Abstract
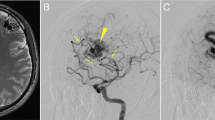
Cerebral cavernous malformations (CCMs) are a neurovascular anomaly that may occur sporadically or be inherited due to autosomal dominant mutations in KRIT1, CCM2 or PDCD10 (refs. 1,2,3,4). Individual lesions are caused by somatic mutations that have been identified in KRIT1, CCM2, PDCD10, MAP3K3 and PIK3CA5,6,7,8,9,10,11. However, the interactions between mutations and their relative contributions to sporadic versus familial cases are unclear. We show that mutations in KRIT1, CCM2, PDCD10 and MAP3K3 are mutually exclusive but may co-occur with mutations in PIK3CA. We also find that MAP3K3 mutations may cause sporadic but not familial CCM. Furthermore, we find identical PIK3CA mutations in CCMs and adjacent developmental venous anomalies (DVAs), a common vascular malformation frequently found in the vicinity of sporadic CCMs12,13,14. However, somatic mutations in MAP3K3 are found only in the CCM. This suggests that sporadic CCMs are derived from cells of the DVA that have acquired an additional mutation in MAP3K3.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data not included in this paper can be accessed through the National Center for Biotechnology Information (DNA sequencing, BioProject accession no. PRJNA802805) or Gene Expression Omnibus (RNA sequencing, accession no. GSE195732). The public datasets used in this study are available at the COSMIC (cancer.sanger.ac.uk/cosmic), dbSNP (ncbi.nlm.nih.gov/snp), 1000 Genomes Project (internationalgenome.org), gnomAD (gnomad.broadinstitute.org), miRWalk 3.0 (mirwalk.umm.uni-heidelberg.de) and DAVID databases (david.ncifcrf.gov).
Code availability
The variant calling software was implemented as part of gonomics, an ongoing effort to develop an open-source genomics platform in the Go programming language.
References
Laberge-le Couteulx, S. et al. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat. Genet. 23, 189–193 (1999).
Sahoo, T. et al. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1). Hum. Mol. Genet. 8, 2325–2333 (1999).
Liquori, C. L. et al. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am. J. Hum. Genet. 73, 1459–1464 (2003).
Bergametti, F. et al. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am. J. Hum. Genet. 76, 42–51 (2005).
Gault, J., Shenkar, R., Recksiek, P. & Awad, I. A. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke 36, 872–874 (2005).
Gault, J. et al. Cerebral cavernous malformations: somatic mutations in vascular endothelial cells. Neurosurgery 65, 138–144 (2009).
Akers, A. L., Johnson, E., Steinberg, G. K., Zabramski, J. M. & Marchuk, D. A. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum. Mol. Genet. 18, 919–930 (2009).
McDonald, D. A. et al. Lesions from patients with sporadic cerebral cavernous malformations harbor somatic mutations in the CCM genes: evidence for a common biochemical pathway for CCM pathogenesis. Hum. Mol. Genet. 23, 4357–4370 (2014).
Ren, A. A. et al. PIK3CA and CCM mutations fuel cavernomas through a cancer-like mechanism. Nature 594, 271–276 (2021).
Weng, J. et al. Somatic MAP3K3 mutation defines a subclass of cerebral cavernous malformation. Am. J. Hum. Genet. 108, 942–950 (2021).
Hong, T. et al. Somatic MAP3K3 and PIK3CA mutations in sporadic cerebral and spinal cord cavernous malformations. Brain 144, 2648–2658 (2021).
Abdulrauf, S. I., Kaynar, M. Y. & Awad, I. A. A comparison of the clinical profile of cavernous malformations with and without associated venous malformations. Neurosurgery 44, 41–46 (1999).
Wurm, G., Schnizer, M. & Fellner, F. A. Cerebral cavernous malformations associated with venous anomalies: surgical considerations. Neurosurgery 57, 42–58 (2005).
Porter, P. J., Willinsky, R. A., Harper, W. & Wallace, M. C. Cerebral cavernous malformations: natural history and prognosis after clinical deterioration with or without hemorrhage. J. Neurosurg. 87, 190–197 (1997).
