. 8, 30 (2017).
Ianni, J. D. et al. Tailored for real-world: a whole slide image classification system validated on uncurated multi-site data emulating the prospective pathology workload. Sci. Rep. 10, 3217 (2020).
Kiani, A. et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit. Med. 3, 23 (2020).
Janowczyk, A. & Madabhushi, A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J. Pathol. Inform. 7, 29 (2016).
Hollon, T. C. et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat. Med. 26, 52–58 (2020).
Chen, P. C. et al. An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat. Med. 25, 1453–1457 (2019).
Gehrung, M. et al. Triage-driven diagnosis of Barrett’s esophagus for early detection of esophageal adenocarcinoma using deep learning. Nat. Med. 27, 833–841 (2021).
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019). This article presents a method for predicting microsatellite instability from WSIs showing that AI can be used in patient screening.
Bychkov, D. et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 8, 3395 (2018).
Kather, J. N. et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. 16, e1002730 (2019).
Leo, P. et al. Computer extracted gland features from H&E predicts prostate cancer recurrence comparably to a genomic companion diagnostic test: a large multi-site study. NPJ Precis. Oncol. 5, 35 (2021).
Wang, X. et al. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci. Rep. 7, 13543 (2017).
Shaban, M. et al. A novel digital score for abundance of tumour infiltrating lymphocytes predicts disease free survival in oral squamous cell carcinoma. Sci. Rep. 9, 13341 (2019).
Kulkarni, P. M. et al. Deep learning based on standard H&E images of primary melanoma tumors identifies patients at risk for visceral recurrence and death. Clin. Cancer Res. 26, 1126–1134 (2020).
Yang, J. et al. Prediction of HER2-positive breast cancer recurrence and metastasis risk from histopathological images and clinical information via multimodal deep learning. Comput. Struct. Biotechnol. J. 20, 333–342 (2022).
Klimov, S. et al. Predicting metastasis risk in pancreatic neuroendocrine tumors using deep learning image analysis. Front. Oncol. 10, 593211 (2021).
Kleppe, A. et al. A clinical decision support system optimising adjuvant chemotherapy for colorectal cancers by integrating deep learning and pathological staging markers: a development and validation study. Lancet Oncol. 23, 1221–1232 (2022).
Courtiol, P. et al. Deep learning-based classification of mesothelioma improves prediction of patient outcome. Nat. Med. 25, 1519–1525 (2019).
Saillard, C. et al. Predicting survival after hepatocellular carcinoma resection using deep learning on histological slides. Hepatology 72, 2000–2013 (2020).
Boehm, K. M., Khosravi, P., Vanguri, R., Gao, J. & Shah, S. P. Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer 22, 114–126 (2022).
Roelofsen, L. M., Kaptein, P. & Thommen, D. S. Multimodal predictors for precision immunotherapy. Immunooncol. Technol. 14, 100071 (2022).
Mobadersany, P. et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl Acad. Sci. USA 115, E2970–E2979 (2018).
Cheerla, A. & Gevaert, O. Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics 35, i446–i454 (2019).
Braman, N. et al. in International Conference on Medical Image Computing and Computer-Assisted Intervention (eds de Bruijne, M. et al.) 667–677 (Springer, 2021).
Chen, R. J. et al. in Proc. IEEE/CVF International Conference on Computer Vision (ICCV) 3995–4005 (IEEE/CVF, 2021).
Foersch, S. et al. Multistain deep learning for prediction of prognosis and therapy response in colorectal cancer. Nat. Med. 29, 430–439 (2023).
Echle, A. et al. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br. J. Cancer 124, 686–696 (2021).
Fu, Y. et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 1, 800–810 (2020).
Wang, S. et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 53, 1800986 (2019).
Loeffler, C. M. L. et al. Artificial intelligence-based detection of FGFR3 mutational status directly from routine histology in bladder cancer: a possible preselection for molecular testing? Eur. Urol. Focus 8, 472–479 (2022).
Schmauch, B. et al. A deep learning model to predict RNA-seq expression of tumours from whole slide images. Nat. Commun. 11, 3877 (2020).
Fremond, S. et al. Interpretable deep learning model to predict the molecular classification of endometrial cancer from haematoxylin and eosin-stained whole-slide images: a combined analysis of the PORTEC randomised trials and clinical cohorts. Lancet Digit. Health 5, e71–e82 (2023).
Hong, R., Liu, W., DeLair, D., Razavian, N. & Fenyö, D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2, 100400 (2021).
