Abstract
Background
IMP-producing Klebsiella spp. (IMPKsp) strains have spread globally, including in China. Currently, the prevalence and genomic characterization of IMPKsp is largely unknown nationwide. Here we aimed to provide a general overview of the phenotypic and genomic characteristics of IMPKsp strains.
Methods
61 IMPKsp strains were obtained from 13 provinces in China during 2016-2021. All strains were tested for their susceptibility to antimicrobial agents by the microdilution broth method and sequenced with Illumina next-generation sequencing. We performed conjugation experiments on thirteen representative strains which were also sequenced by Oxford nanopore sequencing technology to characterize blaIMP-encoding plasmids.
Results
We find that all IMPKsp strains display multidrug-resistant (MDR) phenotypes. All strains belong to 27 different STs. ST307 emerges as a principal IMP-producing sublineage. blaIMP-4 is found to be the major isoform, followed by blaIMP-38. Seven incompatibility types of blaIMP-encoding plasmids are identified, including IncHI5 (32/61, 52.5%), IncN-IncR (10/61, 16.4%), IncFIB(K)-HI1B (7/61, 11.5%), IncN (5/61, 8.2%), IncN-IncFII (2/61, 3.3%), IncFII (1/61, 1.6%) and IncP (1/61, 1.6%). The strains carrying IncHI5 and IncN plasmids belong to diverse ST types, indicating that these two plasmids may play an important role in the transmission of blaIMP genes among Klebsiella spp. strains.
Conclusions
Our results highlight that multi-clonal transmission, multiple genetic environments and plasmid types play a major role in the dissemination process of blaIMP genes among Klebsiella spp. IncHI5 type plasmids have the potential to be the main vectors mediating the spread of the blaIMP genes in Klebsiella spp.
Plain language summary
Antibiotic resistance occurs when bacteria evolve to withstand antibiotic drugs. We are aware that a bacteria called Klebsiella is rapidly becoming resistant to carbapenems, a class of broad-spectrum antibiotics. In this study, we conducted a genetic and microbiological surveillance study across 13 provinces of China to understand factors that contribute to the growing bacterial drug resistance. We find that the way the multiple bacterial types interact with each other and swap certain genetic material may be the main cause of growing resistance. These findings call for close monitoring of genetic evolution as a matter of public health management strategy.
Similar content being viewed by others
Introduction
Klebsiella spp. are Gram-negative bacteria that can cause opportunistic infections and are a leading cause of community-acquired and nosocomial infections1. The emergence and rapid spread of carbapenem-resistant Klebsiella spp. (CRKsp), with high morbidity and mortality rates, represents a major public health threat worldwide2,3,4. Production of carbapenemases is the main mechanism contributing to carbapenem resistance in Enterobacteriaceae5. The class B metallo-β-lactamases (MBLs) Imipenemases (IMPs) are one of the most important carbapenemases and can hydrolyze almost all β-lactams, such as cephalosporins, and carbapenems, but not the monobactams (i.e., aztreonam)2. The first IMP-1 carbapenemase was discovered in Pseudomonas aeruginosa in Japan in 1988, and subsequently, IMPs have been detected in Acinetobacter spp. and members of the Enterobacteriaceae family5,6,7. Outbreaks caused by strains harboring blaIMP genes have been reported in Japan, Australia and China5,6,7,8.At the time of writing, at least 96 isoforms of IMP carbapenemases have been identified (https://www.ncbi.nlm.nih.gov/pathogens/refgene/#IMP). IMP-encoding genes along with other resistance genes are often located within class 1 integrons carried by broad-host-range plasmids, including IncA/C, IncL/M, IncHI2 and IncN plasmids in Enterobacteriaceae5,9,10. In China, the blaIMP genes have been reported in Klebsiella spp. belonging to different high-risk sequence types (ST) ST11, ST15 and ST30711,12. Among these, IMP-4 is the most common variant in IMP-producing Klebsiella spp. (IMPKsp) frequently located on IncN plasmids9. However, detailed information on the plasmids associated with other IMP variants is currently insufficient because Klebsiella spp. producing IMP enzymes have been rare in China. Besides, IMP enzymes often co-exist with the other carbapenemases and ESBLs in Klebsiella spp.13, including SFO-1, a rarely reported ESBL. It was firstly identified in a Enterobacter cloacae strain from Japan in 199914, and can hydrolyze most β-lactams except carbapenems and cephamycins.
In the present study, we have performed a retrospective and descriptive study of resistance phenotypes and genomic epidemiological analysis of 61 clinical IMPKsp isolates from thirteen provinces in China from 2016-2021. We find that multiple clones, genetic environments and plasmid types are involved in disseminating blaIMP genes among Klebsiella spp. These findings provide an improved understanding of the dissemination characteristics of IMPKsp strains.
Methods
Bacterial Isolates
A total of 61 blaIMP-positive Klebsiella spp. isolates were collected from thirteen provinces in China during 2016-2021. Both matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonik GmbH, Bremen, Germany) and 16 S rRNA gene-based sequencing were applied for bacterial identification. Ethical permission for this study was approved by the Ethics Committee from The Second Affiliated Hospital of Zhejiang University, School of Medicine (2020-319).
