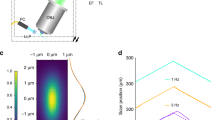
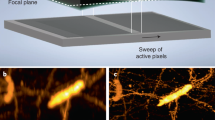
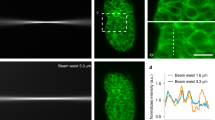
Abstract
Light sheet fluorescence microscopy (LSFM) uses a thin sheet of light to excite only fluorophores within the focal volume. Light sheet microscopes (LSMs) have a true optical sectioning capability and, hence, provide axial resolution, restrict photobleaching and phototoxicity to a fraction of the sample and use cameras to record tens to thousands of images per second. LSMs are used for in-depth analyses of large, optically cleared samples and long-term three-dimensional (3D) observations of live biological specimens at high spatio-temporal resolution. The independently operated illumination and detection trains and the canonical implementations, selective/single plane illumination microscope (SPIM) and digital scanned laser microscope (DSLM), are the basis for many LSM designs. In this Primer, we discuss various applications of LSFM for imaging multicellular specimens, developing vertebrate and invertebrate embryos, brain and heart function, 3D cell culture models, single cells, tissue sections, plants, organismic interaction and entire cleared brains. Further, we describe the combination of LSFM with other imaging approaches to allow for super-resolution or increased penetration depth and the use of sophisticated spatio-temporal manipulations to allow for observations along multiple directions. Finally, we anticipate developments of the field in the near future.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 1 digital issues and online access to articles
$99.00 per year
only $99.00 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Santi, P. A. Light sheet fluorescence microscopy: a review. J. Histochem. Cytochemistry https://doi.org/10.1369/0022155410394857 (2011).
Tomer, R., Khairy, K. & Keller, P. J. Shedding light on the system: studying embryonic development with light sheet microscopy. Curr. Opin. Genet. Dev. 21, 558–565 (2011).
Valm, A. M. et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature https://doi.org/10.1038/nature22369 (2017).
Lu, C. H. et al. Lightsheet localization microscopy enables fast, large-scale, and three-dimensional super-resolution imaging. Commun. Biol. 2, 1–10 (2019).
Sticker, M., Elsässer, R., Neumann, M. & Wolff, H. How to get better fluorescence images with your widefield microscope: a methodology review. Microsc. Today 28, 36–43 (2020).
Cox, I. J. Scanning optical fluorescence microscopy. J. Microsc. 133, 149–154 (1984).
Reynaud, E. G., Kržič, U., Greger, K. & Stelzer, E. H. K. Light sheet-based fluorescence microscopy: more dimensions, more photons, and less photodamage. HFSP J. https://doi.org/10.2976/1.2974980 (2008).
Truong, T. V., Supatto, W., Koos, D. S., Choi, J. M. & Fraser, S. E. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods 8, 757–760 (2011).
Jemielita, M., Taormina, M. J., Delaurier, A., Kimmel, C. B. & Parthasarathy, R. Comparing phototoxicity during the development of a zebrafish craniofacial bone using confocal and light sheet fluorescence microscopy techniques. J. Biophotonics https://doi.org/10.1002/jbio.201200144 (2013).
Power, R. M. & Huisken, J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods https://doi.org/10.1038/nmeth.4224 (2017).
Siedentopf, H. & Zsigmondy, R. Über Sichtbarmachung und Größenbestimmung ultramikoskopischer Teilchen, mit besonderer Anwendung auf Goldrubingläser [German]. Ann. Phys. https://doi.org/10.1002/andp.19023150102 (1902).
Voie, A. H., Burns, D. H. & Spelman, F. A. Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J. Microsc. 170, 229–236 (1993).
Fuchs, E., Jaffe, J., Long, R. & Azam, F. Thin laser light sheet microscope for microbial oceanography. Opt. Express https://doi.org/10.1364/oe.10.000145 (2002).
Resandt, R. W. W. et al. Optical fluorescence microscopy in three dimensions: microtomoscopy. J. Microsc. 138, 29–34 (1985).
Hell, S. & Stelzer, E. H. K. Properties of a 4Pi confocal fluorescence microscope. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 9, 2159–2166 (1992).
Stelzer, E. H. K. & Lindek, S. Fundamental reduction of the observation volume in far-field light microscopy by detection orthogonal to the illumination axis: confocal theta microscopy. Opt. Commun. 111, 536–547 (1994).
Swoger, J., Huisken, J. & Stelzer, E. H. K. Multiple imaging axis microscopy improves resolution for thick-sample applications. Opt. Lett. 28, 1654 (2003).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004). This work describes the first diffraction-limited LSM in the form of the selective/single plane illumination implementation (SPIM), which uses a cylindrical lens to generate a static light sheet.
Stelzer, E. H. K., Enders, S., Huisken, J., Lindek, S. & Swoger, J. H. Microscope with a viewing direction perpendicular to the illumination direction. US Patent 7554725 B2 (2009).
Engelbrecht, C. J. & Stelzer, E. H. Resolution enhancement in a light-sheet-based microscope (SPIM). Opt. Lett. 31, 1477–1479 (2006).
Verveer, P. J. et al. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat. Methods 4, 311–313 (2007).
Swoger, J., Verveer, P., Greger, K., Huisken, J. & Stelzer, E. H. K. K. Multi-view image fusion improves resolution in three-dimensional microscopy. Opt. Express 15, 8029–8042 (2007).
Wohland, T., Shi, X., Sankaran, J. & Stelzer, E. H. K. Single plane illumination fluorescence correlation spectroscopy (SPIM-FCS) probes inhomogeneous three-dimensional environments. Opt. Express 18, 10627–10641 (2010).
Greger, K., Neetz, M. J., Reynaud, E. G. & Stelzer, E. H. K. Three-dimensional fluorescence lifetime imaging with a single plane illumination microscope provides an improved signal to noise ratio. Opt. Express 19, 20743 (2011).
Method of the Year 2014. Nat. Methods 12, 1 (2015).
Fahrbach, F. O., Voigt, F. F., Schmid, B., Helmchen, F. & Huisken, J. Rapid 3D light-sheet microscopy with a tunable lens. Opt. Express 21, 21010–21026 (2013).
Keller, P. J., Schmidt, A. D., Wittbrodt, J. & Stelzer, E. H. K. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008). This work presents the first digital scanned laser light sheet-based fluorescence implementation (DSLM), which used beam scanning to generate a dynamic light sheet.
Keller, P. J. & Stelzer, E. H. K. Digital scanned laser light sheet fluorescence microscopy. Cold Spring Harb. Protoc. 2010, pdb.top78 (2010).
Khonina, S. N., Kazanskiy, N. L., Karpeev, S. V. & Butt, M. A. Bessel beam: significance and applications — a progressive review. Micromachines 11, 997 (2020).
Efremidis, N. K., Chen, Z., Segev, M. & Christodoulides, D. N. Airy beams and accelerating waves: an overview of recent advances. Optica 6, 686 (2019).
Stelzer, E. H. K. Contrast, resolution, pixelation, dynamic range and signal-to-noise ratio: fundamental limits to resolution in fluorescence light microscopy. J. Microsc. https://doi.org/10.1046/j.1365-2818.1998.00290.x (1998).
Olarte, O. E., Andilla, J., Gualda, E. J. & Loza-Alvarez, P. Light-sheet microscopy: a tutorial. Adv. Opt. Photonics https://doi.org/10.1364/aop.10.000111 (2018).
Krzic, U., Gunther, S., Saunders, T. E., Streichan, S. J. & Hufnagel, L. Multiview light-sheet microscope for rapid in toto imaging. Nat. Methods 9, 730–733 (2012).
Preibisch, S. et al. Efficient Bayesian-based multiview deconvolution. Nat. Methods 11, 645–648 (2014).
Huisken, J. & Stainier, D. Y. R. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt. Lett. 32, 2608–2610 (2007).
Tomer, R., Khairy, K., Amat, F. & Keller, P. J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9, 755–763 (2012).
Chhetri, R. K. et al. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat. Methods 12, 1171–1178 (2015).
Wu, Y. et al. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1108494108 (2011).
Wu, Y. et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat. Biotechnol. https://doi.org/10.1038/nbt.2713 (2013).
Wu, Y. et al. Reflective imaging improves spatiotemporal resolution and collection efficiency in light sheet microscopy. Nat. Commun. https://doi.org/10.1038/s41467-017-01250-8 (2017).
McGorty, R. et al. Open-top selective plane illumination microscope for conventionally mounted specimens. Opt. Express 23, 16142–16153 (2015).
Mcgorty, R., Xie, D. & Huang, B. High-NA open-top selective-plane illumination microscopy for biological imaging. Opt. Express https://doi.org/10.1364/oe.25.017798 (2017).
Glaser, A. K. et al. Multi-immersion open-top light-sheet microscope for high-throughput imaging of cleared tissues. Nat. Commun. 10, 2781 (2019).
Gualda, E. J. et al. SPIM-fluid: open source light-sheet based platform for high-throughput imaging. Biomed. Opt. Express 6, 4447 (2015).
Wu, J., Li, J. & Chan, R. K. Y. Y. A light sheet based high throughput 3D- imaging flow cytometer for phytoplankton analysis. Opt. Express 21, 14474–14480 (2013).
Paiè, P., Bragheri, F., Bassi, A. & Osellame, R. Selective plane illumination microscopy on a chip. Lab. Chip https://doi.org/10.1039/c6lc00084c (2016).
Sala, F. et al. High-throughput 3D imaging of single cells with light-sheet fluorescence microscopy on chip. Biomed. Opt. Express https://doi.org/10.1364/boe.393892 (2020).
