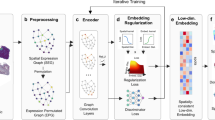
Abstract
Spatial transcriptomics data can provide high-throughput gene expression profiling and the spatial structure of tissues simultaneously. Most studies have relied on only the gene expression information but cannot utilize the spatial information efficiently. Taking advantage of spatial transcriptomics and graph neural networks, we introduce cell clustering for spatial transcriptomics data with graph neural networks, an unsupervised cell clustering method based on graph convolutional networks to improve ab initio cell clustering and discovery of cell subtypes based on curated cell category annotation. On the basis of its application to five in vitro and in vivo spatial datasets, we show that cell clustering for spatial transcriptomics outperforms other spatial clustering approaches on spatial transcriptomics datasets and can clearly identify all four cell cycle phases from multiplexed error-robust fluorescence in situ hybridization data of cultured cells. From enhanced sequential fluorescence in situ hybridization data of brain, cell clustering for spatial transcriptomics finds functional cell subtypes with different micro-environments, which are all validated experimentally, inspiring biological hypotheses about the underlying interactions among the cell state, cell type and micro-environment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data for Figs. 2–6 are available with this manuscript. The datasets utilized in this study can be downloaded from: (1) MERFISH dataset5: https://www.pnas.org/doi/10.1073/pnas.1912459116#supplementary-materials or our Github link: https://github.com/xiaoyeye/CCST/tree/main/dataset; (2) SeqFISH+ dataset35: https://github.com/CaiGroup/seqFISH-PLUS; (3) DLPFC dataset37: http://research.libd.org/globus/jhpce_HumanPilot10x/index.html; (4) 10× Visium spatial transcriptomics data of human breast cancer: https://support.10xgenomics.com/spatial-gene-expression/datasets/1.1.0/V1_Breast_Cancer_Block_A_Section_1. The annotation file can be found on the SEDR32 website: https://github.com/JinmiaoChenLab/SEDR_analyses/tree/master/data/BRCA1.
Code availability
CCST is implemented in Python. The source code and the utilized MERFISH dataset can be downloaded from the supporting website: https://github.com/xiaoyeye/CCST. https://doi.org/10.5281/zenodo.6560643 (ref. 50).
References
Codeluppi, S. et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat. Methods 15, 932–935 (2018).
Moffitt, J. R. & Zhuang, X. RNA imaging with multiplexed error-robust fluorescence in situ hybridization (MERFISH). Methods Enzymol. 572, 1–49 (2016).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl. Acad. Sci. U.S.A. 113, 11046–11051 (2016).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Xia, C., Fan, J., Emanuel, G., Hao, J. & Zhuang, X. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl. Acad. Sci. U.S.A. 116, 19490–19499 (2019).
Eng, C.-H. L., Shah, S., Thomassie, J. & Cai, L. Profiling the transcriptome with RNA SPOTs. Nat. Methods 14, 1153–1155 (2017).
Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M. & Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, aat5691 (2018).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Nichterwitz, S. et al. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 7, 12139 (2016).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 39, 1375–1384 (2021).
Andersson, A. et al. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun. Biol. 3, 565 (2020).
Pal, B. et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat. Commun. 8, 1627 (2017).
Arnol, D., Schapiro, D., Bodenmiller, B., Saez-Rodriguez, J. & Stegle, O. Modeling cell–cell interactions from spatial molecular data with spatial variance component analysis. Cell Rep. 29, 202–211 (2019).
Yuan, Y. & Bar-Joseph, Z. GCNG: graph convolutional networks for inferring gene interaction from spatial transcriptomics data. Genome Biol. 21, 300 (2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Abdelaal, T. et al. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 20, 194 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, P10008 (2008).
Traag, V. A., Waltman, L. & Van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Shekhar, K. et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323 (2016).
Pandey, S., Shekhar, K., Regev, A. & Schier, A. F. Comprehensive identification and spatial mapping of habenular neuronal types using single-cell RNA-seq. Curr. Biol. 28, 1052–1065 (2018).
Zhu, Q., Shah, S., Dries, R., Cai, L. & Yuan, G.-C. Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat. Biotechnol. 36, 1183–1190 (2018).
Stoltzfus, C. R. et al. CytoMAP: a spatial analysis toolbox reveals features of myeloid cell organization in lymphoid tissues. Cell Rep. 31, 107523 (2020).
Dries, R. et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22, 78 (2021).
Pham D., et al. stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell–cell interactions and spatial trajectories within undissociated tissues. Prerint at bioRxiv https://doi.org/10.1101/2020.05.31.125658 (2020).
Teng, H., Yuan, Y. & Bar-Joseph, Z. Clustering spatial transcriptomics data. Bioinformatics 38, 997–1004 (2022).
Hu, J. et al. SpaGCN: integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods 18, 1342–1351 (2021).
Fu, H., et al. Unsupervised spatial embedded deep representation of spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2021.06.15.448542 (2021).