Antimicrobial Susceptibility Testing
The antibiotic susceptibility of all blaIMP-positive Klebsiella spp. isolates to common clinically used antibiotics listed in Table 1 was determined by broth microdilution method. The susceptibility results were interpreted in accordance with the Clinical and Laboratory Standards Institute (CLSI) guideline except tigecycline. The MIC of tigecycline was interpreted following US Food and Drug Administration (FDA) clinical breakpoints. Escherichia coli ATCC 25922 was used as a quality control strain for the antimicrobial susceptibility testing.
Conjugation assay
In vitro conjugation experiments were performed using filter mating method. Rifampin-resistant E. coli EC600 and sodium azide-resistant E. coli J53 were used as the recipient strains, respectively. Transconjugants were selected on LB agar plates containing 0.5 μg/mL meropenem and 600 μg/mL rifampin and LB agar plates containing 0.5 μg/mL meropenem and 300 μg/mL sodium azide. The presence of the blaIMP gene was confirmed by PCR and Sanger sequencing15.
Whole genome sequencing and bioinformatics analysis
Genomic DNA of all blaIMP positive Klebsiella spp. isolates was extracted using a HiPure Bacterial DNA Kit (Magen, China) and sequenced using the HiSeq platform (Illumina, San Diego, CA) with a 2 × 150 bp paired-end sequencing strategy16. Assembly was performed using SPAdes Genome Assembler version 3.11.117. Location of the blaIMP gene was identified by aligning the contigs carrying blaIMP with complete genome sequences in the NCBI database. The genomic DNA of 13 representative Klebsiella spp. isolates with different resistance phenotypes and genetic traits was also sequenced using Oxford Nanopore Technologies MinION platform. The complete genome sequences of these 13 strains were obtained by hybrid assembly of Illumina and nanopore sequencing reads using Unicycler v 0.4.418. The draft genomes of the remaining strains sequenced using Illumina only were aligned to the reference plasmids to deduce the plasmid types and structures. The assembled genome sequences were annotated with RAST server19. Besides, Kleborate v2.0.4 was used to identify the MLSTs, serotyping of KL types, antimicrobial resistance genes and virulence genes20. Plasmid types were identified using PlasmidFinder version 2.121. Insertion sequences (ISs) were identified using ISfinder database. The generation of plasmid map was performed with BRIG v0.95. The analysis of the genetic structure of different blaIMP genes was performed with Easyfig 2.1. Coregenome alignment and single-nucleotide polymorphism (SNP) calling were performed with The Harvest suite, and a core genome phylogenetic tree was constructed using Parsnp22. The phylogenetic tree was visualized and edited using iTOL version 423.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Overview of the bla IMP-positive Klebsiella spp. isolates
A total of 61 blaIMP-positive Klebsiella spp. strains, including 50 K. pneumoniae, 6 K. variicola subsp. variicola, 4 K.quasipneumoniae subsp. quasipneumoniae and 1 K. quasipneumoniae subsp. similipneumoniae were collected across China from 2016-2021. The antimicrobial susceptibility results revealed that all isolates exhibited multidrug resistance phenotypes, with >96.7% of the strains being resistant to meropenem, ceftazidime, cefotaxime, cefmetazole, cefepime, cefoperazone/sulbactam, and ceftazidime/avibactam. Besides, they manifested low to moderate resistance (19.7% to 45.9%) to amikacin, ciprofloxacin, and piperacillin/tazobactam. Only a small number of strains were resistant to colistin (3.3%), and no isolates exhibited resistance to tigecycline (Table 1, Fig. 1a).
a The antimicrobial susceptibility results of strains. CTX: cefotaxime; SCF: cefoperazone/sulbactam; CAV: ceftazidime/avibactam; CAZ: ceftazidime; FEP: cefepime; MEM: meropenem; CMZ: cefmetazole; ETP: ertapenem; ATM: aztreonam; IPM: imipenem; CIP: ciprofloxacin; TZP: piperacillin/tazobactam; AK: amikacin; CST: colistin; TGC: tigecycline. b The distribution of carbapenemases among 61 IMPKsp strains.
Antibiotic resistance genes in IMP-producing Klebsiella spp
In this study, blaIMP genes were the only carbapenemase genes detected in 80.3% of these strains (49/61), with blaIMP-4 (n = 36) being the predominant isoform, followed by blaIMP-38 (n = 12) and blaIMP-1 (n = 1). The remaining 12 strains produced both IMP and other carbapenemases, including IMP-4 and NDM-1(n = 7), IMP-4 and KPC-2 (n = 3), IMP-4 and KPC (n = 1), IMP-8 and KPC-2 (n = 1) (Fig. 1b). The second most prevalent isoform IMP-38 differed from IMP-4 by one amino acid substitution (Ser214Gly). The IMP-8 carbapenemase derived from IMP-2 and had two amino acid substitutions (Arg214Ala and Val206Gly), compared with IMP-2.