Yang, B. et al. Epi-illumination SPIM for volumetric imaging with high spatial-temporal resolution. Nat. Methods 16, 501–504 (2019). This work presents a novel oblique plane microscopy design that enables the collection of fluorescence emissions in high NA to allow for high spatio-temporal resolution and is compatible with common biological sample holders, including multiwell plates.
Dunsby, C. Optically sectioned imaging by oblique plane microscopy. Opt. Express 16, 20306 (2008). This work describes the first oblique light sheet-based microscope, which combines selective plane illumination with oblique imaging by using only one objective.
Bouchard, M. B. et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat. Photonics https://doi.org/10.1038/nphoton.2014.323 (2015).
Voleti, V. et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat. Methods https://doi.org/10.1038/s41592-019-0579-4 (2019).
Kumar, M., Kishore, S., Nasenbeny, J., Mclean, D. L. & Kozorovitskiy, Y. Integrated one- and two-photon scanned oblique plane illumination (SOPi) microscopy for rapid volumetric imaging. Opt. Express https://doi.org/10.1364/OE.26.013027 (2018).
Sapoznik, E. et al. A versatile oblique plane microscope for large-scale and high-resolution imaging of subcellular dynamics. eLife https://doi.org/10.7554/eLife.57681 (2020).
Maioli, V. et al. Time-lapse 3-D measurements of a glucose biosensor in multicellular spheroids by light sheet fluorescence microscopy in commercial 96-well plates. Sci. Rep. https://doi.org/10.1038/srep37777 (2016).
Vaadia, R. D. et al. Characterization of proprioceptive system dynamics in behaving Drosophila larvae using high-speed volumetric microscopy. Curr. Biol. https://doi.org/10.1016/j.cub.2019.01.060 (2019).
Gebhardt, J. C. M. et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat. Methods https://doi.org/10.1038/nmeth.2411 (2013).
Plöschner, M. et al. Multimode fibre: light-sheet microscopy at the tip of a needle. Sci. Rep. https://doi.org/10.1038/srep18050 (2015).
Greiss, F., Deligiannaki, M., Jung, C., Gaul, U. & Braun, D. Single-molecule imaging in living Drosophila embryos with reflected light-Sseet microscopy. Biophys. J. https://doi.org/10.1016/j.bpj.2015.12.035 (2016).
Zagato, E. et al. Microfabricated devices for single objective single plane illumination microscopy (SoSPIM). Opt. Express https://doi.org/10.1364/oe.25.001732 (2017).
Galland, R. et al. 3D high-and super-resolution imaging using single-objective SPIM. Nat. Methods https://doi.org/10.1038/nmeth.3402 (2015).
Meddens, M. B. M. et al. Single objective light-sheet microscopy for high-speed whole-cell 3D super-resolution. Biomed. Opt. Express https://doi.org/10.1364/boe.7.002219 (2016).
Pitrone, P. G. et al. OpenSPIM: an open-access light-sheet microscopy platform. Nat. Methods 10, 598–599 (2013).
Gualda, E. J. et al. OpenSpinMicroscopy: an open-source integrated microscopy platform. Nat. Methods 10, 599–600 (2013).
Stuurman, N., Amdodaj, N. & Vale, R. μManager: open source software for light microscope imaging. Micros. Today https://doi.org/10.1017/s1551929500055541 (2007).
Edelstein, A., Amodaj, N., Hoover, K., Vale, R. & Stuurman, N. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. 92, 14.20.1–14.20.17 (2010).
Edelstein, A. D. et al. Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10 (2014).
Saska, D., Pichler, P., Qian, C., Buckley, C. L. & Lagnado, L. μSPIM Toolset: a software platform for selective plane illumination microscopy. J. Neurosci. Methods https://doi.org/10.1016/j.jneumeth.2020.108952 (2021).
Pinkard, H. et al. Pycro-Manager: open-source software for customized and reproducible microscope control. Nat. Methods https://doi.org/10.1038/s41592-021-01087-6 (2021).
Alamudi, S. H. & Chang, Y. T. Advances in the design of cell-permeable fluorescent probes for applications in live cell imaging. Chem. Commun. 54, 13641–13653 (2018).
Shaner, N. C., Steinbach, P. A. & Tsien, R. Y. A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909 (2005).
Shcherbakova, D. M. et al. Molecular basis of spectral diversity in near-infrared phytochrome-based fluorescent proteins. Chem. Biol. 22, 1540–1551 (2015).
Tran, M. T. N. et al. In vivo image analysis using iRFP transgenic mice. Exp. Anim. 63, 311–319 (2014).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. https://doi.org/10.1038/nbt765 (2003).
Los, G. V. et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008).
Grimm, J. B. et al. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat. Methods https://doi.org/10.1038/nmeth.4403 (2017).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature https://doi.org/10.1038/nature12354 (2013).
Abdelfattah, A. S. et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science https://doi.org/10.1126/science.aav6416 (2019).
Greenwald, E. C., Mehta, S. & Zhang, J. Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 118, 11707–11794 (2018).
Arai, S. et al. RGB-color intensiometric indicators to visualize spatiotemporal dynamics of ATP in single cells. Angew. Chem. Int. Ed. https://doi.org/10.1002/anie.201804304 (2018).
Lindenburg, L. H., Hessels, A. M., Ebberink, E. H. T. M., Arts, R. & Merkx, M. Robust red FRET sensors using self-associating fluorescent domains. ACS Chem. Biol. https://doi.org/10.1021/cb400427b (2013).
Liau, E. S., Yen, Y. P. & Chen, J. A. Visualization of motor axon navigation and quantification of axon arborization in mouse embryos using light sheet fluorescence microscopy. J. Vis. Exp. https://doi.org/10.3791/57546 (2018).
Silvestri, L. et al. Micron-scale resolution optical tomography of entire mouse brains with confocal light sheet microscopy. J. Vis. Exp. https://doi.org/10.3791/50696 (2013).
Ding, Y. et al. Light-sheet fluorescence microscopy for the study of the murine heart. J. Vis. Exp. https://doi.org/10.3791/57769 (2018).
Weber, M., Mickoleit, M. & Huisken, J. Multilayer mounting for long-term light sheet microscopy of zebrafish. J. Vis. Exp. https://doi.org/10.3791/51119 (2014).
Icha, J. et al. Using light sheet fluorescence microscopy to image zebrafish eye development. J. Vis. Exp. https://doi.org/10.3791/53966 (2016).
Lee, J. et al. Light-sheet fluorescence microscopy to capture 4-dimensional images of the effects of modulating shear stress on the developing zebrafish heart. J. Vis. Exp. https://doi.org/10.3791/57763 (2018).
Chardès, C., Mélénec, P., Bertrand, V. & Lenne, P. F. Setting up a simple light sheet microscope for in toto imaging of C. elegans development. J. Vis. Exp. https://doi.org/10.3791/51342 (2014).
Duncan, L. H. et al. Isotropic light-sheet microscopy and automated cell lineage analyses to catalogue Caenorhabditis elegans embryogenesis with subcellular resolution. J. Vis. Exp. 2019, 59533 (2019).
Strobl, F., Klees, S. & Stelzer, E. H. K. Light sheet-based fluorescence microscopy of living or fixed and stained Tribolium castaneum embryos. J. Vis. Exp. https://doi.org/10.3791/55629 (2017).
Ratke, J., Krämer, F. & Strobl, F. Simultaneous live imaging of multiple insect embryos in sample chamber-based light sheet fluorescence microscopes. J. Vis. Exp. https://doi.org/10.3791/61713 (2020).
von Wangenheim, D., Hauschild, R. & Friml, J. Light sheet fluorescence microscopy of plant roots growing on the surface of a gel. J. Vis. Exp. https://doi.org/10.3791/55044 (2017).
Jacob, L., Brito, J. & Thomas, J. L. Three-dimensional imaging of the vertebral lymphatic vasculature and drainage using iDISCO+ and light sheet fluorescence microscopy. J. Vis. Exp. https://doi.org/10.3791/61099 (2020).
Schoppmeyer, R., Zhao, R., Hoth, M. & Qu, B. Light-sheet microscopy for three-dimensional visualization of human immune cells. J. Vis. Exp. https://doi.org/10.3791/57651 (2018).
Rosenberg, J. & Huang, J. Visualizing surface T-cell receptor dynamics four-dimensionally using lattice light-sheet microscopy. J. Vis. Exp. https://doi.org/10.3791/59914 (2019).
Keller, P. J., Schmidt, A. D., Wittbrodt, J. & Stelzer, E. H. K. Digital scanned laser light-sheet fluorescence microscopy (DSLM) of zebrafish and Drosophila embryonic development. Cold Spring Harb. Protoc. 2011, 1235–1243 (2011).
Schmied, C. & Tomancak, P. Sample preparation and mounting of Drosophila embryos for multiview light sheet microscopy. in. Methods Mol. Biol. 1478, 189–202 (2016).
Kaufmann, A., Mickoleit, M., Weber, M. & Huisken, J. Multilayer mounting enables long-term imaging of zebrafish development in a light sheet microscope. Development 139, 3242–3247 (2012). This work describes a method that allows for stacking multiple zebrafish embryos into one sample holder for multi-embryo imaging within a single experiment.
Uribe, V. et al. In vivo analysis of cardiomyocyte proliferation during trabeculation. Development https://doi.org/10.1242/dev.164194 (2018).
de Medeiros, G., Balázs, B. & Hufnagel, L. Light-sheet imaging of mammalian development. Semin. Cell Dev. Biol. https://doi.org/10.1016/j.semcdb.2015.11.001 (2016).
Reichmann, J., Eguren, M., Lin, Y., Schneider, I. & Ellenberg, J. Live imaging of cell division in preimplantation mouse embryos using inverted light-sheet microscopy. Methods Cell Biol. https://doi.org/10.1016/bs.mcb.2018.03.030 (2018).
