Abstract
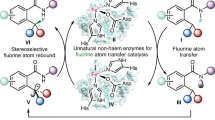
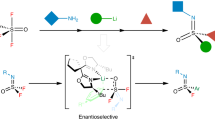
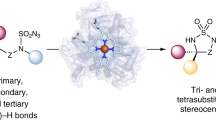
In recent years there has been a surge in the development of methods for the synthesis of organofluorine compounds. However, enzymatic methods for C–F bond formation have been limited to nucleophilic fluoride substitution. Here we report the incorporation of iron-catalysed radical fluorine transfer, a reaction mechanism that is not used in naturally occurring enzymes, into enzymatic catalysis for the development of biocatalytic enantioselective C(sp3)–F bond formation. Using this strategy, we repurposed (S)-2-hydroxypropylphosphonate epoxidase from Streptomyces viridochromogenes (SvHppE) to catalyse an N-fluoroamide-directed C(sp3)–H fluorination. Directed evolution has enabled SvHppE to be optimized, forming diverse chiral benzylic fluoride products with turnover numbers of up to 180 and with excellent enantiocontrol (up to 94% enantiomeric excess). Mechanistic investigations showed that the N–F bond activation is the rate-determining step, and the strong preference for fluorination in the presence of excess NaN3 can be attributed to the spatial proximity of the carbon-centred radical to the iron-bound fluoride.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data needed to evaluate the conclusions in this study are present in the main paper or Supplementary Information. The crystal structure of 1F is available from the Cambridge Crystallographic Data Centre under reference number CCDC 2267386.
Change history
26 April 2024
A Correction to this paper has been published: https://doi.org/10.1038/s44160-024-00547-z
References
Purser, S., Moore, P. R., Swallow, S. & Gouverneur, V. Fluorine in medicinal chemistry. Chem. Soc. Rev. 37, 320–330 (2008).
O’Hagan, D. Understanding organofluorine chemistry. An introduction to the C–F bond. Chem. Soc. Rev. 37, 308–319 (2008).
Inoue, M., Sumii, Y. & Shibata, N. Contribution of organofluorine compounds to pharmaceuticals. ACS Omega 5, 10633–10640 (2020).
Benedetto Tiz, D. et al. New halogen-containing drugs approved by FDA in 2021: an overview on their syntheses and pharmaceutical use. Molecules 27, 1643 (2022).
Britton, R. et al. Contemporary synthetic strategies in organofluorine chemistry. Nat. Rev. Methods Primers 1, 47 (2021).
Furuya, T., Kamlet, A. S. & Ritter, T. Catalysis for fluorination and trifluoromethylation. Nature 473, 470–477 (2011).
Yang, X. Y., Wu, T., Phipps, R. J. & Toste, F. D. Advances in catalytic enantioselective fluorination, mono-, di-, and trifluoromethylation, and trifluoromethylthiolation reactions. Chem. Rev. 115, 826–870 (2015).
Caron, S. Where does the fluorine come from? A review on the challenges associated with the synthesis of organofluorine compounds. Org. Process Res. Dev. 24, 470–480 (2020).
Walker, M. C. & Chang, M. C. Y. Natural and engineered biosynthesis of fluorinated natural products. Chem. Soc. Rev. 43, 6527–6536 (2014).
O’Hagan, D. & Deng, H. Enzymatic fluorination and biotechnological developments of the fluorinase. Chem. Rev. 115, 634–649 (2015).
Vaillancourt, F. H., Yeh, E., Vosburg, D. A., Garneau-Tsodikova, S. & Walsh, C. T. Nature’s inventory of halogenation catalysts: oxidative strategies predominate. Chem. Rev. 106, 3364–3378 (2006).
Agarwal, V. et al. Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse. Chem. Rev. 117, 5619–5674 (2017).
Papadopoulou, A., Meyer, F. & Buller, R. M. Engineering Fe(II)/α-ketoglutarate-dependent halogenases and desaturases. Biochemistry 62, 229–240 (2023).
Dong, C. et al. Crystal structure and mechanism of a bacterial fluorinating enzyme. Nature 427, 561–565 (2004).
O’Hagan, D., Schaffrath, C., Cobb, S. L., Hamilton, J. T. G. & Murphy, C. D. Biosynthesis of an organofluorine molecule—a fluorinase enzyme has been discovered that catalyses carbon–fluorine bond formation. Nature 416, 279 (2002).
Zechel, D. L. et al. Enzymatic synthesis of carbon–fluorine bonds. J. Am. Chem. Soc. 123, 4350–4351 (2001).