In addition to carbapenemase genes, 57 of the 61 blaIMP-positive Klebsiella spp. strains also carried 1–18 other acquired antibiotic resistance genes, mainly including aminoglycoside resistance genes (strA, n = 32; strB, n = 32; aac(6’)-Ib4, n = 19; aac(3)-IId, n = 15), fluoroquinolone resistance genes (qnrS1, n = 24), sulfonamide resistance genes (sul1, n = 22; sul2, n = 21), and genes encoding extended spectrum beta lactamases (ESBLs) (blaTEM-105, n = 27; blaCTX-M-3, n = 18; blaCTX-M-14, n = 13; blaSFO-1, n = 12) (Fig. 2). Only one strain, Z245, carried a recently reported plasmid-borne multidrug resistance gene cluster, tmexCD2-toprJ2, and the minimum inhibitory concentrations (MICs) value of tigecycline for Z245 was 2 µg/mL. R110 was the unique mcr-9.1-positive strain but was susceptible to colistin.
The vertical and horizontal axes represent the strain numbers and antibiotic resistance genes, respectively. Red and white boxes represent the presence and absence of the antibiotic resistance genes in the strains, respectively. Names of antibiotics were displayed below the dashed lines. The genes covered by the line segment from the start point to end point could mediate resistance to labeled antibiotics.
Diversity of MLSTs and phylogenetic analysis of IMP-producing Klebsiella spp
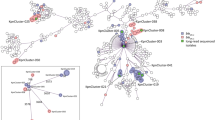
The 61 IMPKsp strains belonged to 27 different STs. The most prevalent ST was ST307 (13/61, 21.3%), followed by ST1779 (10/61, 16.4%), ST20 (7/61, 11.5%) and ST268 (5/61, 8.2%). ST307, ST1779, ST20 and ST268 correspond to capsular locus (KL) 102 types, KL38, KL28 and KL20, respectively. KL20-ST268 K. pneumoniae, often carried virulence factors and was reported to be hypervirulence-associated24,25,26. Similarly, KL20-ST268 K. pneumoniae strains in this study possessed siderophore biosynthetic clusters encoding yersiniabactin, colibactin, aerobactin and/or salmochelin as well as rmpADC or rmpA2 genes encoding regulators of the mucoid phenotype. Besides, KL54-ST29 strains encoded yersiniabactin, salmochelin and RmpADC. KL24-ST29 and ST11-KL47 strains produced yersiniabactin. The remaining IMPKsp strains did not harbor the aforementioned virulence factors. ST11 K. pneumoniae, which was the epidemic clone in China, was only detected in 3.3% (n = 2) of the IMPKsp strains in this study. The two ST11 K. pneumoniae produced both IMP-4 and KPC-2 carbapenemases. In addition, three novel ST types, including ST6238, ST6239 and ST6240, were identified in the present study. The epidemic clones exhibited to be associated with the regions. For instance, ST307 IMPKsp strains were mainly distributed in Hunan province and were primary vectors of the blaIMP-38 gene. ST1779, ST20 and ST268 IMPKsp strains were only detected in specific provinces: Hunan, Guangdong and Hubei, respectively, suggesting the clone dissemination of IMPKsp strains within these regions (Fig. 3).
The phylogenetic relationship analysis based on SNPs of core genomes showed that 61 IMPKsp isolates were clustered into three major clades which were associated with their species. Clade I consisted of K. quasipneumoniae subsp. quasipneumoniae and K. quasipneumoniae subsp. similipneumoniae strains (5/61, 8.2%); clade II consisted of K. variicola subsp. variicola strains (6/61, 9.8%); clade III consisted of K. pneumoniae (50/61, 82.0%). A total of 11 IMPKsp strains from clade I and II were sporadic and isolated from nine provinces. Clade III was further divided into three subgroups (IIIa, IIIb and IIIc), with ST1779 IMPKsp strains (10/50, 20.0%) being the major component of clade IIIa, ST20 (7/50, 14.0%) and ST29 (3/50, 6.0%) IMPKsp strains being the major component of clade IIIb, and ST307 (13/50, 26.0%) and ST268 (5/50, 10%) being the major component of clade IIIc, further suggesting a phenomenon of clone dissemination of IMPKsp strains (Fig. 3).
Characteristics of bla IMP-carrying plasmids and genetic environments of bla IMP
Plasmid replicon typing revealed that 58 blaIMP-carrying plasmids belonged to seven Inc types and 3 IMPKsp strains carried non-typable blaIMP-encoding plasmids. The IncHI5 (32/61, 52.5%) and IncN-IncR (10/61, 16.4%) were the predominant plasmid types, followed by IncFIB(K)-IncHI1B (7/61, 11.5%), IncN (5/61, 8.2%), IncN-IncFII (2/61, 3.3%), IncFII (1/61, 1.6%) and IncP (1/61, 1.6%) (Fig. 4a). The conjugation experiments were conducted on all thirteen representative strains sequenced by Oxford nanopore sequencing technology. The results revealed that, with the exception of IncFIB(K)-IncHI1B, each type of blaIMP-carrying plasmids from six representative strains could be transferred to the recipient E. coli EC600 at discrepant conjugation frequencies (1.5 × 10−6−3.2 × 10−3). The IncN-IncR plasmid pIMP-R215-3 had the highest conjugation efficiency (3.2 × 10−3), but the IncHI5 plasmid pIMP-T405 exhibited the lowest conjugation efficiency. And the conjugation experiments were failed in the remaining seven representative isolates encoded IncHI5 blaIMP-positive plasmids, using EC600 or J53 as the recipient.
a Sankey diagram consisted of the phylogroups, MLST types, Inc types, and the genetic environment of blaIMP genes in sequence. The width of the line is proportional to the number of isolates. b The alignment of genetic environment structures of blaIMP genes among IMPKsp strains. Homologous regions are shaded in gray.
