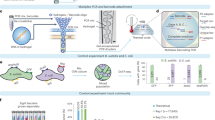
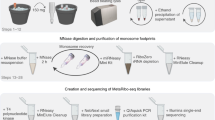
Abstract
Existing genomic sequencing data can be used to study host–microbiome ecosystems; however, distinguishing signals that originate from truly present microbes from contaminating species and artifacts is a substantial and often prohibitive challenge. Here we show that emerging sequencing technologies definitely capture reads from present microbes. We developed SAHMI, a computational resource to identify truly present microbial nucleic acids, as well as filter contaminants and spurious false-positive taxonomic assignments from standard transcriptomic sequencing of mammalian tissues. In benchmark studies, SAHMI correctly identifies known microbial infections present in diverse tissues, and we validate SAHMI’s enrichment for correctly classified, truly present species using multiple orthogonal computational experiments. The application of SAHMI to single-cell and spatial genomic data thus enables co-detection of somatic cells and microorganisms and joint analysis of host–microbiome ecosystems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The cell lines microbiome negative control dataset is available in Supplementary Table 4. Other data are available on our Github: https://github.com/sjdlabgroup/SAHMI and at Zenodo: https://doi.org/10.5281/zenodo.7017103 (ref. 24). The following infection datasets were analyzed in this manuscript: COVID-19 (GSE145926), M. leprae (GSE151528 and GSE167889), gastric samples (GSE134520), Salmonella (GSE79363), Candida (GSE111731), M. tuberculosis (GSE167232) and HIV (GSE111727). The following human reference genome was used: hg19 (PRJNA31257). Source Data are provided with this paper.
Code availability
The SAHMI pipeline is available on our Github (https://github.com/sjdlabgroup/SAHMI) and at Zenodo (https://doi.org/10.5281/zenodo.7017103)24.
References
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).
Poore, G. D. et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579, 567–574 (2020).
Breitwieser, F. P., Baker, D. N. & Salzberg, S. L. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 19, 198 (2018).
Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 257 (2019).
Liao, M. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020).
Ma, F. et al. The cellular architecture of the antimicrobial response network in human leprosy granulomas. Nat. Immunol. 22, 839–850 (2021).
Zhang, P. et al. Dissecting the single-cell transcriptome network underlying gastric premalignant lesions and early gastric cancer. Cell Rep. 27, 1934–1947.e5 (2019).
Saliba, A. E. et al. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat. Microbiol. 2, 16206 (2016).
Muñoz, J. F. et al. Coordinated host-pathogen transcriptional dynamics revealed using sorted subpopulations and single macrophages infected with Candida albicans. Nat. Commun. 10, 1607 (2019).
Pisu, D. et al. Single cell analysis of M. tuberculosis phenotype and macrophage lineages in the infected lung. J. Exp. Med. 218, e20210615 (2021).
Golumbeanu, M. et al. Single-cell RNA-seq reveals transcriptional heterogeneity in latent and reactivated HIV-infected cells. Cell Rep. 23, 942–950 (2018).
Wyler, E. et al. Single-cell RNA-sequencing of herpes simplex virus 1-infected cells connects NRF2 activation to an antiviral program. Nat. Commun. 10, 4878 (2019).
Sims, D., Sudbery, I., Ilott, N. E., Heger, A. & Ponting, C. P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 15, 121–132 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Byrd, A. L., Belkaid, Y. & Segre, J. A. The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155 (2018).
Jin, H. et al. mBodyMap: a curated database for microbes across human body and their associations with health and diseases. Nucleic Acids Res. 50, D808–D816 (2022).
Ghaddar, B. et al. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell 40, 1240–1253.e5 (2022).
Riquelme, E. et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 178, 795–806.e12 (2019).
Jia, Y. et al. Sequencing introduced false positive rare taxa lead to biased microbial community diversity, assembly, and interaction interpretation in amplicon studies. Environ. Microbiome 17, 43 (2022).
Stuart, T. et al. Comprehensive Integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Franzén, O., Gan, L.-M. & Björkegren, J. L. M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, baz046 (2019).
sjdlabgroup. sjdlabgroup/SAHMI: SAHMI v1.0 (v1.0). Zenodo https://doi.org/10.5281/zenodo.7017103 (2022).
Acknowledgements
We acknowledge the Office of Advanced Research Computing (OARC) at Rutgers, The State University of New Jersey for providing access to the Amarel cluster (https://it.rutgers.edu/oarc). We also acknowledge grant support from National Institutes of Health grants R21CA248122, R01GM129066 and R35GM149224 (S.D.), National Institutes of Health grant U01AI22285 (M.J.B.), Sergei Zlinkoff Foundation (M.J.B.), Canadian Institute for Advanced Research (M.J.B.), and National Institutes of Health, National Center for Advancing Translational Sciences, Rutgers Clinical and Translational Science Award TL1TR003019 (B.G.).
Author information
Authors and Affiliations
Contributions
B.G. and S.D. conceived the study. B.G. designed and performed all data analyses. B.G., M.J.B. and S.D. interpreted the data, and wrote and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.J.B. declares that he serves on the Scientific Advisory Board of Micronoma, Inc. B.G. and S.D. have jointly filed PCT patent applications PCT/US2022/025829 and PCT/US2022/025832.
Peer review
Peer review information
Nature Computational Science thanks Anders B. Dohlman, Thomas S.B. Schmidt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ananya Rastogi, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Sections 1–3 and Figs. 1–3.
Supplementary Table 1
Summary of benchmark studies analyzed.
Supplementary Table 2
SAHMI results for benchmark datasets.
Supplementary Table 3
Summary of cell-line experiments analyzed.
Supplementary Table 4
Cell lines microbiome reference.
Source data
Source Data Fig. 1
Numerical source data for all panels in Fig. 1.
Source Data Fig. 2
Numerical source data for all panels in Fig. 2.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghaddar, B., Blaser, M.J. & De, S. Denoising sparse microbial signals from single-cell sequencing of mammalian host tissues. Nat Comput Sci 3, 741–747 (2023). https://doi.org/10.1038/s43588-023-00507-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-023-00507-1