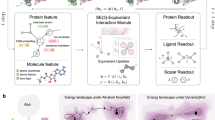
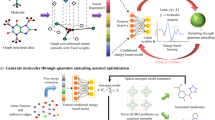
Abstract
Data-driven deep learning algorithms provide accurate prediction of high-level quantum-chemical molecular properties. However, their inputs must be constrained to the same quantum-chemical level of geometric relaxation as the training dataset, limiting their flexibility. Adopting alternative cost-effective conformation generative methods introduces domain-shift problems, deteriorating prediction accuracy. Here we propose a deep contrastive learning-based domain-adaptation method called Local Atomic environment Contrastive Learning (LACL). LACL learns to alleviate the disparities in distribution between the two geometric conformations by comparing different conformation-generation methods. We found that LACL forms a domain-agnostic latent space that encapsulates the semantics of an atom’s local atomic environment. LACL achieves quantum-chemical accuracy while circumventing the geometric relaxation bottleneck and could enable future application scenarios such as inverse molecular engineering and large-scale screening. Our approach is also generalizable from small organic molecules to long chains of biological and pharmacological molecules.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The preprocessed data in this work for reproducing the results are available on figshare at https://doi.org/10.6084/m9.figshare.24445129 (ref. 52). The model checkpoints used in this work for reproducing the results are available on GitHub at https://github.com/parkyjmit/LACL and figshare at https://doi.org/10.6084/m9.figshare.24456802 (ref. 53). Source data are provided with this paper.
Code availability
The Python code capsule of this work including the training script for reproducing the results is available on GitHub at https://github.com/parkyjmit/LACL and figshare at https://doi.org/10.6084/m9.figshare.24456802 (ref. 53).
References
Olivecrona, M., Blaschke, T., Engkvist, O. & Chen, H. Molecular de-novo design through deep reinforcement learning. J. Cheminform. 9, 48 (2017).
Jeon, W. & Kim, D. Autonomous molecule generation using reinforcement learning and docking to develop potential novel inhibitors. Sci. Rep. 10, 22104 (2020).
Reker, D. & Schneider, G. Active-learning strategies in computer-assisted drug discovery. Drug Discov. Today 20, 458–465 (2015).
De Cao, N. & Kipf, T. MolGAN: an implicit generative model for small molecular graphs. Preprint at arXiv https://doi.org/10.48550/arXiv.1805.11973 (2018).
Guo, M. et al. Data-efficient graph grammar learning for molecular generation. Preprint at arXiv https://doi.org/10.48550/arXiv.2203.08031 (2022).
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. In Proc. 34th International Conference on Machine Learning, Proc. Machine Learning Research Vol. 70 (eds Precup, D. & Teh, Y. W.) 1263–1272 (PMLR, 2017).
Unke, O. T. & Meuwly, M. PhysNet: a neural network for predicting energies, forces, dipole moments, and partial charges. J. Chem. Theory Comput. 15, 3678–3693 (2019).
Schütt, K. T., Sauceda, H. E., Kindermans, P.-J., Tkatchenko, A. & Müller, K.-R. SchNet—a deep learning architecture for molecules and materials. J. Chem. Phys. 148, 241722 (2018).
Klicpera, J., Groß, J. & Günnemann, S. Directional message passing for molecular graphs. In International Conference on Learning Representations Vol. 8 (2020).
Klicpera, J., Giri, S., Margraf, J. T. & Günnemann, S. Fast and uncertainty-aware directional message passing for non-equilibrium molecules. Preprint at https://arxiv.org/abs/2011.14115 (2020).
Choudhary, K. & DeCost, B. Atomistic line graph neural network for improved materials property predictions. npj Comput. Mater. 7, 185 (2021).
Schütt, K., Unke, O. & Gastegger, M. Equivariant message passing for the prediction of tensorial properties and molecular spectra. In Proc. 38th International Conference on Machine Learning, Proc. Machine Learning Research Vol. 139 (eds Meila, M. & Zhang, T.) 9377–9388 (PMLR, 2021).
Lim, J. et al. Predicting drug–target interaction using a novel graph neural network with 3D structure-embedded graph representation. J. Chem. Inf. Model. 59, 3981–3988 (2019).
Fout, A., Byrd, J., Shariat, B. & Ben-Hur, A. Protein interface prediction using graph convolutional networks. Adv. Neural Inf. Process. Syst. 30, 6530–6539 (2017).
Tang, B. et al. A self-attention based message passing neural network for predicting molecular lipophilicity and aqueous solubility. J. Cheminform. 12, 15 (2020).
Ramakrishnan, R., Dral, P. O., Rupp, M. & Von Lilienfeld, O. A. Quantum chemistry structures and properties of 134 kilo molecules. Sci. Data 1, 140022 (2014).