Sequence alignment suggested that thirteen blaIMP-encoding plasmids aligned well to pA708-1 from other report27 (Supplementary Fig. 1a); eleven blaIMP-encoding plasmids aligned well to pIMP-S225-3 (Supplementary Fig. 1b); nine blaIMP-encoding plasmids aligned well to pIMP-R215-3 (Supplementary Fig. 1c); six blaIMP-encoding plasmids aligned well to pIMP-GD1954-1 (Supplementary Fig. 1d); four blaIMP-encoding plasmids aligned to pIMP-2028 (Supplementary Fig. 1e); one blaIMP-encoding plasmids aligned well to pIMP-GD1972-1 and one blaIMP-encoding plasmid aligned well to pIMP-K210045 (Supplementary Fig. 1f, g).
Plasmid pIMP-2028 isolated from strain 2028 carrying blaIMP-4 is a representative IncN plasmid, which is 50,137 bp in size and has an average GC content of 50.24%. It was highly similar to plasmid pIMP-FJ1503 from C. freundii (accession no. KU051710; 100% nucleotide identity at 93% query coverage) and pIMP-ECL14-57 from a clinical E. coli isolate (accession no. MH727565; 99.89% nucleotide identity and 94% query coverage) (Supplementary Fig. 2a). pIMP-2028 had the intact ISKpn19 element but lost qnrS1, which located upstream of ISKpn19 in plasmid pIMP-FJ1503 and pIMP-ECL14-57. The blaIMP-4 with a truncated Intl1 gene upstream was embedded in class 1 integron In823, which was present in a transposon-like structure IS6100-mobC-ltrA-blaIMP-4 -ΔIntI1-IS26 (Fig. 4b). The IncN-IncR type plasmid pIMP-R215-3 was 105,677 bp in size and had GC content of 52.51% and 124 open reading frames. It showed 100% identity and 40% query coverage identity with pC52_002 (accession no. CP042547), 100% identity and 42% query coverage identity with pIMP-FJ1503 (accession no. KU051710), 99.98% identity and 40% query coverage identity with pk9 (accession no. CP049891) (Supplementary Fig. 2c). The genetic environment of blaIMP-4 gene on plasmid pIMP-R215-3 was the same as that of pIMP-2028.
The plasmid pIMP-K210045 was identified as a novel hybrid plasmid simultaneously carrying blaIMP-4 and blaKPC-2. It had 189,882 bp in length, and contained 252 open reading frames with an average GC content of 52.91%. The plasmid pIMP-K210045 carried IncN and IncFII family plasmid replication initiator RepA. Comparative genomic analysis showed that the sequence of pIMP-K210045 was composed of two main parts containing blaIMP-4 and blaKPC-2 genes, respectively. The two parts came from two different types of plasmids: the blaIMP-4 region shared 100% identity and 33% query coverage identity with pIMP-FJ1503. The genetic environment surrounding the blaIMP-4 genes was IS6100-mobC-ltrA-blaIMP-4-ΔIntI1-IS26 (Fig. 4b); the blaKPC-2 region shared 100% identity and 73% query coverage with pKPC-2-KP65 (accession no. CP044259) of K. pneumoniae (Supplementary Fig. 2d). The formation of hybrid plasmid was presumably linked to the presence of IS26.
The plasmid pIMP-GD1954-1 was 263,080 bp in length with a GC content of 51.58% and showed 99.96% identity and 82% query coverage identity with a hybrid plasmid pAZS099-NDM-IMP (accession no. CP086762) carrying both blaIMP-4 and blaNDM-1 in a ST20-KL28 K. pneumoniae (Supplementary Fig. 2e). The blaIMP-4 gene was located in the genetic environment IS6100-mobC-ltrA-blaIMP-4-ΔIntI1-IS26 (Fig. 4b). The remnant region of pAZS099-NDM-IMP with blaNDM-1 gene was also detected in the form of a circular IncX3-type plasmid in the strain GD1954-1 and named pNDM-GD1954-1. Further analysis showed that IS6-like element IS26 family transposase led to the formation of two single plasmids originating from pAZS099-NDM-IMP. It is known that IncX3-type plasmid is an easily transmissible vector. And in the filter-mating assay in this study, only transconjugants with blaNDM-1 gene was obtained and no transconjugants carrying blaIMP-4 gene was obtained.