Cros, A., Alfaro-Espinoza, G., De Maria, A., Wirth, N. T. & Nikel, P. I. Synthetic metabolism for biohalogenation. Curr. Opin. Biotechnol. 74, 180–193 (2022).
Cheng, X. & Ma, L. Enzymatic synthesis of fluorinated compounds. Appl. Microbiol. Biotechnol. 105, 8033–8058 (2021).
Eustáquio, A. S., O’Hagan, D. & Moore, B. S. Engineering fluorometabolite production: fluorinase expression in salinispora tropica yields fluorosalinosporamide. J. Nat. Prod. 73, 378–382 (2010).
Liu, W. & Groves, J. T. Manganese catalyzed C–H halogenation. Acc. Chem. Res. 48, 1727–1735 (2015).
Panda, C., Anny-Nzekwue, O., Doyle, L. M., Gericke, R. & McDonald, A. R. Evidence for a high-valent iron-fluoride that mediates oxidative C(sp3)–H fluorination. JACS Au 3, 919–928 (2023).
Farley, G. W., Siegler, M. A. & Goldberg, D. P. Halogen transfer to carbon radicals by high-valent iron chloride and iron fluoride corroles. Inorg. Chem. 60, 17288–17302 (2021).
Bower, J. K., Cypcar, A. D., Henriquez, B., Stieber, S. C. E. & Zhang, S. C(sp3)–H fluorination with a copper(II)/(III) redox couple. J. Am. Chem. Soc. 142, 8514–8521 (2020).
Huang, X., Liu, W., Hooker, J. M. & Groves, J. T. Targeted fluorination with the fluoride ion by manganese-catalyzed decarboxylation. Angew. Chem. Int. Ed. 54, 5241–5245 (2015).
Huang, X. et al. Late stage benzylic C–H fluorination with [18F]fluoride for PET imaging. J. Am. Chem. Soc. 136, 6842–6845 (2014).
Liu, W. & Groves, J. T. Manganese-catalyzed oxidative benzylic C–H fluorination by fluoride ions. Angew. Chem. Int. Ed. 52, 6024–6027 (2013).
Liu, W. et al. Oxidative aliphatic C–H fluorination with fluoride ion catalyzed by a manganese porphyrin. Science 337, 1322–1325 (2012).
Pinter, E. N., Bingham, J. E., AbuSalim, D. I. & Cook, S. P. N-directed fluorination of unactivated Csp3–H bonds. Chem. Sci. 11, 1102–1106 (2020).
Groendyke, B. J., AbuSalim, D. I. & Cook, S. P. Iron-catalyzed, fluoroamide-directed C–H fluorination. J. Am. Chem. Soc. 138, 12771–12774 (2016).
Hintz, H. et al. Copper-catalyzed electrochemical C–H fluorination. Chem Catal. 3, 100491 (2023).
Rui, J. et al. Directed evolution of nonheme iron enzymes to access abiological radical-relay C(sp3)–H azidation. Science 376, 869–874 (2022).
Liu, P. et al. Protein purification and function assignment of the epoxidase catalyzing the formation of fosfomycin. J. Am. Chem. Soc. 123, 4619–4620 (2001).
Wang, C. et al. Evidence that the fosfomycin-producing epoxidase, HppE, is a non-heme-iron peroxidase. Science 342, 991–995 (2013).
Mitchell, A. J. et al. Structure-guided reprogramming of a hydroxylase to halogenate its small molecule substrate. Biochemistry 56, 441–444 (2017).
Gomez, C. A., Mondal, D., Du, Q., Chan, N. & Lewis, J. C. Directed evolution of an iron(II)- and α-ketoglutarate-dependent dioxygenase for site-selective azidation of unactivated aliphatic C–H bonds. Angew. Chem. Int. Ed. 62, e202301370 (2023).
Neugebauer, M. E. et al. Reaction pathway engineering converts a radical hydroxylase into a halogenase. Nat. Chem. Biol. 18, 171–179 (2022).
Papadopoulou, A. et al. Re-programming and optimization of a l-proline cis-4-hydroxylase for the cis-3-halogenation of its native substrate. ChemCatChem 13, 3914–3919 (2021).
Olivares, P., Ulrich, E. C., Chekan, J. R., van der Donk, W. A. & Nair, S. K. Characterization of two late-stage enzymes involved in fosfomycin biosynthesis in pseudomonads. ACS Chem. Biol. 12, 456–463 (2017).
Yun, D. et al. Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis. J. Am. Chem. Soc. 133, 11262–11269 (2011).