Becke, A. D. Density-functional thermochemistry. I. The effect of the exchange-only gradient correction. J. Chem. Phys. 96, 2155–2160 (1992).
Lee, C., Yang, W. & Parr, R. G. Development of the Colle–Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 37, 785 (1988).
Ditchfield, R., Hehre, W. J. & Pople, J. A. Self-consistent molecular-orbital methods. IX. An extended Gaussian-type basis for molecular-orbital studies of organic molecules. J. Chem. Phys. 54, 724–728 (1971).
Frisch, M. J., Pople, J. A. & Binkley, J. S. Self-consistent molecular orbital methods 25. Supplementary functions for Gaussian basis sets. J. Chem. Phys. 80, 3265–3269 (1984).
Hehre, W. J., Ditchfield, R. & Pople, J. A. Self-consistent molecular orbital methods. XII. Further extensions of Gaussian-type basis sets for use in molecular orbital studies of organic molecules. J. Chem. Phys. 56, 2257–2261 (1972).
Krishnan, R., Binkley, J. S., Seeger, R. & Pople, J. A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 72, 650–654 (1980).
Halgren, T. A. MMFF VI. MMFF94s option for energy minimization studies. J. Comput. Chem. 20, 720–729 (1999).
Lemm, D., von Rudorff, G. F. & von Lilienfeld, O. A. Machine learning based energy-free structure predictions of molecules, transition states, and solids. Nat. Commun. 12, 4468 (2021).
Xu, M. et al. GeoDiff: a geometric diffusion model for molecular conformation generation. In International Conference on Learning Representations Vol. 10 (2022).
Luo, S., Shi, C., Xu, M. & Tang, J. Predicting molecular conformation via dynamic graph score matching. Adv. Neural Inf. Process. Syst. 34, 19784–19795 (2021).
Zhu, J. et al. Direct molecular conformation generation. Transactions on Machine Learning Research (2022).
Lemm, D., von Rudorff, G. F. & von Lilienfeld, O. A. Machine learning based energy-free structure predictions of molecules, transition states, and solids. Nat. Commun. 12, 4468 (2021).
Mansimov, E., Mahmood, O., Kang, S. & Cho, K. Molecular geometry prediction using a deep generative graph neural network. Sci. Rep. 9, 20381 (2019).
Isert, C., Atz, K., Jiménez-Luna, J. & Schneider, G. QMugs, quantum mechanical properties of drug-like molecules. Sci. Data 9, 273 (2022).
Ganin, Y. & Lempitsky, V. Unsupervised domain adaptation by backpropagation. In Proc. 32nd International Conference on Machine Learning, Proc. Machine Learning Research Vol. 37 (eds Bach, F. & Blei, D.) 1180–1189 (PMLR, 2015).
Chen, Z., Li, X. & Bruna, J. Supervised community detection with line graph neural networks. In International Conference on Learning Representations Vol. 5 (2017).
Thakoor, S. et al. Large-scale representation learning on graphs via bootstrapping. In International Conference on Learning Representations Vol. 10 (2022).
Xu, M., Luo, S., Bengio, Y., Peng, J. & Tang, J. Learning neural generative dynamics for molecular conformation generation. In International Conference on Learning Representations Vol. 9 (2021).
Van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988).
Weininger, D., Weininger, A. & Weininger, J. L. SMILES. 2. Algorithm for generation of unique smiles notation. J. Chem. Inf. Comput. Sci. 29, 97–101 (1989).
Riniker, S. & Landrum, G. A. Better informed distance geometry: using what we know to improve conformation generation. J. Chem. Inf. Model. 55, 2562–2574 (2015).
Landrum, G. RDKit: open-source cheminformatics. http://www.rdkit.org (2006).
Hsu, T. et al. Efficient and interpretable graph network representation for angle-dependent properties applied to optical spectroscopy. npj Comput. Mater. 8, 151 (2022).
Kaundinya, P. R., Choudhary, K. & Kalidindi, S. R. Prediction of the electron density of states for crystalline compounds with atomistic line graph neural networks (ALIGNN). JOM 74, 1395–1405 (2022).
Fang, X. et al. Geometry-enhanced molecular representation learning for property prediction. Nat. Mach. Intell. 4, 127–134 (2022).
Xie, T. & Grossman, J. C. Crystal graph convolutional neural networks for an accurate and interpretable prediction of material properties. Phys. Rev. Lett. 120, 145301 (2018).
Choudhary, K. et al. The joint automated repository for various integrated simulations (JARVIS) for data-driven materials design. npj Comput. Mater. 6, 173 (2020).