The IncFII plasmid pIMP-ZJ578 was 45,535 bp long and had an average GC content of 49.31%, encoding 57 ORFs. This plasmid showed 99.99% nucleotide identity and 99% query coverage with p16005813B (MK036884), which was isolated from clinical Leclercia adecarboxylata in Ningbo, China (Supplementary Fig. 2f). The blaIMP-8 was located on a class 1 integron In655 (tniA-tniB-tniQ-tniR-aacA4-blaIMP-8-ΔIntI1) (Fig. 4b), which was carried by Tn6505 derived from Tn1696. The Tn6505 of pIMP-ZJ578 differed from that of p16005813B by insertion of 45 nucleotides in merT gene.
The pIMP-20R25 was an IncP-1β type plasmid encoding blaIMP-1 gene and had a circularly closed 61,282 bp DNA sequence consisting of 68 ORFs, with an average GC content of 63.14%. Sequence analysis showed that the pIMP-20R25 was highly homologous to another blaIMP-1-carrying IncP-1β type plasmid pNXM63-IMP (accession no. NZ_MW150990; coverage, 88%; identity, 99.99%) found in Morganella morganii and plasmid pIMP4-ECL352 (accession no. CP083711; coverage, 95%; identity, 99.99%) (Supplementary Fig. 2b). The blaIMP-1 was located in a novel genetic environment of ant1-blaIMP-1-ΔIntI1 (Fig. 4b), which was firstly identified to be adjacent to ant1, a streptomycin resistance gene. The existence of two putative transfer regions belonging to the IncP-1β plasmid backbone made it self-transmissible easily. So far, IMP-1-encoding IncP-1β plasmids were only detected in Achromobacter xylosoxidans and M. morganii28,29. This is the first report of K. variicola subsp. variicola harboring a conjugative blaIMP-1-positive IncP-1β plasmid.
The seven plasmids including pIMP-GD1972-1, pIMP-P407-1, pIMP-R110, pIMP-S225-3, pIMP-T226, pIMP-T405, and pIMP-Z245 belonged to IncHI5 incompatibility group in this study. The plasmids varied in size from about 223 kb to nearly 377 kb and the number of predicted ORFs varied from 241 to 424. The seven plasmids described above carried blaIMP-4 gene except that pIMP-S225-3 carried blaIMP-38 gene. IncHI plasmids are important vectors of genes encoding for antibiotic resistance, such as β-lactams including carbapenems, aminoglycosides, and quinolones. In this study, IncHI5 plasmid harbored more antibiotic resistance genes than other types of plasmids, leading to multi-drug resistance and facilitating survival under different antimicrobial pressures. According to the previous report, blaIMP genes were generally located in the transposons or transposon-like structures on the antibiotic resistance islands designated ARI-A. In the present work, the blaIMP-4 gene was possessed by a class I integron within Tn1696-Related ARI-A with the gene cassette of ISCR1-sul1- qacEΔ1-ltrA-blaIMP-4 -IntI1-tnpR-tnpA-IS5075 on pIMP-P407-1, pIMP-T226 and pIMP-T405, consistent with that of p11219-IMP (accession no. MF344561) (Supplementary Fig. 3b-d). The genetic environment of the blaIMP-4 gene was ΔcatB3-aacA4-ltrA-blaIMP-4-IntI1-tnpR-tnpA-IS5075 on pIMP-R110 and pIMP-Z245, consistent with that of pA708-1(accession no. CP026369) (Fig. 4b, Supplementary Fig. 3f,g). The ARI-A islands were not found in pIMP-GD1972-1 (Supplementary Fig. 3e). The blaIMP-4 gene of pIMP-GD1972-1was located in the genetic environment IS6100-mobC-ltrA-blaIMP-4-ΔIntI1-IS26 (Fig. 4b). The pIMP-S225-3 showed 100% nucleotide identity and 82% query coverage with pA324-IMP (accession no. MF344566) (Supplementary Fig. 3a). They both had the intact Tn1696 derivatives-Tn6382 bearing blaCTX-M-3 and blaTEM-1B genes, and the blaIMP-38 gene was also located within Tn6382 with the gene cassette of ΔcatB3-aacA4-qacG2-blaIMP-38- IntI1 (Fig. 4b).
Together, these results suggested the plasmid types and genetic environment of blaIMP genes were diversified and there was a certain correlation between them. Most of the blaIMP genes located on the IncHI5 plasmids were associated with the ARI-A islands. IncHI5 plasmid was the predominant plasmid type and carried complex antibiotic resistance genes. The strains carrying IncHI5 plasmids belonged to diverse ST types, indicating that they played an important role in the transmission of blaIMP genes among Klebsiella spp. strains.
Discussion
The ongoing epidemic of carbapenemase-producing Klebsiella spp. has emerged as a mounting menace for human health worldwide1. However, the prevalence and genomic charaterization of IMPKsp isolates are less studied nationwide due to that KPC, OXA-48-like and NDM enzymes are the major carbapenemases in CRKP strains in China, and they have been widely studied4. The prevalence of IMP in Klebsiella spp. is relatively low and often ignored. Large-scale studies on the “ancient” carbapenemase IMP in K. pneumoniae are still lacking.This descriptive work investigated molecular and epidemiological characteristics of clinical IMPKsp strains in China and revealed that the genetic background of blaIMP genes was diverse and IncHI5 plasmid has the potential to be another important vector mediating transmission of blaIMP.