Ichikawa, T. et al. Live imaging of whole mouse embryos during gastrulation: migration analyses of epiblast and mesodermal cells. PLoS ONE 8, e64506 (2013).
Ichikawa, T. et al. Live imaging and quantitative analysis of gastrulation in mouse embryos using light-sheet microscopy and 3D tracking tools. Nat. Protoc. 9, 575–585 (2014).
McDole, K. et al. In toto imaging and reconstruction of post-implantation mouse development at the single-cell level. Cell https://doi.org/10.1016/j.cell.2018.09.031 (2018). This work demonstrates the use of light sheet microscopy for imaging the developing mouse embryo over the course of 2 days from gastrulation to early organogenesis, and reconstructs dynamic, lineage-based cell fate maps.
Pampaloni, F., Ansari, N. & Stelzer, E. H. K. High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res. https://doi.org/10.1007/s00441-013-1589-7 (2013).
Hötte, K. et al. Ultra-thin fluorocarbon foils optimise multiscale imaging of three-dimensional native and optically cleared specimens. Sci. Rep. https://doi.org/10.1038/s41598-019-53380-2 (2019).
Pampaloni, F. et al. Tissue-culture light sheet fluorescence microscopy (TC-LSFM) allows long-term imaging of three-dimensional cell cultures under controlled conditions. Integr. Biol. https://doi.org/10.1039/c4ib00121d (2014).
Flood, P., Page, H. & Reynaud, E. G. Using hydrogels in microscopy: a tutorial. Micron https://doi.org/10.1016/j.micron.2016.02.002 (2016).
Ettinger, A. & Wittmann, T. Fluorescence live cell imaging. Methods Cell Biol. https://doi.org/10.1016/B978-0-12-420138-5.00005-7 (2014).
Von Wangenheim, D., Daum, G., Lohmann, J. U., Stelzer, E. K. & Maizel, A. Live imaging of Arabidopsis development. Methods Mol. Biol. https://doi.org/10.1007/978-1-62703-580-4_28 (2014).
Ovecka, M. et al. Preparation of plants for developmental and cellular imaging by light-sheet microscopy. Nat. Protoc. https://doi.org/10.1038/nprot.2015.081 (2015).
Strobl, F., Schmitz, A. & Stelzer, E. H. K. Live imaging of Tribolium castaneum embryonic development using light-sheet-based fluorescence microscopy. Nat. Protoc. 10, 1486–1507 (2015). This work describes a two-step calibration routine for light sheet-based microscopes (primarily DSLM-based set-ups) with a comprehensively illustrated troubleshooting guide.
Gao, L. Extend the field of view of selective plan illumination microscopy by tiling the excitation light sheet. Opt. Express 23, 6102 (2015).
Yanlu, C. et al. A versatile tiling light sheet microscope for imaging of cleared tissues. Cell Rep. 33, 108349 (2020).
Sancataldo, G. et al. Flexible multi-beam light-sheet fluorescence microscope for live imaging without striping artifacts. Front. Neuroanat. https://doi.org/10.3389/fnana.2019.00007 (2019).
Fahrbach, F. O. & Rohrbach, A. A line scanned light-sheet microscope with phase shaped self-reconstructing beams. Opt. Express 18, 24229 (2010). This work proposes the first implementation of LSFM with Bessel beams, which in principle can avoid some of the trade-offs of conventional Gaussian beams.
Fahrbach, F. O. & Rohrbach, A. Propagation stability of self-reconstructing Bessel beams enables contrast-enhanced imaging in thick media. Nat. Commun. 3, 632 (2012).
Müllenbroich, M. C. et al. Bessel beam illumination reduces random and systematic errors in quantitative functional studies using light-sheet microscopy. Front. Cell. Neurosci. https://doi.org/10.3389/fncel.2018.00315 (2018).
Salili, S. M., Harrington, M. & Durian, D. J. Note: Eliminating stripe artifacts in light-sheet fluorescence imaging. Rev. Sci. Instrum. https://doi.org/10.1063/1.5016546 (2018).
Liang, X. et al. Stripe artifact elimination based on nonsubsampled contourlet transform for light sheet fluorescence microscopy. J. Biomed. Opt. https://doi.org/10.1117/1.jbo.21.10.106005 (2016).
Keller, P. J. et al. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat. Methods 7, 637–642 (2010).
Neil, M. A. A., Juškaitis, R. & Wilson, T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt. Lett. https://doi.org/10.1364/ol.22.001905 (1997).
Silvestri, L., Bria, A., Sacconi, L., Iannello, G. & Pavone, F. S. Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain. Opt. Express 20, 20582 (2012).
Baumgart, E. & Kubitscheck, U. Scanned light sheet microscopy with confocal slit detection. Opt. Express 20, 21805–21814 (2012).
Medeiros, G. D. et al. Confocal multiview light-sheet microscopy. Nat. Commun. https://doi.org/10.1038/ncomms9881 (2015).
Gavryusev, V. et al. Dual-beam confocal light-sheet microscopy via flexible acousto-optic deflector. J. Biomed. Opt. https://doi.org/10.1117/1.jbo.24.10.106504 (2019).
Jahr, W., Schmid, B., Schmied, C., Fahrbach, F. O. & Huisken, J. Hyperspectral light sheet microscopy. Nat. Commun. 6, 7990 (2015).
Lavagnino, Z. et al. Snapshot hyperspectral light-sheet imaging of signal transduction in live pancreatic islets. Biophys. J. https://doi.org/10.1016/j.bpj.2016.06.014 (2016).
Jones, R. R., Hooper, D. C., Zhang, L., Wolverson, D. & Valev, V. K. Raman techniques: fundamentals and frontiers. Nanoscale Res. Lett. 14, 1–34 (2019).
Rocha-Mendoza, I. et al. Rapid spontaneous Raman light sheet microscopy using cw-lasers and tunable filters. Biomed. Opt. Express https://doi.org/10.1364/boe.6.003449 (2015).
Müller, W., Kielhorn, M., Schmitt, M., Popp, J. & Heintzmann, R. Light sheet Raman micro-spectroscopy. Optica https://doi.org/10.1364/optica.3.000452 (2016).
Yu, L. et al. A comprehensive review of fluorescence correlation spectroscopy. Front. Phys. 9, 110 (2021).
Capoulade, J., Wachsmuth, M., Hufnagel, L. & Knop, M. Quantitative fluorescence imaging of protein diffusion and interaction in living cells. Nat. Biotechnol. https://doi.org/10.1038/nbt.1928 (2011).
Struntz, P. & Weiss, M. Multiplexed measurement of protein diffusion in Caenorhabditis elegans embryos with SPIM-FCS. J. Phys. D. Appl. Phys. https://doi.org/10.1088/0022-3727/49/4/044002 (2015).
Singh, A. P. et al. 3D protein dynamics in the cell nucleus. Biophys. J. https://doi.org/10.1016/j.bpj.2016.11.3196 (2017).
Krieger, J. W., Singh, A. P., Garbe, C. S., Wohland, T. & Langowski, J. Dual-color fluorescence cross-correlation spectroscopy on a single plane illumination microscope (SPIM-FCCS). Opt. Express https://doi.org/10.1364/oe.22.002358 (2014).
Krieger, J. W. et al. Imaging fluorescence (cross-)correlation spectroscopy in live cells and organisms. Nat. Protoc. https://doi.org/10.1038/nprot.2015.100 (2015).
Buchholz, J. et al. Widefield high frame rate single-photon SPAD imagers for SPIM-FCS. Biophys. J. https://doi.org/10.1016/j.bpj.2018.04.029 (2018).
Datta, R., Heaster, T. M., Sharick, J. T., Gillette, A. A. & Skala, M. C. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt. 25, 1 (2020).
Mitchell, C. A. et al. Functional in vivo imaging using fluorescence lifetime light-sheet microscopy. Opt. Lett. https://doi.org/10.1364/ol.42.001269 (2017).
Ulku, A. et al. Wide-field time-gated SPAD imager for phasor-based FLIM applications. Methods Appl. Fluoresc. https://doi.org/10.1088/2050-6120/ab6ed7 (2020).
Oleksiievets, N. et al. Wide-field fluorescence lifetime imaging of single molecules. J. Phys. Chem. A https://doi.org/10.1021/acs.jpca.0c01513 (2020).
Reynaud, E. G., Peychl, J., Huisken, J. & Tomancak, P. Guide to light-sheet microscopy for adventurous biologists. Nat. Methods 12, 30–34 (2015).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Schindelin, J., Rueden, C. T., Hiner, M. C. & Eliceiri, K. W. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. https://doi.org/10.1002/mrd.22489 (2015).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics https://doi.org/10.1186/s12859-017-1934-z (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Amat, F. et al. Efficient processing and analysis of large-scale light-sheet microscopy data. Nat. Protoc. 10, 1679–1696 (2015). This work provides a comprehensive protocol for the processing and analysis of light sheet-based data in the terabyte range, including advice for data compression, multi-view fusion automated cell tracking and visualization.
Huisman, M. et al. A perspective on microscopy metadata: data provenance and quality control. Preprint at https://arxiv.org/abs/1910.11370 (2019).
Gao, R. et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science https://doi.org/10.1126/science.aau8302 (2019).
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009).
Hörl, D. et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods https://doi.org/10.1038/s41592-019-0501-0 (2019).
Guo, M. et al. Rapid image deconvolution and multiview fusion for optical microscopy. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0560-x (2020).
Preibisch, S., Saalfeld, S., Schindelin, J. & Tomancak, P. Software for bead-based registration of selective plane illumination microscopy data. Nat. Methods 7, 418–419 (2010).
Wallace, W., Schaefer, L. H. & Swedlow, J. R. A workingperson’s guide to deconvolution in light microscopy. Biotechniques 31, 1076–1097 (2001).
