Abstract
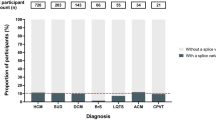
High-proportion spliced-in titin truncating variants (hiPSI TTNtvs) have been associated with an increased risk of atrial fibrillation, dilated cardiomyopathy (DCM) and heart failure in individuals of European ancestry1. However, similar data in individuals of African ancestry are lacking. Here we examined the association of hiPSI TTNtvs with atrial fibrillation, DCM and heart failure in individuals of African ancestry using data from the All of Us Research Program. Among 38,154 individuals of African ancestry, 169 (0.4%) individuals carried a hiPSI TTNtv. hiPSI TTNtv carriers were at a higher risk of developing atrial fibrillation (adjusted hazard ratio (HRadj) 2.42, 95% confidence interval (CI) 1.52–3.85), DCM (HRadj 2.82, 95% CI 1.81–4.39) and heart failure (HRadj 2.07, 95% CI 1.43–3.00) compared with noncarriers. The association of hiPSI TTNtvs with atrial fibrillation, DCM and heart failure was similar in individuals of African ancestry and those of European ancestry. Therefore, genetic testing for hiPSI TTNtvs may permit early identification of carriers and support preventive measures to reduce the likelihood of heart failure development both in individuals of European ancestry and in individuals of African ancestry.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
This study used the All of Us Research Program Controlled Tier Dataset version 7 (C2022Q4R9 Curated Data Repository), which is available in the All of Us Researcher Workbench. Data used for the current study are available to approved users of the All of Us Researcher Workbench (https://workbench.researchallofus.org/login). Controlled tier access provides access to genetic data on the All of Us Researcher Workbench and requires the completion of additional training.
Code availability
Code utilized in this study will be made available to All of Us Researcher Workbench users on contacting the study team.
References
Haggerty, C. M. et al. Genomics-first evaluation of heart disease associated with titin-truncating variants. Circulation 140, 42–54 (2019).
Santiago, C. F., Huttner, I. G. & Fatkin, D. Titin-related cardiomyopathy: is it a distinct disease? Curr. Cardiol. Rep. 24, 1069–1075 (2022).
Schafer, S. et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nat. Genet. 49, 46–53 (2017).
Herman, D. S. et al. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 366, 619–628 (2012).
Akinrinade, O., Alastalo, T. P. & Koskenvuo, J. W. Relevance of truncating titin mutations in dilated cardiomyopathy. Clin. Genet. 90, 49–54 (2016).
Akinrinade, O., Koskenvuo, J. W. & Alastalo, T. P. Prevalence of titin truncating variants in general population. PLoS ONE 10, e0145284 (2015).
Roberts, A. M. et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci. Transl. Med. 7, 270ra276 (2015).
Huggins, G. S. et al. Prevalence and cumulative risk of familial idiopathic dilated cardiomyopathy. JAMA 327, 454–463, (2022).
Tsao, C. W. et al. Heart Disease and Stroke Statistics-2023 Update: a report from the American Heart Association. Circulation 147, e93–e621 (2023).
All of Us Research Program Investigators et al. The “All of Us” Research Program. N. Engl. J. Med. 381, 668–676 (2019).
Genomic research data quality report. All of Us Research Program https://support.researchallofus.org/hc/en-us/articles/4617899955092-All-of-Us-Genomic-Quality-Report (2023).
Acknowledgements
The AoURP is supported by the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549; 1 OT2 OD026554; 1 OT2 OD026557; 1 OT2 OD026556; 1 OT2 OD026550; 1 OT2 OD 026552; 1 OT2 OD026553; 1 OT2 OD026548; 1 OT2 OD026551; 1 OT2 OD026555; IAA #: AOD 16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205; 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277; 3 OT2 OD025315; 1 OT2 OD025337; 1 OT2 OD025276. In addition, the AoURP would not be possible without the partnership of its participants. P.A. is supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) awards (R01HL160982, R01HL163852, R01HL163081 and K23HL146887).
Author information
Authors and Affiliations
Contributions
P.A. conceptualized and designed the study. P.A., N.S.S., A.P., N.P., P.L. and G.A. acquired, analyzed or interpreted data. P.A., N.S.S., A.P., N.P., P.L. and G.A. drafted the paper. All authors performed critical revisions of the paper. A.P. did the statistical analysis. P.A. and P.L. were responsible for supervision.
Corresponding author
Ethics declarations
Competing interests
P.A. reports grant support from Merck Sharp & Dohme LLC and Bristol-Myers Squibb and consulting income from Bristol-Myers Squibb, which are all unrelated to this work. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shetty, N.S., Pampana, A., Patel, N. et al. High-proportion spliced-in titin truncating variants in African and European ancestry in the All of Us Research Program. Nat Cardiovasc Res 3, 140–144 (2024). https://doi.org/10.1038/s44161-023-00417-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44161-023-00417-5