Only one IMPKsp strain was carbapenem-susceptible. A previous study reported that the transcription factor ArdK encoded by plasmids inhibited the expression of the IMP-6 metallo-β-lactamase, conferring a carbapenem-susceptible phenotype in the blaIMP-6-positive E. coli strain30. Further research is needed to explain the mechanism of carbapenem susceptibility of IMP-positive isolates.
The IMP-type MBL was first identified in Klebsiella spp. from Wuhan in China in 200831. Since then, sporadic cases of IMPKsp infections have been frequently reported nationwide. And there have also been sporadic epidemics caused by IMPKsp strains. IMP-38-producing ST307 K. pneumoniae strains and IMP-4-producing ST2253 K. pneumoniae infected neonatal patients in the neonatal ward in Shandong and Hunan province, respectively11,32. This again is consistent with our findings that there was a regional spread of ST307 IMPKsp strains carrying IMP-38 carbapenemases in Hunan province. K. pneumoniae ST307 has emerged as an antimicrobial-resistant high-risk clone during the 1990s and has been associated with the transmission of various carbapenemases worldwide11,33,34,35. Control measures are urgently required to prevent the transmission rates of ST307 K. pneumoniae producing IMP carbapenemase. In this study, another epidemic clone IMP-4-producing ST1779 K. pneumoniae was first detected in Hunan province. Though this new ST1779 was not relevant to other well-known epidemic IMP-4 producing K. pneumoniae clones reported in China, this novel ST might become a high-risk clone that needed close attention. ST20 was the one of the most common K. pneumoniae ST type in China and was the third most frequently observed clone in the present study. ST20 K. pneumoniae was the common vector of IMP-4 and NDM-1 carbapenemases36. Recently, Jia et al. have first reported a novel hybrid plasmid coharboring IMP-4 and NDM-1 carbapenemases in ST20 K. pneumoniae37. But in our study, the blaIMP-4 and blaNDM-1 genes were located on two different plasmids in ST20 K. pneumoniae, indicating that this ST could also pose a potential threat to human health. These reflected the vertical transmission of blaIMP genes. Therefore, it is necessary to monitor the spread of specific epidemic clones in specific regions to combat this gene.
In this study, plasmid spread was the main transmission mode of blaIMP-4 gene, though clonal spread, and integron spread were also observed. IncHI5 was the predominant type of blaIMP-harboring plasmid in the isolates investigated and involved a variety of ST clones in the study, suggesting the potential of horizontal transfer of IncHI5-type plasmids among Klebsiella spp. Though this type of plasmids has broad host range, the low transformation efficiency or non-transferability of plasmids has been observed in this study, probably due to the nonoptimal conjugation condition, which required further study. So far, blaIMP-4 and blaIMP-38 genes have been found to reside on IncHI5 plasmids in three previous studies from China11,38, which is consistent with our findings. A previous study has reported that another subgroup of IncHI plasmid, IncHI2 plasmid has led to the high prevalence of blaIMP-4 gene, which encoded the major carbapenemases in carbapenemase-producing Enterobacteriaceae (CPE) in Queensland39. Similarly, extra vigilance is required that blaIMP gene may be maintained in the Klebsiella spp. population by circulating on IncHI5 plasmids or via clonal expansion in China.
Synthesizing across previous researches, the IncN plasmids were the major carrier of blaIMP gene among Klebsiella spp. and play an important role in transmission of carbapenemases9,13,40. And in our study, the broad-host-range and conjugative IncN plasmid was detected in diverse ST types, and the fusion events between IncN and other types of plasmids including IncR and IncFII plasmids were observed, indicating a potential route for the evolution of IncN plasmids.
In addition to the variety of plasmid types, class 1 integrons linked to autonomously transferable genetic structures surrounding blaIMP genes were also divergent, and were the vehicles for dissemination of blaIMP genes9, contributing to the diversity of plasmids in this study. Though the structure IS6100-mobC-ltrA-blaIMP-4-ΔIntI1-IS26 was highly conserved, the plasticity of genetic contexts of blaIMP genes in IncHI5 plasmid was observed in the study. Multiple insertional elements, mainly IS26 and IS5075, were involved in the genetic structure alterations of blaIMP gene.