Becker, K. et al. Deconvolution of light sheet microscopy recordings. Sci. Rep. https://doi.org/10.1038/s41598-019-53875-y (2019).
Verveer, P. J. et al. Restoration of light sheet multi-view data with the huygens fusion and deconvolution wizard. Micros. Today https://doi.org/10.1017/s1551929518000846 (2018).
Long, F., Zhou, J. & Peng, H. Visualization and analysis of 3D microscopic images. PLoS Comput. Biol. 8, e1002519 (2012).
Pietzsch, T., Saalfeld, S., Preibisch, S. & Tomancak, P. BigDataViewer: visualization and processing for large image data sets. Nat. Methods 12, 481–483 (2015).
Pietzsch, T., Preibisch, S., Tomančák, P. & Saalfeld, S. Img lib 2-generic image processing in Java. Bioinformatics https://doi.org/10.1093/bioinformatics/bts543 (2012).
Preusser, F. et al. FRC-QE: a robust and comparable 3D microscopy image quality metric for cleared organoids. Bioinformatics https://doi.org/10.1093/bioinformatics/btab160 (2021).
Peng, H., Ruan, Z., Long, F., Simpson, J. H. & Myers, E. W. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. https://doi.org/10.1038/nbt.1612 (2010).
Bria, A. & Iannello, G. TeraStitcher — a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinformatics https://doi.org/10.1186/1471-2105-13-316 (2012).
Royer, L. A. et al. ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nat. Methods 12, 480–481 (2015).
Schmid, B. et al. 3Dscript: animating 3D/4D microscopy data using a natural-language-based syntax. Nat. Methods https://doi.org/10.1038/s41592-019-0359-1 (2019).
Bria, A., Iannello, G., Onofri, L. & Peng, H. TeraFly: real-time three-dimensional visualization and annotation of terabytes of multidimensional volumetric images. Nat. Methods https://doi.org/10.1038/nmeth.3767 (2016).
Günther U. et al. Scenery: flexible virtual reality visualization on the Java VM. 2019 IEEE Visualization Conference (VIS) 2019, 1–5, https://doi.org/10.1109/VISUAL.2019.8933605 (2019).
Pettersen, E. F. et al. UCSF Chimera — a visualization system for exploratory research and analysis. J. Comput. Chem. https://doi.org/10.1002/jcc.20084 (2004).
Fritz-Laylin, L. K. et al. Actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. eLife https://doi.org/10.7554/eLife.26990 (2017).
Cheeseman, B. L., Günther, U., Gonciarz, K., Susik, M. & Sbalzarini, I. F. Adaptive particle representation of fluorescence microscopy images. Nat. Commun. https://doi.org/10.1038/s41467-018-07390-9 (2018).
Vladimirov, N. npy2bdv: writing numpy arrays to Fiji/BigDataViewer HDF5 files. ZENODO https://doi.org/10.5281/ZENODO.3971783 (2020).
Beati, I., Andreica, E. & Majer, P. ImarisWriter: open source software for storage of large images in blockwise multi-resolution format. Preprint at https://arxiv.org/abs/2008.10311 (2020).
Balázs, B., Deschamps, J., Albert, M., Ries, J. & Hufnagel, L. A real-time compression library for microscopy images. Preprint at bioRxiv https://doi.org/10.1101/164624 (2017).
Wolff, C. et al. Multi-view light-sheet imaging and tracking with the MaMuT software reveals the cell lineage of a direct developing arthropod limb. eLife https://doi.org/10.7554/eLife.34410 (2018). This work demonstrates the tracking of individual lineages in developing Parhyale limbs using multi-view light-sheet microscopy and an open-source Fiji-based tracking plug-in called MaMuT.
Amat, F. et al. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods 11, 951–958 (2014).
Maling-Mayor, C. et al. Automated reconstruction of whole-embryo cell lineages by learning from sparse annotations. Preprint at bioRxiv https://doi.org/10.1101/2021.07.28.454016 (2021).
Haase, R. et al. CLIJ: GPU-accelerated image processing for everyone. Nat. Methods https://doi.org/10.1038/s41592-019-0650-1 (2020).
Haase, R. et al. Interactive design of GPU-accelerated image data flow graphs and cross-platform deployment using multi-lingual code generation. Preprint at bioRxiv https://doi.org/10.1101/2020.11.19.386565 (2020).
Belthangady, C. & Royer, L. A. Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Nat. Methods https://doi.org/10.1038/s41592-019-0458-z (2019).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods https://doi.org/10.1038/s41592-019-0582-9 (2019).
Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with star-convex polygons. Lecture Notes Comput. Sci. https://doi.org/10.1007/978-3-030-00934-2_30 (2018).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods https://doi.org/10.1038/s41592-020-01018-x (2021).
Sugawara, K., Cevrim, C. & Averof, M. Tracking cell lineages in 3D by incremental deep learning. Preprint at bioRxiv https://doi.org/10.1101/2021.02.26.432552 (2021).
Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods https://doi.org/10.1038/s41592-018-0216-7 (2018).
Wagner, A., Regev, A. & Yosef, N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. https://doi.org/10.1038/nbt.3711 (2016).
Waschke, J. et al. linus: Conveniently explore, share, and present large-scale biological trajectory data from a web browser. Preprint at bioRxiv https://doi.org/10.1101/2020.04.17.043323 (2021).
Pampaloni, F., Knuppertz, L., Hamann, A., Osiewacz, H. D. & Stelzer, E. H. K. Three-dimensional live imaging of filamentous fungi with light sheet-based fluorescence microscopy (LSFM). Methods Mol. Biol. 1563, 19–31 (2017).
Amich, J. et al. Three-dimensional light sheet fluorescence microscopy of lungs to dissect local host immune-aspergillus fumigatus interactions. mBio https://doi.org/10.1128/mBio.02752-19 (2020).
Qin, B. et al. Cell position fates and collective fountain flow in bacterial biofilms revealed by light-sheet microscopy. Science https://doi.org/10.1126/science.abb8501 (2020).
Zhang, M. et al. Non-invasive single-cell morphometry in living bacterial biofilms. Nat. Commun. 11, 6151 (2020).
Bhagwat, A. R., Le Sage, V. & Lakdawala, S. S. Live imaging of influenza viral ribonucleoproteins using light-sheet microscopy. Methods Mol. Biol. 1836, 303–327 (2018).
Mascheroni, L. et al. Combining sample expansion and light sheet microscopy for the volumetric imaging of virus-infected cells with super-resolution. Biomed. Opt. Express https://doi.org/10.1364/boe.399404 (2020).
Liao, P. et al. Three-dimensional digital PCR through light-sheet imaging of optically cleared emulsion. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.2002448117 (2020).
Attardi, A. et al. Neuromesodermal progenitors are a conserved source of spinal cord with divergent growth dynamics. Development https://doi.org/10.1242/dev.166728 (2018).
Wan, Y. et al. Single-cell reconstruction of emerging population activity in an entire developing circuit. Cell https://doi.org/10.1016/j.cell.2019.08.039 (2019).
Daetwyler, S., Gunther, U., Modes, C. D., Harrington, K. & Huisken, J. Multi-sample SPIM image acquisition, processing and analysis of vascular growth in zebrafish. Development https://doi.org/10.1242/dev.173757 (2019).
Wan, Y. et al. Single-cell reconstruction of emerging population activity in an entire developing circuit. Cell 179, 355–372.e23 (2019).
Rozbicki, E. et al. Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nat. Cell Biol. 17, 397–408 (2015).
Goḿez-Gaviro, M. V. et al. Optimized CUBIC protocol for three-dimensional imaging of chicken embryos at single-cell resolution. Development https://doi.org/10.1242/dev.145805 (2017).
Strnad, P. et al. Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat. Methods 13, 139–142 (2016).
Wu, C. et al. Comparison and combination of rotational imaging optical coherence tomography and selective plane illumination microscopy for embryonic study. Biomed. Opt. Express https://doi.org/10.1364/boe.8.004629 (2017).
Fu, Q., Martin, B. L., Matus, D. Q. & Gao, L. Imaging multicellular specimens with real-time optimized tiling light-sheet selective plane illumination microscopy. Nat. Commun. 7, 1–10 (2016).
Chen, B.-C. B. C. et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998–1–1257998–13 (2014). This work introduces lattice light sheets, where multiple Bessel beams interfere coherently to tailor the properties of a light sheet, resulting in high-resolution 3D imaging and reduced phototoxicity compared with line-scanned Bessel beams.
Stegmaier, J. et al. Real-time three-dimensional cell segmentation in large-scale microscopy data of developing embryos. Dev. Cell 36, 225–240 (2016).
Streichan, S. J., Lefebvre, M. F., Noll, N., Wieschaus, E. F. & Shraiman, B. I. Global morphogenetic flow is accurately predicted by the spatial distribution of myosin motors. eLife https://doi.org/10.7554/eLife.27454 (2018).
Strobl, F. & Stelzer, E. H. K. Non-invasive long-term fluorescence live imaging of Tribolium castaneum embryos. Development 141, 2331–2338 (2014).
Münster, S. et al. Attachment of the blastoderm to the vitelline envelope affects gastrulation of insects. Nature https://doi.org/10.1038/s41586-019-1044-3 (2019).
Hilbrant, M., Horn, T., Koelzer, S. & Panfilio, K. A. The beetle amnion and serosa functionally interact as apposed epithelia. eLife 5, e13834 (2016).
Li, F. et al. Insect genomes: progress and challenges. Insect Mol. Biol. 28, 739–758 (2019).
Keller, P. J. J. & Ahrens, M. B. B. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 85, 462–483 (2015).
Hillman, E. M. C., Voleti, V., Li, W. & Yu, H. Light-sheet microscopy in neuroscience. Annu. Rev. Neurosci. https://doi.org/10.1146/annurev-neuro-070918-050357 (2019).