Chen Y., Zhou S., Li M., Zhao F., & Qi J. STEEL enables high-resolution delineation of spatiotemporal transcriptomic data. Preprint at research square https://doi.org/10.21203/rs.3.rs-1240258/v1 (2022).
Kipf T. N. & Welling M. Semi-supervised classification with graph convolutional networks. In Proc. International Conference on Learning Representations (2017). https://openreview.net/forum?id=SJU4ayYgl
Eng, C.-H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019).
Veličković P., et al. Deep graph infomax. In Proc. International Conference on Learning Representations (2019). https://openreview.net/forum?id=rklz9iAcKQ
Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021).
Donjerkovic, D. & Scott, D. W. Regulation of the G1 phase of the mammalian cell cycle. Cell Res. 284, C349–364 (2000).
Tripathi, V. et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 9, e1003368 (2013).
Wang, J. et al. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed. Pharmacother. 68, 557–564 (2014).
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A. & Giraudat, J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303 (2001).
Mahdessian, D. et al. Spatiotemporal dissection of the cell cycle with single-cell proteogenomics. Nature 590, 649–654 (2021).
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Cheng, C. et al. Cloning, expression and characterization of a novel human VMP gene. Mol. Biol. Rep. 29, 281–286 (2002).
Li, S. et al. Endothelial cell-derived GABA signaling modulates neuronal migration and postnatal behavior. Cell Res. 28, 221–248 (2018).
Russ, A. P. et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature 404, 95–99 (2000).
Taberner, L., Bañón, A. & Alsina, B. Sensory neuroblast quiescence depends on vascular cytoneme contacts and sensory neuronal differentiation requires initiation of blood flow. Cell Rep. 32, 107903 (2020).
Hie, B., Bryson, B. & Berger, B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol. 37, 685–691 (2019).
Li J., Chen S., Pan X., Yuan Y., & Shen H.-B. Cell clustering for spatial transcriptomics data with graph neural networks. Zenodo https://doi.org/10.5281/zenodo.6560643 (2022).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (no. 61725302 to H.S.), 62073219 (to H.S.), 62103262 (to Y.Y.) and 61903248 (to X.P.) and the Shanghai Pujiang Programme (no. 21PJ1407700 to Y.Y.).
Author information
Authors and Affiliations
Contributions
H.S. and Y.Y. conceived and supervised the study. Y.Y. designed experiments. J.L. developed the computational model and conducted data analysis. Y.Y., H.S. and X.P. provided advice on data analysis. Y.Y. and S.C. proposed the proper computational model. J.L. drafted the manuscript. Y.Y. and H.S. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Xin Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Handling editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Comparison on sample 151676 of DLPFC.
Annotation and cluster labels obtained by CCST and prior methods on sample 151676 of DLPFC. Metrics including ARI, FMI and NMI are annotated on the bottom of each figure. Numbers in the legend refer to cluster labels.
Extended Data Fig. 2 Comparison on 10x Visium spatial transcriptomics data of human breast cancer.
Annotation and cluster labels obtained by CCST and prior methods on 10x Visium spatial transcriptomics data of human breast cancer. Metrics including ARI, FMI and NMI are annotated on the bottom of each figure. Numbers in the legend refer to cluster labels.
Supplementary information
Supplementary information Comparison on 10x Visium spatial transcriptomics data of human breast cancer.
Supplementary Figs. 1–30, Sections 1–15 and Tables 1–4.
Supplementary Data 1. Comparison on 10x Visium spatial transcriptomics data of human breast cancer.
The top 200 significantly DE genes of each cell group obtained by CCST on the MERFISH dataset.
Source data
Source Data Fig. 2.
Statistical source data of each subfigure is listed in each sheet.
Source Data Fig. 3.
Statistical source data of each subfigure is listed in each sheet.
Source Data Fig. 4.
Raw data of boxplots is listed in each sheet.
Source Data Fig. 5.
Statistical source data of each subfigure is listed in each sheet.
Source Data Fig. 6.
Statistical source data of each subfigure is listed in each sheet.
Source Data Extended Data Fig. 1.
Cell cluster labels.
Source Data Extended Data Fig. 2.
Cell cluster labels.
Rights and permissions
About this article
Cite this article
Li, J., Chen, S., Pan, X. et al. Cell clustering for spatial transcriptomics data with graph neural networks. Nat Comput Sci 2, 399–408 (2022). https://doi.org/10.1038/s43588-022-00266-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-022-00266-5
This article is cited by
-
PROST: quantitative identification of spatially variable genes and domain detection in spatial transcriptomics
Nature Communications (2024)
-
MENDER: fast and scalable tissue structure identification in spatial omics data
Nature Communications (2024)
-
Benchmarking spatial clustering methods with spatially resolved transcriptomics data
Nature Methods (2024)
-
Mapping cell-to-tissue graphs across human placenta histology whole slide images using deep learning with HAPPY
Nature Communications (2024)
-
SODB facilitates comprehensive exploration of spatial omics data
Nature Methods (2023)