Higgins, L. J., Yan, F., Liu, P. H., Liu, H. W. & Drennan, C. L. Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme. Nature 437, 838–844 (2005).
Tarantino, G. & Hammond, C. Catalytic C(sp3)–F bond formation: recent achievements and pertaining challenges. Green Chem. 22, 5195–5209 (2020).
Matthews, M. L. et al. Substrate positioning controls the partition between halogenation and hydroxylation in the aliphatic halogenase, SyrB2. Proc. Natl Acad. Sci. USA 106, 17723–17728 (2009).
Kulik, H. J. & Drennan, C. L. Substrate placement influences reactivity in non-heme Fe(II) halogenases and hydroxylases. J. Biol. Chem. 288, 11233–11241 (2013).
Kissman, E. N. et al. Biocatalytic control of site-selectivity and chain length-selectivity in radical amino acid halogenases. Proc. Natl Acad. Sci. USA 120, e2214512120 (2023).
Chan, N. H. et al. Non-native anionic ligand binding and reactivity in engineered variants of the Fe(II)- and α-ketoglutarate-dependent oxygenase, SadA. Inorg. Chem. 61, 14477–14485 (2022).
Dai, L. et al. Structural and functional insights into a nonheme iron- and α-ketoglutarate-dependent halogenase that catalyzes chlorination of nucleotide substrates. Appl. Environ. Microbiol. 88, e02497–02421 (2022).
Kastner, D. W., Nandy, A., Mehmood, R. & Kulik, H. J. Mechanistic insights into substrate positioning that distinguish non-heme Fe(II)/α-ketoglutarate-dependent halogenases and hydroxylases. ACS Catal. 13, 2489–2501 (2023).
Acknowledgements
We thank M. Greenberg for helpful discussion and comments on the manuscript. We thank T. Yuan from H. Xiao’s group at Rice University for help on obtaining optical rotation data. We thank M. A. Siegler and JHU X-ray Crystallography Facility for analytical support. Financial support was provided by the Johns Hopkins University and National Institute for General Medical Sciences R00GM129419 and R35GM147639 (to X.H.). This work was also supported by the Spanish MICINN (Ministerio de Ciencia e Innovación) PID2019-111300GA-I00, PID2022-141676NB-I00 and TED2021-130173B-C42 projects (to M.G.-B.), and the Ramón y Cajal programme via the RYC 2020-028628-I fellowship (to M.G.-B.), the Generalitat de Catalunya (2021SGR00623 project to M.G.-B.), the Spanish MIU (Ministerio de Universidades) predoctoral fellowship FPU18/02380 (to J.S.), the National Natural Science Foundation of China (21978272) (to Y.Y.) and the Fundamental Research Funds for the Provincial Universities of Zhejiang (RF-C2022006) (to Y.Y.).
Author information
Authors and Affiliations
Contributions
X.H. conceived and directed the project. X.H., Z.C. and Q.Z. designed the experiments. Q.Z. and Z.C. performed screening of initial enzyme activity. Q.Z. and Z.C. performed directed evolution and results analysis. Q.Z., Z.C., J.R., Q.E.Y. and N.T.J. performed substrate scope study. M.G.-B. conceived and directed the computational modelling studies. J.S. and X.C. performed DFT calculations under the guidance of M.G.-B. and Y.Y. J.S. performed MD simulations under the guidance of M.G.-B. X.H. and Q.Z. wrote the manuscript with input from all other authors; Y.Y. and M.G.-B. wrote the computational sections.
Corresponding authors
Ethics declarations
Competing interests
A provisional patent application (no. PCT/US2023/022431) covering enantioselective biocatalytic C–F bond formation has been filed through the Johns Hopkins University with Q.Z., X.H., Z.C. and J.R. as inventors. Authors J.S., N.T.J., Q.E.Y., X.C., Y.Y. and M.G.-B. declare no competing interests.
Peer review
Peer review information
Nature Synthesis thanks Kyle Biegasiewicz, Anna Fryszkowska, Hans Senn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Thomas West, in collaboration with the Nature Synthesis team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1–9, Figs. 1–16, discussion, and further experimental and computational details.
Supplementary Data 1
Crystallographic data for compound 1F, CCDC 2267386.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, Q., Chen, Z., Soler, J. et al. Engineering non-haem iron enzymes for enantioselective C(sp3)–F bond formation via radical fluorine transfer. Nat. Synth (2024). https://doi.org/10.1038/s44160-024-00507-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44160-024-00507-7