Simpson, J. H. & Looger, L. L. Functional imaging and optogenetics in Drosophila. Genetics https://doi.org/10.1534/genetics.117.300228 (2018).
Ahrens, M. B., Orger, M. B., Robson, D. N., Li, J. M. & Keller, P. J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10, 413–420 (2013).
Lemon, W. C. et al. Whole-central nervous system functional imaging in larval Drosophila. Nat. Commun. 6, 7924 (2015).
Arrenberg, A. B., Stainier, D. Y. R., Baier, H. & Huisken, J. Optogenetic control of cardiac function. Science 330, 971–974 (2010).
Mickoleit, M. et al. High-resolution reconstruction of the beating zebrafish heart. Nat. Methods 11, 919–922 (2014).
Taylor, J. M. et al. Adaptive prospective optical gating enables day-long 3D time-lapse imaging of the beating embryonic zebrafish heart. Nat. Commun. https://doi.org/10.1038/s41467-019-13112-6 (2019).
Ding, Y. et al. Multiscale light-sheet for rapid imaging of cardiopulmonary system. JCI Insight https://doi.org/10.1172/jci.insight.121396 (2018).
Pampaloni, F., Reynaud, E. G. & Stelzer, E. H. K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 8, 839–845 (2007).
Simian, M. & Bissell, M. J. Organoids: a historical perspective of thinking in three dimensions. J. Cell Biol. https://doi.org/10.1083/jcb.201610056 (2017).
Lebreton, F. et al. Insulin-producing organoids engineered from islet and amniotic epithelial cells to treat diabetes. Nat. Commun. https://doi.org/10.1038/s41467-019-12472-3 (2019).
Lorenzo, C. et al. Live cell division dynamics monitoring in 3D large spheroid tumor models using light sheet microscopy. Cell Div. https://doi.org/10.1186/1747-1028-6-22 (2011).
Medeios, G. De et al. Multiscale light-sheet organoid imaging framework. Preprint at bioRxiv https://doi.org/10.1101/2021.05.12.443427 (2021).
Glaser, A. K. et al. Multidirectional digital scanned light-sheet microscopy enables uniform fluorescence excitation and contrast-enhanced imaging. Sci. Rep. https://doi.org/10.1038/s41598-018-32367-5 (2018).
Andilla, J. et al. Imaging tissue-mimic with light sheet microscopy: a comparative guideline. Sci. Rep. https://doi.org/10.1038/srep44939 (2017).
Schmitz, A., Fischer, S. C., Mattheyer, C., Pampaloni, F. & Stelzer, E. H. K. Multiscale image analysis reveals structural heterogeneity of the cell microenvironment in homotypic spheroids. Sci. Rep. https://doi.org/10.1038/srep43693 (2017).
Schöneberg, J. et al. 4D cell biology: big data image analytics and lattice light-sheet imaging reveal dynamics of clathrin-mediated endocytosis in stem cell–derived intestinal organoids. Mol. Biol. Cell https://doi.org/10.1091/mbc.E18-06-0375 (2018).
Dean, K. M. et al. Imaging subcellular dynamics with fast and light-efficient volumetrically parallelized microscopy. Optica https://doi.org/10.1364/optica.4.000263 (2017).
Chen, Y. et al. Rapid pathology of lumpectomy margins with open-top light-sheet (OTLS) microscopy. Biomed. Opt. Express https://doi.org/10.1364/boe.10.001257 (2019).
Glaser, A. K. et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-017-0084 (2017).
Maizel, A., Von Wangenheim, D., Federici, F., Haseloff, J. & Stelzer, E. H. K. High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant. J. 68, 377–385 (2011).
Ovečka, M. et al. Multiscale imaging of plant development by light-sheet fluorescence microscopy. Nat. Plants https://doi.org/10.1038/s41477-018-0238-2 (2018).
Sena, G., Frentz, Z., Birnbaum, K. D. & Leibler, S. Quantitation of cellular dynamics in growing Arabidopsis roots with light sheet microscopy. PLoS ONE https://doi.org/10.1371/journal.pone.0021303 (2011).
Von Wangenheim, D. et al. Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Curr. Biol. 26, 439–449 (2016).
Yan, J., Wang, B. & Zhou, Y. A root penetration model of Arabidopsis thaliana in phytagel medium with different strength. J. Plant. Res. https://doi.org/10.1007/s10265-017-0926-4 (2017).
Roué, J. et al. Root cap size and shape influence responses to the physical strength of the growth medium in Arabidopsis thaliana primary roots. J. Exp. Bot. https://doi.org/10.1093/jxb/erz418 (2020).
Candeo, A., Doccula, F. G., Valentini, G., Bassi, A. & Costa, A. Light sheet fluorescence microscopy quantifies calcium oscillations in root hairs of Arabidopsis thaliana. Plant. Cell Physiol. https://doi.org/10.1093/pcp/pcx045 (2017).
Valuchova, S. et al. Imaging plant germline differentiation within Arabidopsis flowers by light sheet microscopy. eLife https://doi.org/10.7554/eLife.52546 (2020).
Tichá, M. et al. Advanced microscopy reveals complex developmental and subcellular localization patterns of ANNEXIN 1 in Arabidopsis. Front. Plant. Sci. https://doi.org/10.3389/fpls.2020.01153 (2020).
Höhn, S., Honerkamp-Smith, A. R., Haas, P. A., Trong, P. K. & Goldstein, R. E. Dynamics of a Volvox embryo turning itself inside out. Phys. Rev. Lett. https://doi.org/10.1103/PhysRevLett.114.178101 (2015).
Lichtenberg, M., Trampe, E. C. L., Vogelmann, T. C. & Kühl, M. Light sheet microscopy imaging of light absorption and photosynthesis distribution in plant tissue. Plant. Physiol. https://doi.org/10.1104/pp.17.00820 (2017).
Truhaut, R. Ecotoxicology: objectives, principles and perspectives. Ecotoxicol. Environ. Saf. 1, 151–173 (1977).
Inglese, J. et al. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 3, 466–479 (2007).
Chandler, G. T. & Volz, D. C. Semiquantitative confocal laser scanning microscopy applied to marine invertebrate ecotoxicology. Mar. Biotechnol. 6, 128–137 (2004).
Nancharaiah, Y. V., Rajadurai, M. & Venugopalan, V. P. Single cell level microalgal ecotoxicity assessment by confocal microscopy and digital image analysis. Environ. Sci. Technol. 41, 2617–2621 (2007).
Scott, G. R. & Sloman, K. A. The effects of environmental pollutants on complex fish behaviour: integrating behavioural and physiological indicators of toxicity. Aquat. Toxicol. 68, 369–392 (2004).
Peterson, E. K. et al. Integrative behavioral ecotoxicology: bringing together fields to establish new insight to behavioral ecology, toxicology, and conservation. Curr. Zool. 63, 185 (2017).
Bae, M. J. & Park, Y. S. Biological early warning system based on the responses of aquatic organisms to disturbances: a review. Sci. Total. Environ. 466–467, 635–649 (2014).
Dell, A. I. et al. Automated image-based tracking and its application in ecology. Trends Ecol. Evol. https://doi.org/10.1016/j.tree.2014.05.004 (2014).
Cong, L. et al. Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio). eLife https://doi.org/10.7554/eLife.28158 (2017).
Mougi, A. The roles of amensalistic and commensalistic interactions in large ecological network stability. Sci. Rep. 6, 1–6 (2016).
Taormina, M. J. et al. Investigating bacterial–animal symbioses with light sheet microscopy. Biol. Bull. 223, 7–20 (2012).
Jemielita, M. et al. Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio 5, 1751–1765 (2014).
Wiles, T. J. et al. Host gut motility promotes competitive exclusion within a model intestinal microbiota. PLoS Biol. 14, e1002517 (2016).
Logan, S. L. et al. The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc. Natl Acad. Sci. USA 115, E3779–E3787 (2018).
Schlomann, B. H., Wiles, T. J., Wall, E. S., Guillemin, K. & Parthasarathy, R. Sublethal antibiotics collapse gut bacterial populations by enhancing aggregation and expulsion. Proc. Natl Acad. Sci. USA 116, 21392–21400 (2019).
Niz, M. D. et al. 3D imaging of undissected optically cleared Anopheles stephensi mosquitoes infected with Plasmodium parasites. PLoS ONE 15, e0238134 (2019).
Liu, C., Cheng, S. H. & Lin, S. Illuminating the dark depths inside coral. Cell. Microbiol. 22, e13122 (2020).
Hamill, P. Unit Test Frameworks: Tools for High-Quality Software Development (O’Reilly Media, 2004).
Marqués, G., Pengo, T. & Sanders, M. A. Imaging methods are vastly underreported in biomedical research. eLife 9, 1–10 (2020).
Wilkinson, M. D. et al. Comment: The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 1–9 (2016).
Linkert, M. et al. Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777–782 (2010).
Swedlow, J. R., Goldberg, I., Brauner, E. & Sorger, P. K. Informatics and quantitative analysis in biological imaging. Science 300, 100–102 (2003).
Allan, C. et al. OMERO: flexible, model-driven data management for experimental biology. Nat. Methods https://doi.org/10.1038/nmeth.1896 (2012).
Williams, E. et al. Image Data Resource: a bioimage data integration and publication platform. Nat. Methods https://doi.org/10.1038/nmeth.4326 (2017).
Sarkans, U. et al. The BioStudies database — one stop shop for all data supporting a life sciences study. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx965 (2018).
Saalfeld, S., Cardona, A., Hartenstein, V. & Tomančák, P. CATMAID: Collaborative Annotation Toolkit for Massive Amounts of Image Data. Bioinformatics https://doi.org/10.1093/bioinformatics/btp266 (2009).