Shamai, G. et al. Deep learning-based image analysis predicts PD-L1 status from H&E-stained histopathology images in breast cancer. Nat. Commun. 13, 6753 (2022).
Naik, N. et al. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat. Commun. 11, 5727 (2020).
He, B. et al. Integrating spatial gene expression and breast tumour morphology via deep learning. Nat. Biomed. Eng. 4, 827–834 (2020).
Acosta, P. H. et al. Intratumoral resolution of driver gene mutation heterogeneity in renal cancer using deep learning. Cancer Res. 82, 2792–2806 (2022).
Song, A. H., Williamson, D. F. & Mahmood, F. Investigating morphologic correlates of driver gene mutation heterogeneity via deep learning. Cancer Res. 82, 2672–2673 (2022).
Harder, N. et al. Automatic discovery of image-based signatures for ipilimumab response prediction in malignant melanoma. Sci. Rep. 9, 7449 (2019).
Hu, J. et al. Using deep learning to predict anti-PD-1 response in melanoma and lung cancer patients from histopathology images. Transl. Oncol. 14, 100921 (2021).
Berry, S. et al. Analysis of multispectral imaging with the astropath platform informs efficacy of PD-1 blockade. Science 372, eaba2609 (2021).
Johannet, P. et al. Using machine learning algorithms to predict immunotherapy response in patients with advanced melanoma. Clin. Cancer Res. 27, 131–140 (2021).
Farahmand, S. et al. Deep learning trained on hematoxylin and eosin tumor region of interest predicts HER2 status and trastuzumab treatment response in HER2+ breast cancer. Mod. Pathol. 35, 44–51 (2022).
Bychkov, D. et al. Deep learning identifies morphological features in breast cancer predictive of cancer ERBB2 status and trastuzumab treatment efficacy. Sci. Rep. 11, 4037 (2021).
Li, F. et al. Deep learning-based predictive biomarker of pathological complete response to neoadjuvant chemotherapy from histological images in breast cancer. J. Transl. Med. 19, 348 (2021).
Sammut, S.-J. et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature 601, 623–629 (2022).
Vanguri, R. S. et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat. Cancer 3, 1151–1164 (2022).
Liu, R. et al. Systematic pan-cancer analysis of mutation–treatment interactions using large real-world clinicogenomics data. Nat. Med. 28, 1656–1661 (2022).
Bera, K., Schalper, K. A., Rimm, D. L., Velcheti, V. & Madabhushi, A. Artificial intelligence in digital pathology — new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 16, 703–715 (2019).
Niazi, M. K. K., Parwani, A. V. & Gurcan, M. N. Digital pathology and artificial intelligence. Lancet Oncol. 20, e253–e261 (2019).
Kleppe, A. et al. Designing deep learning studies in cancer diagnostics. Nat. Rev. Cancer 21, 199–211 (2021).
Van der Laak, J., Litjens, G. & Ciompi, F. Deep learning in histopathology: the path to the clinic. Nat. Med. 27, 775–784 (2021).
Baxi, V., Edwards, R., Montalto, M. & Saha, S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod. Pathol. 35, 23–32 (2022).
Hedge, N. et al. Similar image search for histopathology: SMILY. NPJ Digit. Med. 2, 56 (2019).
Kalra, S. et al. Yottixel — an image search engine for large archives of histopathology whole slide images. Med. Image Anal. 65, 101757 (2020).
Chen, C. et al. Fast and scalable search of whole-slide images via self-supervised deep learning. Nat. Biomed. Eng. 6, 1420–1434 (2022).
Lipkova, J. et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell 40, 1095–1110 (2022).
Singer, D. S. A new phase of the Cancer Moonshot to end cancer as we know it. Nat. Med. 28, 1345–1347 (2022).
Kiemen, A. L. et al. CODA: quantitative 3D reconstruction of large tissues at cellular resolution. Nat. Methods 19, 1490–1499 (2022).
Glaser, A. K. et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng. 1, 0084 (2017).
Fereidouni, F. et al. Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat. Biomed. Eng. 1, 957–966 (2017).
Katsamenis, O. L. et al. X-ray micro-computed tomography for nondestructive three-dimensional (3D) X-ray histology. Am. J. Pathol. 189, 1608–1620 (2019).
Xie, W. et al. Prostate cancer risk stratification via non-destructive 3D pathology with deep learning assisted gland analysis. Cancer Res. 82, 334–345 (2022).
Allam, M., Cai, S. & Coskun, A. F. Multiplex bioimaging of single-cell spatial profiles for precision cancer diagnostics and therapeutics. NPJ Precis. Oncol. 4, 11 (2020).