Satorras, V. G., Hoogeboom, E. & Welling, M. E(n) equivariant graph neural networks. In Proc. 38th International Conference on Machine Learning, Proc. Machine Learning Research Vol. 139 (eds Meila, M. & Zhang, T.) 9323–9332 (PMLR, 2021).
Batzner, S. et al. E(3)-equivariant graph neural networks for data-efficient and accurate interatomic potentials. Nat. Commun. 13, 2453 (2022).
Sun, Q. et al. PySCF: the Python-based simulations of chemistry framework. Wiley Interdiscip. Rev. Comput. Mol. Sci. 8, e1340 (2018).
Bannwarth, C., Ehlert, S. & Grimme, S. GFN2-xtTB—an accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J. Chem. Theory Comput. 15, 1652–1671 (2019).
Larsen, A. H. et al. The atomic simulation environment—a Python library for working with atoms. J. Phys. Condensed Matter 29, 273002 (2017).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 32, 8026–8037 (2019).
Wang, M. Y. Deep graph library: towards efficient and scalable deep learning on graphs. In International Conference on Learning Representations Vol. 7 (2019).
Park, Y. J., Kim, H., Jo, J. & Yoon, S. sharedata-to-reproduce-lacl. figshare https://doi.org/10.6084/m9.figshare.24445129 (2023).
Park, Y. J., Kim, H., Jo, J. & Yoon, S. LACL. figshare https://doi.org/10.6084/m9.figshare.24456802 (2023).
Acknowledgements
Y.J.P. was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korean government, Ministry of Science and ICT (MSIT) (no. 2021R1A6A3A01086766). The 05-Neuron supercomputer was provided by the Korea Institute of Science and Technology Information (KISTI) National Supercomputing Center for Y.J.P. Y.J.P., H.K., J.J. and S.Y. were supported by Institute of Information and Communications Technology Planning and Evaluation (IITP) grant funded by the Korea government (MSIT) (2021-0-01343: Artificial Intelligence Graduate School Program (Seoul National University)), National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (2022R1A3B1077720) and the BK21 FOUR program of the Education and Research Program for Future ICT Pioneers, Seoul National University in 2023. We express our gratitude to J. Im at the Chemical Data-driven Research Center in the Korea Research Institute of Chemical Technology (KRICT) for his valuable insights and discussion on the content of this paper.
Author information
Authors and Affiliations
Contributions
Y.J.P. conceived the study. Y.J.P. and S.Y. supervised the research. Y.J.P. designed and implemented the deep learning framework. Y.J.P., H.K. and J.J. conducted benchmarks and case studies. All authors participated in the preparation (writing and drawing) of the paper and the analysis of experimental results. All authors reviewed and edited the submitted version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Comparison between a backbone ALIGNN and the LACL model from the point of view of accuracy and inference time.
(a) Parity plot of trained ALIGNN and LACL model for various regression targets in QM9 dataset. Molecular conformation data from both the DFT domain and the CGCF domain is used as a test dataset. μ is the dipole moment. (b) Comparison of computation time between an ALIGNN model with DFT geometric relaxation and the LACL model with MMFF geometric relaxation. Geometric relaxations running on two 24-core Intel Cascade Lake i9 CPUs and GNN architectures running on a single NVIDIA RTX3090 graphics processing unit (GPU). The bar indicates the mean of computation time and the error bar indicates the standard deviation. Five samples were collected for runtime calculation, except When the number of heavy atoms was one (three samples).
Extended Data Fig. 2 2-D t-SNE visualization of trained node and graph representations from both ALIGNN and LACL model for bandgap and internal energy at 0 K (U0) regression in QM9 dataset.
(a) 2-D t-SNE visualization of trained representations of the local atomic environment from LACL model for bandgap regression in QM9 dataset. Molecular conformation data from both DFT and CGCF domains are used as the test dataset. Orange, sky blue, green, yellow, and blue-colored point indicates hydrogen, carbon, nitrogen, oxygen, and fluorine, respectively. To visualize the node representation of different molecules, several example molecules are shown. The atom surrounded by a green circle is a nitrogen atom belonging to the cyanyl group. The atom surrounded by the purple circle is the oxygen atom included in the ring. The hydrogen, carbon, nitrogen, and oxygen atoms in molecules are indicated as white, gray, purple, and red color, respectively. (b) t-SNE visualization for trained node (atom-level) and graph (molecule-level) representations. Representations are visualized for each level, feature, model, and domain.
Supplementary information
Supplementary Information
Supplementary Sections 1–9, Figs. 1–10 and Tables 1–4.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Park, Y.J., Kim, H., Jo, J. et al. Deep contrastive learning of molecular conformation for efficient property prediction. Nat Comput Sci 3, 1015–1022 (2023). https://doi.org/10.1038/s43588-023-00560-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-023-00560-w