Stelzer, E. H. K. Light-sheet fluorescence microscopy for quantitative biology. Nat. Methods 12, 23–26 (2015).
Dean, K. M., Roudot, P., Welf, E. S., Danuser, G. & Fiolka, R. Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophys. J. https://doi.org/10.1016/j.bpj.2015.05.013 (2015). This work presents ASLM, which can achieve 390-nm isotropic resolution and high optical sectioning strength over a large field of view by using a remote focusing technique.
Kim, B. et al. Open-top axially swept light-sheet microscopy. Biomed. Opt. Express https://doi.org/10.1364/boe.419030 (2021).
Voigt, F. F. et al. The mesoSPIM initiative: open-source light-sheet microscopes for imaging cleared tissue. Nat. Methods https://doi.org/10.1038/s41592-019-0554-0 (2019).
Chakraborty, T. et al. Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nat. Methods https://doi.org/10.1038/s41592-019-0615-4 (2019).
Planchon, T. A. et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods https://doi.org/10.1038/nmeth.1586 (2011).
Vettenburg, T. et al. Light-sheet microscopy using an Airy beam. Nat. Methods 11, 541–544 (2014).
Chang, B. J. et al. Universal light-sheet generation with field synthesis. Nat. Methods https://doi.org/10.1038/s41592-019-0327-9 (2019).
Remacha, E., Friedrich, L., Vermot, J. & Fahrbach, F. O. How to define and optimize axial resolution in light-sheet microscopy: a simulation-based approach. Biomed. Opt. Express https://doi.org/10.1364/boe.11.000008 (2020).
Tang, J. & Han, K. Y. Instantaneous non-diffracting light-sheet generation by controlling spatial coherence. Opt. Commun. https://doi.org/10.1016/j.optcom.2020.126154 (2020).
Gustafsson, M. G. L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. https://doi.org/10.1046/j.1365-2818.2000.00710.x (2000). This work introduces SIM, a method that can double the resolving power of a microscope.
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science https://doi.org/10.1126/science.1127344 (2006).
Hell, S. W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. https://doi.org/10.1364/ol.19.000780 (1994). This work presents STED microscopy, the first description of a super-resolution microscopy technique.
Klar, T. A., Jakobs, S., Dyba, M., Egner, A. & Hell, S. W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.97.15.8206 (2000).
Gustafsson, M. G. L. et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. https://doi.org/10.1529/biophysj.107.120345 (2008).
Chang, B. J., Meza, V. D. P. & Stelzer, E. H. K. csiLSFM combines light-sheet fluorescence microscopy and coherent structured illumination for a lateral resolution below 100 nm. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1609278114 (2017).
Huang, B., Jones, S. A., Brandenburg, B. & Zhuang, X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods https://doi.org/10.1038/nmeth.1274 (2008).
Cella Zanacchi, F. et al. Live-cell 3D super-resolution imaging in thick biological samples. Nat. Methods https://doi.org/10.1038/nmeth.1744 (2011).
Legant, W. R. et al. High-density three-dimensional localization microscopy across large volumes. Nat. Methods 13, 359–365 (2016).
Gustavsson, A. K., Petrov, P. N., Lee, M. Y., Shechtman, Y. & Moerner, W. E. 3D single-molecule super-resolution microscopy with a tilted light sheet. Nat. Commun. https://doi.org/10.1038/s41467-017-02563-4 (2018).
Kim, J. et al. Oblique-plane single-molecule localization microscopy for tissues and small intact animals. Nat. Methods https://doi.org/10.1038/s41592-019-0510-z (2019).
Friedrich, M., Gan, Q., Ermolayev, V. & Harms, G. S. STED-SPIM: stimulated emission depletion improves sheet illumination microscopy resolution. Biophys. J. 100, L43–L45 (2011).
Gohn-Kreuz, C. & Rohrbach, A. Light-sheet generation in inhomogeneous media using self-reconstructing beams and the STED-principle. Opt. Express https://doi.org/10.1364/oe.24.005855 (2016).
Hernández, J. M., Buisson, A., Wang, I. & Vial, J.-C. Improved optical slicing by stimulated emission depletion light sheet microscopy. Biomed. Opt. Express https://doi.org/10.1364/boe.379646 (2020).
Hoyer, P. et al. Breaking the diffraction limit of light-sheet fluorescence microscopy by RESOLFT. Proc. Natl Acad. Sci. USA 113, 3442–3446 (2016).
Richardson, D. S. S. & Lichtman, J. W. W. Clarifying tissue clearing. Cell 162, 246–257 (2015).
Ariel, P. A beginner’s guide to tissue clearing. Int. J. Biochem. Cell Biol. https://doi.org/10.1016/j.biocel.2016.12.009 (2017).
Genina, E. A., Bashkatov, A. N., Sinichkin, Y. P., Yanina, I. Y. & Tuchin, V. V. Optical clearing of biological tissues: prospects of application in medical diagnostics and phototherapy. J. Biomed. Photonics Eng. https://doi.org/10.18287/jbpe-2015-1-1-22 (2015).
Costa, E. C., Silva, D. N., Moreira, A. F. & Correia, I. J. Optical clearing methods: an overview of the techniques used for the imaging of 3D spheroids. Biotechnol. Bioeng. https://doi.org/10.1002/bit.27105 (2019).
Gómez-Gaviro, M. V., Sanderson, D., Ripoll, J. & Desco, M. Biomedical applications of tissue clearing and three-dimensional imaging in health and disease. iScience https://doi.org/10.1016/j.isci.2020.101432 (2020).
Costantini, I., Cicchi, R., Silvestri, L., Vanzi, F. & Pavone, F. S. In-vivo and ex-vivo optical clearing methods for biological tissues: review. Biomed. Opt. Express https://doi.org/10.1364/boe.10.005251 (2019).
Dekkers, J. F. et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. https://doi.org/10.1038/s41596-019-0160-8 (2019).
Azaripour, A. et al. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog. Histochem. Cytochem. 51, 9–23 (2016).
Dodt, H.-U. U. et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007).
Tomer, R. & Deisseroth, K. Rapid high-resolution brain mapping with CLARITY optimized light sheet microscopy (COLM). Microsc. Microanal. https://doi.org/10.1017/s1431927615004389 (2015).
Migliori, B. et al. Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol. https://doi.org/10.1186/s12915-018-0521-8 (2018).
Glaser, A. K., Bishop, K. W., Barner, L. A., Serafin, R. B. & Liu, J. T. C. A hybrid open-top light-sheet microscope for multi-scale imaging of cleared tissues. Preprint at bioRxiv https://doi.org/10.1101/2020.05.06.081745 (2021).
Mano, T. et al. Whole-brain analysis of cells and circuits by tissue clearing and light-sheet microscopy. J. Neurosci. https://doi.org/10.1523/JNEUROSCI.1677-18.2018 (2018).
Ueda, H. R. et al. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. https://doi.org/10.1038/s41583-019-0250-1 (2020).
Pan, C. et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods https://doi.org/10.1038/nmeth.3964 (2016).
Chung, K. & Deisseroth, K. CLARITY for mapping the nervous system. Nat. Methods https://doi.org/10.1038/nmeth.2481 (2013).
Chen, F., Tillberg, P. W. & Boyden, E. S. Expansion microscopy. Science https://doi.org/10.1126/science.1260088 (2015). This work introduces expansion microscopy, a super-resolution technique that is based on physically expanding the sample.
Chang, J. B. et al. Iterative expansion microscopy. Nat. Methods https://doi.org/10.1038/nmeth.4261 (2017).
Bridges, W. B. et al. Coherent optical adaptive techniques. Appl. Opt. https://doi.org/10.1364/ao.13.000291 (1974).
Buffington, A., Crawford, F. S., Muller, R. A., Schwemin, A. J. & Smits, R. G. Correction of atmospheric distortion with an image-sharpening telescope. J. Opt. Soc. Am. https://doi.org/10.1364/josa.67.000298 (1977).
Le Gargasson, J. F., Glanc, M. & Léna, P. Retinal imaging with adaptive optics. Comptes Rendus l’Academie des. Sci. IV Phys. Astrophys. 2, 1131–1138 (2001).
Booth, M. J. Adaptive optical microscopy: the ongoing quest for a perfect image. Light: Sci. Appl. https://doi.org/10.1038/lsa.2014.46 (2014).
Booth, M. J. Adaptive optics in microscopy. Philos. Trans. R. Soc. A: Math.Phys. Eng. Sci. 365, 2829–2843 (2007).
Dalgarno, H. I. C. et al. Wavefront corrected light sheet microscopy in turbid media. Appl. Phys. Lett. https://doi.org/10.1063/1.4710527 (2012).
Jorand, R. et al. Deep and clear optical imaging of thick inhomogeneous samples. PLoS ONE https://doi.org/10.1371/journal.pone.0035795 (2012).
Bourgenot, C., Saunter, C. D., Taylor, J. M., Girkin, J. M. & Love, G. D. 3D adaptive optics in a light sheet microscope. Opt. Express https://doi.org/10.1364/oe.20.013252 (2012).
Wilding, D., Pozzi, P., Soloviev, O., Vdovin, G. & Verhaegen, M. Adaptive illumination based on direct wavefront sensing in a light-sheet fluorescence microscope. Opt. Express https://doi.org/10.1364/oe.24.024896 (2016).
Liu, T. L. et al. Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science https://doi.org/10.1126/science.aaq1392 (2018).
Nishizaki, Y. et al. Deep learning wavefront sensing. Opt. Express https://doi.org/10.1364/oe.27.000240 (2019).
Saha, D. et al. Practical sensorless aberration estimation for 3D microscopy with deep learning. Opt. Express 28, 20738–26040 (2020).
Krishnan, A. P. et al. Optical aberration correction via phase diversity and deep learning. Preprint at bioRxiv https://doi.org/10.1101/2020.04.05.026567 (2020).