Wu, Z. et al. Graph deep learning for the characterization of tumour microenvironments from spatial protein profiles in tissue specimens. Nat. Biomed. Eng. 6, 1435–1448 (2022).
Wang, Y. et al. Cell graph neural networks enable the precise prediction of patient survival in gastric cancer. NPJ Precis. Oncol. 6, 45 (2022).
Lu, M. Y., Sater, H. A. & Mahmood, F. Multiplex computational pathology for treatment response prediction. Cancer Cell 39, 1053–1055 (2021).
Price, W. N. & Cohen, I. G. Privacy in the age of medical big data. Nat. Med. 25, 37–43 (2019).
Shmatko, A., Ghaffari Laleh, N., Gerstung, M. & Kather, J. N. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat. Cancer 3, 1026–1038 (2022).
Zhang, A., Xing, L., Zou, J. & Wu, J. C. Shifting machine learning for healthcare from development to deployment and from models to data. Nat. Biomed. Eng. 6, 1330–1345 (2022).
Lu, M. Y. et al. Federated learning for computational pathology on gigapixel whole slide images. Med. Image Anal. 76, 102298 (2022).
Adnan, M., Kalra, S., Cresswell, J. C., Taylor, G. W. & Tizhoosh, H. R. Federated learning and differential privacy for medical image analysis. Sci. Rep. 12, 1953 (2022).
Ogier du Terrail, J. et al. Federated learning for predicting histological response to neoadjuvant chemotherapy in triple-negative breast cancer. Nat. Med. 29, 135–146 (2023).
Parisi, G. I., Kemker, R., Part, J. L., Kanan, C. & Wermter, S. Continual lifelong learning with neural networks: a review. Neural Netw. 113, 54–71 (2019).
Saldanha, O. L. et al. Swarm learning for decentralized artificial intelligence in cancer histopathology. Nat. Med. 28, 1232–1239 (2022).
Obermeyer, Z., Powers, B., Vogeli, C. & Mullainathan, S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 366, 447–453 (2019).
Seyyed-Kalantari, L., Zhang, H., McDermott, M., Chen, I. Y. & Ghassemi, M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat. Med. 27, 2176–2182 (2021).
Ricci Lara, M. A., Echeveste, R. & Ferrante, E. Addressing fairness in artificial intelligence for medical imaging. Nat. Commun. 13, 4581 (2022).
Chen, R. J. et al. Algorithmic fairness in artificial intelligence for medicine and healthcare. Nat. Biomed. Eng. 7, 719–742 (2023).
Chen, R. J., Lu, M. Y., Chen, T. Y., Williamson, D. F. & Mahmood, F. Synthetic data in machine learning for medicine and healthcare. Nat. Biomed. Eng. 5, 493–497 (2021).
Krasanakis, E., Spyromitros-Xioufis, E., Papadopoulos, S. & Kompatsiaris, Y. in Proc. 2018 World Wide Web Conference 853–862 (2018).
Jiang, H. & Nachum, O. in Proc. Twenty Third International Conference on Artificial Intelligence and Statistics (eds Chiappa, S. & Calandra, R.) 702–712 (PMLR, 2020).
Li, B., Li, Y. & Eliceiri, K. W. Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. Conf. Comput. Vis. Pattern Recognit. Workshops 2021, 14318–14328 (2021).
Huang, Z. et al. in International Conference on Medical Image Computing and Computer-Assisted Intervention (eds de Bruijne, M. et al.) 561–570 (Springer, 2021).
Abbet, C., Zlobec, I., Bozorgtabar, B. & Thiran, J.-P. in International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Martel, A. L. et al.) 480–489 (Springer, 2020).
Kang, M. et al. in Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 3344–3354 (IEEE, 2023).
Azizi, S. et al. Robust and data-efficient generalization of self-supervised machine learning for diagnostic imaging. Nat. Biomed. Eng. 7, 756–779 (2023).
He, K. et al. Masked autoencoders are scalable vision learners. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 15979–15988 (2022).
Lu, M. Y. et al. in Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition, 19764–19775 (IEEE, 2023).
Tellez, D. et al. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 58, 101544 (2019).
Howard, F. M. et al. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat. Commun. 12, 4423 (2021).
Fort, S., Hu, H. & Lakshminarayanan, B. Deep ensembles: a loss landscape perspective. Preprint at arXiv https://doi.org/10.48550/arXiv.1912.02757 (2019).
Guo, C., Pleiss, G., Sun, Y. & Weinberger, K. in Proc. 34th International Conference on Machine Learning 1321–1330 (PMLR, 2017).