The blaSFO-1 gene was usually detected in Enterobacter spp. in China and has been reported only once in association with blaIMP-4 gene in K. pneumoniae41,42. But our study revealed that the blaSFO-1 gene is not as uncommon as previously considered. Besides, among the 12 blaSFO-1-positive IMPKsp strains, we found the coexistence of blaSFO-1 gene and blaIMP-4 on the IncHI5 plasmids in 11 strains. The 11 strains belonged to six different ST types, suggesting the potential for horizontal transfer of the blaSFO-1 gene along with the IncHI5 plasmids. Ai et al. has reported that mcr-9, blaNDM−1 and a rare gene blaSFO−1 were detected simultaneously on the same IncHI2-ST1 plasmid43. This plasmid type often led to the spread of carbapenemase-encoding genes and the evolution of complex resistance phenotypes, so did IncHI5 plasmid. The prevalence of blaSFO-1-positive strains might be underestimated, and more international surveillance of blaSFO-1-positive strains and IncHI plasmids should be adopted to prevent their dissemination. Besides, colistin and tigecyline were recognized as the last line of defense against severe infections, but the coexistence of blaIMP genes with colistin resistance gene mcr-9.1 or tigecycline resistance gene tmexCD2-toprJ2, was observed in this study, aggravating the problem of antibiotic resistance. Though the mcr-9.1 did not confer resistance to colistin in this study, continued vigilance was needed to the “silent” transmission of mcr-9.1, especially to strains with qseB/qseC two-component system, which could regulate expression of mcr-9.1 gene44.
There are some limitations in this descriptive work. 61 IMP-positive isolates were collected from 13 provinces rather than all provinces in China. No positive strains were detected in some provinces. Phenotypic and genomic characteristics of 61 IMP-positive isolates could represent most but not all regions of China.
In conclusion, this descriptive work elucidated the phenotypic and genotypic characteristics of plasmids containing blaIMP genes in Klebsiella spp. The IMPKsp isolates exhibited high sequence diversity. The dissemination of blaIMP genes was driven by the multiple genetic environment and plasmid types. Compared with previous studies, IncHI5-type plasmids may be developed into epidemic types in Klebsiella spp. in China. Class 1 integrons still play an important role in the dissemination of blaIMP genes. The primary epidemic clones have regional differences in several provinces in China, suggesting that stringent monitoring and appropriate actions are needed.
References
Wyres, K., Lam, M. & Holt, K. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359 (2020).
van Duin, D. et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet. Infectious Dis. 20, 731–741 (2020).
Grundmann, H. et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17, 153–163 (2017).
Zhang, R. et al. Nationwide Surveillance of Clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19, 98–106 (2017).
Matsumura, Y. et al. Global Molecular Epidemiology of IMP-Producing Enterobacteriaceae. Antimicrobial Agents Chemother. 61, https://doi.org/10.1128/aac.02729-16 (2017).
Sidjabat, H. E. et al. Dominance of IMP-4-producing enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob. Agents Chemother. 59, 4059–4066 (2015).
Chu, Y. W. et al. IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45, 710–714 (2001).
Fukigai, S. et al. Nosocomial outbreak of genetically related IMP-1 beta-lactamase-producing Klebsiella pneumoniae in a general hospital in Japan. Int. J. Antimicrob. Agents 29, 306–310 (2007).
Liu, W. et al. Molecular characterization of blaIMP-4-carrying enterobacterales in Henan Province of China. Front Microbiol 12, 626160 (2021).
Ho, P. L. et al. pIMP-PH114 carrying bla IMP-4 in a Klebsiella pneumoniae strain is closely related to other multidrug-resistant IncA/C2 plasmids. Curr. Microbiol. 68, 227–232 (2014).
Wang, S. et al. IMP-38-Producing High-Risk Sequence Type 307 Klebsiella pneumoniae Strains from a Neonatal Unit in China. 5, https://doi.org/10.1128/mSphere.00407-20 (2020).
Lai, K. et al. Molecular characterization of clinical IMP-producing Klebsiella pneumoniae isolates from a Chinese Tertiary Hospital. Ann. Clin. Microbiol. Antimicrob. 16, 42 (2017).
Wang, J. et al. First report of Klebsiella oxytoca strain simultaneously producing NDM-1, IMP-4, and KPC-2 Carbapenemases. Antimicrob. Agents Chemother. 61, https://doi.org/10.1128/aac.00877-17 (2017).
Matsumoto, Y. & Inoue, M. Characterization of SFO-1, a plasmid-mediated inducible class A beta-lactamase from Enterobacter cloacae. Antimicrob. Agents Chemother. 43, 307–313 (1999).
Lv, L. et al. Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including Tigecycline, in Klebsiella pneumoniae. Mbio 11, e02930–19 (2020).
Li, R. et al. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7, gix132 (2018).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Overbeek, R. et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214 (2013).
Lam, M. M. et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12, 1–16 (2021).
Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Treangen, T. J., Ondov, B. D., Koren, S. & Phillippy, A. M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524 (2014).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–w259 (2019).
Matono, T., Morita, M., Nakao, N., Teshima, Y. & Ohnishi, M. Genomic insights into virulence factors affecting tissue-invasive Klebsiella pneumoniae infection. Ann. Clin. Microbiol. Antimicrob. 21, 2 (2022).
Zhu, J., Jiang, X., Zhao, L. & Li, M. An outbreak of ST859-K19 Carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese Teaching Hospital. mSystems 7, e0129721 (2022).
Shen, P. et al. Hypervirulence markers among non-ST11 strains of Carbapenem- and multidrug-resistant Klebsiella pneumoniae isolated from patients with bloodstream infections. Front. Microbiol. 11, 1199 (2020).