Masson, A. et al. High-resolution in-depth imaging of optically cleared thick samples using an adaptive SPIM. Sci. Rep. https://doi.org/10.1038/srep16898 (2015).
Benninger, R. K. P. & Piston, D. W. Two-photon excitation microscopy for the study of living cells and tissues. Curr. Protoc. Cell Biol. 59, 4.11.1–4.11.24 (2013).
Svoboda, K. & Yasuda, R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron https://doi.org/10.1016/j.neuron.2006.05.019 (2006).
Helmchen, F. & Denk, W. Deep tissue two-photon microscopy. Nat. Methods https://doi.org/10.1038/nmeth818 (2005).
Lavagnino, Z., Cella Zanacchi, F., Ronzitti, E. & Diaspro, A. Two-photon excitation selective plane illumination microscopy (2PE-SPIM) of highly scattering samples: characterization and application. Opt. Express 21, 5998 (2013).
Keller, P. J. & Dodt, H. U. Light sheet microscopy of living or cleared specimens. Curr. Opin. Neurobiol. https://doi.org/10.1016/j.conb.2011.08.003 (2012).
Lavagnino, Z. et al. 4D (x–y–z–t) imaging of thick biological samples by means of two-photon inverted selective plane illumination microscopy (2PE-iSPIM). Sci. Rep. https://doi.org/10.1038/srep23923 (2016).
Mahou, P., Vermot, J., Beaurepaire, E. & Supatto, W. Multicolor two-photon light-sheet microscopy. Nat. Methods https://doi.org/10.1038/nmeth.2963 (2014).
Cella Zanacchi, F., Lavagnino, Z., Faretta, M., Furia, L. & Diaspro, A. Light-sheet confined super-resolution using two-photon photoactivation. PLoS ONE https://doi.org/10.1371/journal.pone.0067667 (2013).
Welf, E. S. et al. Quantitative multiscale cell imaging in controlled 3D microenvironments. Dev. Cell https://doi.org/10.1016/j.devcel.2016.01.022 (2016).
Dean, K. M. & Fiolka, R. Lossless three-dimensional parallelization in digitally scanned light-sheet fluorescence microscopy. Sci. Rep. https://doi.org/10.1038/s41598-017-08113-8 (2017).
Wagner, N. et al. Instantaneous isotropic volumetric imaging of fast biological processes. Nat. Methods 16, 497–500 (2019).
Ren, Y. X. et al. Parallelized volumetric fluorescence microscopy with a reconfigurable coded incoherent light-sheet array. Light. Sci. Appl. https://doi.org/10.1038/s41377-020-0245-8 (2020).
Sheppard, C. J. R. et al. Pixel reassignment in image scanning microscopy: a re-evaluation. J. Opt. Soc. Am. A https://doi.org/10.1364/josaa.37.000154 (2020).
Müller, C. B. & Enderlein, J. Image scanning microscopy. Phys. Rev. Lett. https://doi.org/10.1103/PhysRevLett.104.198101 (2010).
Hoffman, D. P., Slavitt, I. & Fitzpatrick, C. A. The promise and peril of deep learning in microscopy. Nat. Methods https://doi.org/10.1038/s41592-020-01035-w (2021).
Scherf, N. & Huisken, J. The smart and gentle microscope. Nat. Biotechnol. 33, 815–818 (2015).
Royer, L. A. et al. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. https://doi.org/10.1038/nbt.3708 (2016). This work describes an automated framework (Autopilot) that integrates into multi-view light sheet-based devices in order to control the microscope’s various degrees of freedom to optimize spatial resolution within a sample and throughout time.
Power, R. M. & Huisken, J. Adaptable, illumination patterning light sheet microscopy. Sci. Rep. https://doi.org/10.1038/s41598-018-28036-2 (2018).
He, J. & Huisken, J. Image quality guided smart rotation improves coverage in microscopy. Nat. Commun. https://doi.org/10.1038/s41467-019-13821-y (2020).
Heinrich, L. et al. Automatic whole cell organelle segmentation in volumetric electron microscopy. Preprint at bioRxiv https://doi.org/10.1101/2020.11.14.382143 (2020).
Vergara, H. M. et al. Whole-body integration of gene expression and single-cell morphology. Cell https://doi.org/10.1016/j.cell.2021.07.017 (2021).
von Chamier, L. et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat. Commun. https://doi.org/10.1038/s41467-021-22518-0 (2021).
Heinrich, L. et al. Whole-cell organelle segmentation in volume electron microscopy. Nature https://doi.org/10.1038/s41586-021-03977-3 (2021).
Strobl, F., Schmitz, A. & Stelzer, E. H. K. Improving your four-dimensional image: traveling through a decade of light-sheet-based fluorescence microscopy research. Nat. Protoc. 12, 1103–1109 (2017).
Piccinini, F. et al. Advanced cell classifier: user-friendly machine-learning-based software for discovering phenotypes in high-content imaging data. Cell Syst. https://doi.org/10.1016/j.cels.2017.05.012 (2017).
Moen, E. et al. Deep learning for cellular image analysis. Nat. Methods https://doi.org/10.1038/s41592-019-0403-1 (2019).
Ladoux, B. & Mège, R. M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. https://doi.org/10.1038/nrm.2017.98 (2017).
Chatterjee, K., Pratiwi, F. W., Wu, F. C. M., Chen, P. & Chen, B. C. Recent progress in light sheet microscopy for biological applications. Appl. Spectrosc. https://doi.org/10.1177/0003702818778851 (2018).
Heddleston, J. M. & Chew, T. L. Light sheet microscopes: novel imaging toolbox for visualizing life’s processes. Int. J. Biochem. Cell Biol. https://doi.org/10.1016/j.biocel.2016.10.002 (2016).
Girkin, J. M. & Carvalho, M. T. The light-sheet microscopy revolution. J. Opt. https://doi.org/10.1088/2040-8986/aab58a (2018).
Elisa, Z. et al. Technical implementations of light sheet microscopy. Microscopy Res. Tech. https://doi.org/10.1002/jemt.22981 (2018).
Albert-Smet, I. et al. Applications of light-sheet microscopy in microdevices. Front. Neuroanatomy https://doi.org/10.3389/fnana.2019.00001 (2019).
Madrid-Wolff, J. & Forero-Shelton, M. Protocol for the design and assembly of a light sheet light field microscope. Methods Protoc. https://doi.org/10.3390/mps2030056 (2019).
De Vos, W. H. et al. Invited Review Article: Advanced light microscopy for biological space research. Rev. Sci. Instrum. https://doi.org/10.1063/1.4898123 (2014).
Daetwyler, S. & Huisken, J. Fast fluorescence microscopy with light sheets. Biol. Bull. 231, 14–25 (2016).
Gualda, E. J., Pereira, H., Martins, G. G., Gardner, R. & Moreno, N. Three-dimensional imaging flow cytometry through light-sheet fluorescence microscopy. Cytometry A https://doi.org/10.1002/cyto.a.23046 (2017).
Royer, L. A., Lemon, W. C., Chhetri, R. K. & Keller, P. J. A practical guide to adaptive light-sheet microscopy. Nat. Protoc. https://doi.org/10.1038/s41596-018-0043-4 (2018).
Lemon, W. C. & McDole, K. Live-cell imaging in the era of too many microscopes. Curr. Opin. Cell Biol. https://doi.org/10.1016/j.ceb.2020.04.008 (2020).
Hu, Y. S., Zimmerley, M., Li, Y., Watters, R. & Cang, H. Single-molecule super-resolution light-sheet microscopy. ChemPhysChem https://doi.org/10.1002/cphc.201300732 (2014).
Tang, J., Ren, J. & Han, K. Y. Fluorescence imaging with tailored light. Nanophotonics https://doi.org/10.1515/nanoph-2019-0227 (2019).
Amat, F. & Keller, P. J. Towards comprehensive cell lineage reconstructions in complex organisms using light-sheet microscopy. Dev. Growth Differ. 55, 563–578 (2013).
Huisken, J. & Stainier, D. Y. R. Selective plane illumination microscopy techniques in developmental biology. Development https://doi.org/10.1242/dev.022426 (2009).
Weber, M. & Huisken, J. Light sheet microscopy for real-time developmental biology. Curr. Opin. Genet. Dev. 21, 566–572 (2011).
Huisken, J. Slicing embryos gently with laser light sheets. BioEssays 34, 406–411 (2012).
Wan, Y., McDole, K. & Keller, P. J. Light-sheet microscopy and its potential for understanding developmental processes. Annu. Rev. Cell Dev. Biol. https://doi.org/10.1146/annurev-cellbio-100818-125311 (2019).
Corsetti, S., Gunn-Moore, F. & Dholakia, K. Light sheet fluorescence microscopy for neuroscience. J. Neurosci. Methods https://doi.org/10.1016/j.jneumeth.2018.07.011 (2019).
Ueda, H. R. et al. Whole-brain profiling of cells and circuits in mammals by tissue clearing and light-sheet microscopy. Neuron https://doi.org/10.1016/j.neuron.2020.03.004 (2020).
Tomer, R., Khairy, K. & Keller, P. J. Light sheet microscopy in cell biology. Methods Mol. Biol. 931, 123–137 (2012).
Ding, Y. et al. Light-sheet imaging to elucidate cardiovascular injury and repair. Curr. Cardiol. Rep. https://doi.org/10.1007/s11886-018-0979-6 (2018).
Poola, P. K., Afzal, M. I., Yoo, Y., Kim, K. H. & Chung, E. Light sheet microscopy for histopathology applications. Biomed. Eng. Lett. https://doi.org/10.1007/s13534-019-00122-y (2019).