Brinjikji, W., El-Rida El-Masri, A., Wald, J. T. & Lanzino, G. Prevalence of developmental venous anomalies increases with age. Stroke 48, 1997–1999 (2017).
Linscott, L. L., Leach, J. L., Jones, B. V. & Abruzzo, T. A. Developmental venous anomalies of the brain in children—imaging spectrum and update. Pediatr. Radiol. 46, 394–406 (2016).
Gökçe, E., Acu, B., Beyhan, M., Celikyay, F. & Celikyay, R. Magnetic resonance imaging findings of developmental venous anomalies. Clin. Neuroradiol. 24, 135–143 (2014).
Dammann, P. et al. Correlation of the venous angioarchitecture of multiple cerebral cavernous malformations with familial or sporadic disease: a susceptibility-weighted imaging study with 7-Tesla MRI. J. Neurosurg. 126, 570–577 (2017).
Zhou, Z. et al. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev. Cell 32, 168–180 (2015).
Couto, J. A. et al. A somatic MAP3K3 mutation is associated with verrucous venous malformation. Am. J. Hum. Genet. 96, 480–486 (2015).
Al-Qattan, M. M. et al. Late-onset multiple venous malformations confined to the upper limb: link to somatic MAP3K3 mutations. J. Hand Surg. Eur. Vol. 45, 1023–1027 (2020).
Xu, L. et al. Clonal evolution and changes in two AML patients detected with a novel single-cell DNA sequencing platform. Sci. Rep. 9, 11119 (2019).
Bizzotto, S. et al. Landmarks of human embryonic development inscribed in somatic mutations. Science 371, 1249–1253 (2021).
Mori, M. A., Ludwig, R. G., Garcia-Martin, R., Brandão, B. B. & Kahn, C. R. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 30, 656–673 (2019).
Yang, C. et al. Exosome miR-134-5p restrains breast cancer progression via regulating PI3K/AKT pathway by targeting ARHGAP1. J. Obstet. Gynaecol. Res. 47, 4037–4048 (2021).
Yuan, H. et al. MicroRNA let-7c-5p promotes osteogenic differentiation of dental pulp stem cells by inhibiting lipopolysaccharide-induced inflammation via HMGA2/PI3K/Akt signal blockade. Clin. Exp. Pharmacol. Physiol. 46, 389–397 (2019).
Li, Y., Li, P. & Wang, N. Effect of let-7c on the PI3K/Akt/FoxO signaling pathway in hepatocellular carcinoma. Oncol. Lett. 21, 96 (2021).
Srivastava, S. P., Hedayat, A. F., Kanasaki, K. & Goodwin, J. E. microRNA crosstalk influences epithelial-to-mesenchymal, endothelial-to-mesenchymal, and macrophage-to-mesenchymal transitions in the kidney. Front. Pharmacol. 10, 904 (2019).
Yin, H. et al. MiR-148a-3p regulates skeletal muscle satellite cell differentiation and apoptosis via the PI3K/AKT signaling pathway by targeting Meox2. Front. Genet. 11, 512 (2020).
Cimino, D. et al. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 27, 1223–1235 (2013).
Huo, L. et al. miR-128-3p inhibits glioma cell proliferation and differentiation by targeting NPTX1 through IRS-1/PI3K/AKT signaling pathway. Exp. Ther. Med. 17, 2921–2930 (2019).
Correia de Sousa, M., Gjorgjieva, M., Dolicka, D., Sobolewski, C. & Foti, M. Deciphering miRNAs’ action through miRNA editing. Int. J. Mol. Sci. 20, 6249 (2019).
Detter, M. R., Snellings, D. A. & Marchuk, D. A. Cerebral cavernous malformations develop through clonal expansion of mutant endothelial cells. Circ. Res. 123, 1143–1151 (2018).
Malinverno, M. et al. Endothelial cell clonal expansion in the development of cerebral cavernous malformations. Nat. Commun. 10, 2761 (2019).