Liang, Q. et al. Sequencing and genomic diversity analysis of IncHI5 plasmids. Front. Microbiol. 9, 3318 (2018).
Chen, Z. et al. IMP-1 encoded by a novel Tn402-like class 1 integron in clinical Achromobacter xylosoxidans, China. Sci. Rep. 4, 7212 (2014).
Xiang, G. et al. Clinical Molecular and Genomic Epidemiology of Morganella morganii in China. Front. Microbiol. 12, 744291 (2021).
Segawa, T. et al. The plasmid-encoded transcription factor ArdK contributes to the repression of the IMP-6 metallo-β-lactamase gene blaIMP-6, leading to a carbapenem-susceptible phenotype in the blaIMP-6-positive Escherichia coli strain A56-1S. PLoS ONE 13, e0208976 (2018).
Mendes, R. E. et al. Carbapenem-resistant isolates of Klebsiella pneumoniae in China and detection of a conjugative plasmid (blaKPC-2 plus qnrB4) and a blaIMP-4 gene. Antimicrob. Agents Chemother. 52, 798–799 (2008).
Bai, Y., Shao, C., Hao, Y., Wang, Y. & Jin, Y. Using whole genome sequencing to trace, control and characterize a hospital infection of IMP-4-producing Klebsiella pneumoniae ST2253 in a Neonatal Unit in a Tertiary Hospital, China. Frontiers in public health 9, 755252 (2021).
Strydom, K. A. et al. Klebsiella pneumoniae ST307 with OXA-181: threat of a high-risk clone and promiscuous plasmid in a resource-constrained healthcare setting. J. Antimicrob. Chemother. 75, 896–902 (2020).
Patil, S. et al. Emergence of ST307 Klebsiella pneumoniae Co-producing CTX-M with SHV and KPC from paediatric patients at Shenzhen Children’s Hospital, China. Infection and drug resistance 14, 3581–3588 (2021).
Haller, S. et al. Extensively drug-resistant Klebsiella pneumoniae ST307 outbreak, north-eastern Germany, June to October 2019. Euro Surveill 24, https://doi.org/10.2807/1560-7917.es.2019.24.50.1900734 (2019).
Pei, N. et al. Large-scale genomic epidemiology of Klebsiella pneumoniae identified clone divergence with hypervirulent plus antimicrobial-resistant characteristics causing within-ward strain transmissions. Microbiol. Spectrum 10, e0269821 (2022).
Jia, X. et al. Coexistence of bla (NDM-1) and bla (IMP-4) in one novel hybrid plasmid confers transferable carbapenem resistance in an ST20-K28 Klebsiella pneumoniae. Front. Microbiol. 13, 891807 (2022).
Liu, L., Feng, Y., Long, H., McNally, A. & Zong, Z. Sequence Type 273 Carbapenem-Resistant Klebsiella pneumoniae Carrying bla(NDM-1) and bla(IMP-4). Antimicrob. Agents Chemother. 62, https://doi.org/10.1128/aac.00160-18 (2018).
Roberts, L. W. et al. Genomic analysis of carbapenemase-producing Enterobacteriaceae in Queensland reveals widespread transmission of bla(IMP-4) on an IncHI2 plasmid. Microb. Genom. 6, https://doi.org/10.1099/mgen.0.000321 (2020).
Sun, M., Xiao, W. & Xu, Q. IncN1 ST7 epidemic plasmid carrying blaIMP-4 in One ST85-type Klebsiella oxytoca clinical isolate with porin deficiency. Infect Drug Resist. 14, 3827–3835 (2021).
Zhou, K. et al. Dissemination of a ‘rare’ extended-spectrum β-lactamase gene bla(SFO-1) mediated by epidemic clones of carbapenemase-producing Enterobacter hormaechei in China. Int. J. Antimicrob. Agents 56, 106079 (2020).
Zhou, K. et al. A novel Tn1696-like composite transposon (Tn6404) harboring bla (IMP-4) in a Klebsiella pneumoniae isolate carrying a rare ESBL gene bla (SFO-1). Sci. Rep. 7, 17321 (2017).
Ai, W. et al. First report of coexistence of bla (SFO-1) and bla (NDM-1) β-lactamase genes as well as colistin resistance gene mcr-9 in a transferrable plasmid of a clinical isolate of enterobacter hormaechei. Front. Microbiol. 12, 676113 (2021).
Kieffer, N. et al. mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob. Agents Chemother. 63, https://doi.org/10.1128/aac.00965-19 (2019).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82272392, 22193064 and 82072341).
Author information
Authors and Affiliations
Contributions
C.L. performed the experiments, bioinformatic analysis and draft the manuscript. N.D. helped with bioinformatic analysis. Y.Z. and Q.S. helped with strain collection. Y.H. and C.C. helped draft the manuscript. G.C. and R.Z. supervised the project and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Adam Jenney and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, C., Dong, N., Zhang, Y. et al. Phenotypic and genomic characteristics of clinical IMP-producing Klebsiella spp. Isolates in China. Commun Med 4, 25 (2024). https://doi.org/10.1038/s43856-024-00439-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-024-00439-5