Pocevičiūtė, M., Eilertsen, G., Jarkman, S. & Lundström, C. Generalisation effects of predictive uncertainty estimation in deep learning for digital pathology. Sci. Rep. 12, 8329 (2022).
Dolezal, J. M. et al. Uncertainty-informed deep learning models enable high-confidence predictions for digital histopathology. Nat. Commun. 13, 6572 (2022).
Linmans, J., Elfwing, S., van der Laak, J. & Litjens, G. Predictive uncertainty estimation for out-of-distribution detection in digital pathology. Med. Image Anal. 83, 102655 (2023).
Olsson, H. et al. Estimating diagnostic uncertainty in artificial intelligence assisted pathology using conformal prediction. Nat. Commun. 13, 7761 (2022).
Castro, D. C., Walker, I. & Glocker, B. Causality matters in medical imaging. Nat. Commun. 11, 3673 (2020).
Roux, L. et al. Mitosis detection in breast cancer histological images an ICPR 2012 contest. J. Pathol. Inform. 4, 8 (2013).
Veta, M. et al. Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge. Med. Image Anal. 54, 111–121 (2019).
Aubreville, M. et al. Mitosis domain generalization in histopathology images - the MIDOG challenge. Med. Image Anal. 84, 102699 (2023).
Bandi, P. et al. From detection of individual metastases to classification of lymph node status at the patient level: the CAMELYON17 challenge. IEEE Trans. Med. Imaging 38, 550–560 (2018).
Aresta, G. et al. BACH: Grand challenge on breast cancer histology images. Med. Image Anal. 56, 122–139 (2019).
Verma, R. et al. MoNuSAC2020: a multi-organ nuclei segmentation and classification challenge. IEEE Trans. Med. Imaging 40, 3413–3423 (2021).
Moulin, P., Grünberg, K., Barale-Thomas, E. & der Laak, J. V. IMI — Bigpicture: a central repository for digital pathology. Toxicol. Pathol. 49, 711–713 (2021).
Jennings, C. N. et al. Bridging the gap with the UK genomics pathology imaging collection. Nat. Med. 28, 1107–1108 (2022).
Wagner, S. J. et al. Make deep learning algorithms in computational pathology more reproducible and reusable. Nat. Med. 28, 1744–1746 (2022).
Gilbert, B. et al. Openslide. GitHub https://github.com/openslide/openslide-python/ (2020).
Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Rosenthal, J. et al. Building tools for machine learning and artificial intelligence in cancer research: best practices and a case study with the pathML toolkit for computational pathology. Mol. Cancer Res. 20, 202–206 (2022).
Beezley, J. et al. Histomicstk. GitHub https://github.com/DigitalSlideArchive/HistomicsTK (2021).
Pocock, J. et al. TIAToolbox as an end-to-end library for advanced tissue image analytics. Commun. Med. 2, 120 (2022).
Raciti, P. et al. Clinical validation of artificial intelligence — augmented pathology diagnosis demonstrates significant gains in diagnostic accuracy in prostate cancer detection. Arch. Pathol. Lab. Med. (2022).
Lennerz, J. K., Green, U., Williamson, D. F. & Mahmood, F. A unifying force for the realization of medical AI. NPJ Digit. Med. 5, 172 (2022).
Acknowledgements
The authors thank R. J. Chen for feedback on the manuscript and K. Yost for helping with the figures. This work was supported in part by the National Institute of General Medical Sciences (NIGMS) R35GM138216 (to F.M.), Brigham President’s Fund, Brigham and Women’s Hospital (BWH) and Massachusetts General Hospital (MGH) Pathology, BWH Precision Medicine Program. M.Y.L. was additionally supported by the Tau Beta Pi Fellowship and the Siebel Foundation.
Author information
Authors and Affiliations
Contributions
A.H.S., G.J. and F.M. contributed to conceptualization and outlining the review. All authors contributed to writing, reviewing and editing. F.M. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Bioengineering thanks Håvard Danielsen, who co-reviewed with Andreas Kleppe; Tingying Peng, who co-reviewed with Sophia Wagner; and Amaro Taylor-Weiner, who co-reviewed with Archit Khosla, for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Artificial Intelligence and Machine Learning Software as a Medical Device Action Plan: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Song, A.H., Jaume, G., Williamson, D.F.K. et al. Artificial intelligence for digital and computational pathology. Nat Rev Bioeng 1, 930–949 (2023). https://doi.org/10.1038/s44222-023-00096-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44222-023-00096-8