Turchinovich, A., Samatov, T. R., Tonevitsky, A. G. & Burwinkel, B. Circulating miRNAs: cell–cell communication function? Front. Genet. 4, 119 (2013).
Roux, A. et al. High prevalence of developmental venous anomaly in diffuse intrinsic pontine gliomas: a pediatric control study. Neurosurgery 86, 517–523 (2020).
Brinjikji, W. et al. Facial venous malformations are associated with cerebral developmental venous anomalies. AJNR Am. J. Neuroradiol. 39, 2103–2107 (2018).
Tan, W.-H. et al. The spectrum of vascular anomalies in patients with PTEN mutations: implications for diagnosis and management. J. Med. Genet. 44, 594–602 (2007).
Dhamija, R. et al. Neuroimaging abnormalities in patients with Cowden syndrome: retrospective single-center study. Neurol. Clin. Pract. 8, 207–213 (2018).
Rasalkar, D. D. & Paunipagar, B. K. Developmental venous anomaly associated with cortical dysplasia. Pediatr. Radiol. 40, S165 (2010).
Akers, A. et al. Synopsis of guidelines for the Clinical Management of Cerebral Cavernous Malformations: consensus recommendations based on systematic literature review by the Angioma Alliance Scientific Advisory Board Clinical Experts Panel. Neurosurgery 80, 665–680 (2017).
Rueda, A. et al. sRNAtoolbox: an integrated collection of small RNA research tools. Nucleic Acids Res. 43, W467–W473 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Sticht, C., De La Torre, C., Parveen, A. & Gretz, N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE 13, e0206239 (2018).
Acknowledgements
We thank the patients who donated tissue for this study. We thank the Angioma Alliance, Barrow Neurological Institute and University of Chicago for patient enrollment and sample collection. Nucleus sorting was performed in the Duke Human Vaccine Institute Research Flow Cytometry Shared Resource Facility. We thank Duke University School of Medicine for the use of the Sequencing and Genomic Technologies Shared Resource for library preparation and sequencing. These studies were supported by National Institutes of Health grant nos. P01NS092521 (D.A.M., I.A.A., M.L.K.) and F31HL152738 (D.A.S.).
Author information
Authors and Affiliations
Contributions
D.A.S. designed and performed the genetic studies of human CCM lesions and wrote the manuscript. I.A.A. performed the surgical resection of the CCM and DVA samples used in this study. R.G., R.L., A.S., S.R., Y.L., C.C. and I.A.A. performed the plasma miRNA sequencing and analysis. A.A.R. and M.L.K. assisted with experimental design. R.G., I.A.A. and D.A.M. designed the experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
I.A.A. is Chairman of the scientific advisory board for the Angioma Alliance and provides expert opinions related to clinical care of CCMs. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks Matteo Malinverno, Murat Gunel and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Snellings, D.A., Girard, R., Lightle, R. et al. Developmental venous anomalies are a genetic primer for cerebral cavernous malformations. Nat Cardiovasc Res 1, 246–252 (2022). https://doi.org/10.1038/s44161-022-00035-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44161-022-00035-7
This article is cited by
-
Discovery and Characterization of Ephrin B2 and EphB4 Dysregulation and Novel Mutations in Cerebral Cavernous Malformations: In Vitro and Patient-Derived Evidence of Ephrin-Mediated Endothelial Cell Pathophysiology
Cellular and Molecular Neurobiology (2024)
-
Transcriptomic signatures of individual cell types in cerebral cavernous malformation
Cell Communication and Signaling (2024)
-
Impact of heterozygous ALK1 mutations on the transcriptomic response to BMP9 and BMP10 in endothelial cells from hereditary hemorrhagic telangiectasia and pulmonary arterial hypertension donors
Angiogenesis (2024)
-
Single-nucleus DNA sequencing reveals hidden somatic loss-of-heterozygosity in Cerebral Cavernous Malformations
Nature Communications (2023)
-
Angioarchitecture and genetic variants of spinal cord cavernous malformations and associated developmental venous anomalies: a case report
Child's Nervous System (2023)