Berthet, B. B. & Maizel, A. Light sheet microscopy and live imaging of plants. J. Microsc. 263, 158–164 (2016).
Komis, G., Novák, D., Ovečka, M., Šamajová, O. & Šamaj, J. Advances in imaging plant cell dynamics. Plant. Physiol. https://doi.org/10.1104/pp.17.00962 (2018).
Parthasarathy, R. Monitoring microbial communities using light sheet fluorescence microscopy. Curr. Opin. Microbiol. https://doi.org/10.1016/j.mib.2017.11.008 (2018).
Joseph, J. L. & Christensen, C. M. Disruptive technologies: catching the wave. J. Prod. Innov. Manag. https://doi.org/10.1016/0737-6782(96)81091-5 (1996).
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. https://doi.org/10.1038/nrg.2016.49 (2016).
Greger, K., Swoger, J. & Stelzer, E. H. K. Basic building units and properties of a fluorescence single plane illumination microscope. Rev. Sci. Instrum. 78, 023705 (2007).
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995).
Schetelig, M. F. et al. Site-specific recombination for the modification of transgenic strains of the Mediterranean fruit fly Ceratitis capitata. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.0907264106 (2009).
Nakamura, T. et al. Imaging of transgenic cricket embryos reveals cell movements consistent with a syncytial patterning mechanism. Curr. Biol. 20, 1641–1647 (2010).
Misof, B. et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767 (2014).
Acknowledgements
The Ceratitis, Apis and Gryllus live imaging data were acquired by a cooperation of F.S. and E.H.K.S. with M. F. Schetelig (Justus-Liebig-Universität, Gießen, Germany), P. Siefert and B. Grünewald (Institut für Bienenkunde, Oberursel, Germany) and T. Mito (University of Tokushima, Japan), respectively. The human prostate biopsy images were kindly provided by A. K. Glaser and J. T. C. Liu (Department of Mechanical Engineering, University of Washington, USA) and N. P. Reder and L. D. True (Department of Pathology, University of Washington, USA). F.S. and E.H.K.S. thank S. Plath for his assistance in generating the computer-assisted design schemes.
Author information
Authors and Affiliations
Contributions
Introduction (E.H.K.S.); Experimentation (E.H.K.S., F.S., B.-J.C., K.M. and R.F.); Results (F.S., F.P., S.P., K.M. and R.F.); Applications (E.H.K.S., F.S., B.-J.C., K.M. and R.F.); Reproducibility and data deposition (E.H.K.S., F.P., S.P. and K.M.); Limitations and optimizations (B.-J.C., S.P., K.M., S.P. and R.F.); Outlook (E.H.K.S., F.S., S.P., F.P., K.M. and R.F.); Overview of the Primer (E.H.K.S., F.S., B.-J.C., K.M., R.F., F.P. and S.P.).
Corresponding authors
Ethics declarations
Competing interests
E.H.K.S. has shares in related patents. The other authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Methods Primers thanks J. Liu, who co-reviewed with A. Glaser; P. Mondal; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
2014 Nature Methods Method of the Year: https://www.nature.com/collections/cktrchwtrc
3Dscript: https://imagej.net/3Dscript
μManager: https://micro-manager.org
μSPIM: https://uspim.org/
BigDataViewer: https://imagej.net/BigDataViewer
BigStitcher: https://imagej.net/BigStitcher
BigVolumeViewer: https://github.com/tpietzsch/jogl-minimal
Bio-Formats: https://github.com/ome/bioformats
BioStudies: https://www.ebi.ac.uk/biostudies
B3D framework: https://git.embl.de/balazs/B3D
Caltech file naming convention worksheet: https://resolver.caltech.edu/CaltechAUTHORS:20200601-161923247
CARE: https://github.com/CSBDeep/CSBDeep
Cellpose: https://github.com/MouseLand/cellpose
Cell Organelle Segmentation by Electron Microscopy: https://www.janelia.org/project-team/cosem
ClearVolume: https://clearvolume.github.io
CLIJ: https://clij.github.io
Collaborative Annotation Toolkit for Massive Amounts of Image Data (CATMAID): https://catmaid.readthedocs.io
Conda: https://docs.conda.io/en/latest
Dask: https://dask.org
Docker: https://www.docker.com
FAIR principles: https://www.go-fair.org/fair-principles
Fiji: https://fiji.sc
FlyLight: https://www.janelia.org/project-team/flylight
FPbase: https://www.fpbase.org/
FPvis: https://www.fpvis.org/
Git: https://git-scm.com
GZIP: https://www.gzip.org/
HDF5 file format: https://www.hdfgroup.org/solutions/hdf5
Ilastik: https://www.ilastik.org/
Image Data Resource: https://idr.openmicroscopy.org
ImageJ: https://imagej.nih.gov/ij
ImarisWriter: https://github.com/imaris/ImarisWriter
ImgLib2: https://imagej.net/ImgLib2
Insight toolkit: https://itk.org
Keller lab block file format: https://bitbucket.org/fernandoamat/keller-lab-block-filetype/src/master
Linus: https://gitlab.com/imb-dev/linus
LZ4: https://github.com/lz4/lz4
MaMuT: http://imagej.net/MaMuT
Mastodon: https://github.com/mastodon-sc/mastodon
Maven: https://maven.apache.org
N5 file format: https://github.com/saalfeldlab/n5
napari: https://github.com/napari/napari
Nature Methods LSFM collection: https://www.nature.com/collections/shjqwygmvh
Neuroglancer: https://github.com/google/neuroglancer
Npy2bdv: https://github.com/nvladimus/npy2bdv
Open Microscopy Environment Consortium: https://www.openmicroscopy.org
Open Microscopy Environment Remote Objects: https://www.openmicroscopy.org/omero
OpenSPIM: https://openspim.org
OpenSpinMicroscopy: https://sites.google.com/site/openspinmicroscopy/
Pycro-Manager: https://github.com/micro-manager/pycro-manager
Scenery: https://github.com/scenerygraphics/scenery
Singularity: https://sylabs.io/docs
Spark: http://sparkjava.com
StarDist: https://github.com/mpicbg-csbd/stardist
TeraFly: https://github.com/abria/TeraStitcher/wiki/TeraFly
TeraStitcher: https://github.com/abria/TeraStitcher
Tracking with Gaussian mixture models: https://www.janelia.org/lab/keller-lab/software/fast-accurate-reconstruction-cell-lineages-large-scale-fluorescence
UCSF Chimera: https://www.cgl.ucsf.edu/chimera/
Vaa3D: https://alleninstitute.org/what-we-do/brain-science/research/products-tools/vaa3d
Visualization toolkit: https://vtk.org
xarray: http://xarray.pydata.org/en/stable
ZARR file format: https://zarr.readthedocs.io/en/stable
Glossary
- Focal volume
-
The planar volume in front of an objective, from which a sharp image can be obtained. It is proportional to the depth of field of the detection objective and the field of view.
- Voxels
-
Portmanteau term of ‘volumes’ and ‘elements’, referring to single points of a three-dimensional grid.
- Diffraction limit
-
The maximum achievable optical/spatial resolution of an image recorded with an optical microscope, equivalent to about half the wavelength of the illumination light.
- Illumination train
-
The optical path used for forming the light sheet and excitation of fluorophores.
- Detection train
-
The optical path used for collection of the emission signal.
- Coherent illumination
-
Illumination with light composed of wave sources of the same frequency, waveform and phase; scattered laser light retains the original phase.
- Numerical aperture
-
(NA). A refraction index-based dimensionless number that states the maximum half-angle across which an optical element, such as an objective, emits and collects light. The NA influences both the lateral and the axial resolutions.
- Gaussian beams
-
Pencil-shaped diffracting focused beams whose planar profiles perpendicular to the beam axis are described with two-dimensional Gaussian functions.
- Bessel beams
-
Non-diffracting, self-reconstructing beams generated with an axicon or a spatial light modulator.
- Airy beams
-
Non-diffracting, self-reconstructing beams generated with a spatial light modulator.
- Free working distance
-
The distance between the front surface of an objective and the centre of the focal volume.
- Volume of view
-
The volume covered by the field of view (along the x axis and y axis) and the free working distance of the detection objective (along the z axis).
- Lateral resolution
-
Spatial resolution along the x axis and y axis.
- Oversampling and undersampling
-
The collection of redundant and less than available spatial/temporal information, respectively.
- Axial resolution
-
Spatial resolution along the z axis.
- Point spread functions
-
The images of point sources, often used to describe the response or the resolution of an imaging system.
- High-content imaging
-
Collection of a large amount of complementary information from the same specimen.
- Air objectives
-
Objective lenses that operate in air or vacuum; these cannot achieve the high and very high numerical apertures of water or oil immersion objectives.
- Water-dipping objectives
-
Objective lenses that operate in aqueous or organic media; these can have higher numerical apertures (NAs) than air objectives, but not the very high NAs of oil objectives.
- Isotropic resolution
-
Identical resolution along the x axis, y axis and z axis.
- Oviparous
-
Describes a metazoan species that lays eggs with no or partial embryonic development in the parent.
- Fluorinated ethylene propylene
-
(FEP). A synthetic material with a refractive index close to that of water (1.33), available as threads, foils and tubes.
- Viviparous
-
Describes a metazoan species in which the embryos develop within the body of the parent.
- Explantation
-
In developmental biology, the process of removing a developing embryo from the uterus of the parent for experimentation.
- Pre-implantation
-
In developmental biology, the time period between fertilization of the oocyte and implantation of the blastocyst into the uterine wall.
- Phytagel
-
Water-soluble anionic polysaccharide of bacterial origin with similar biophysical and optical properties to agarose.
- Rolling shutter
-
The line-wise recording of scientific complementary metal–oxide–semiconductor sensors; in light sheet fluorescence microscopy it is synchronized with line-wise illumination to record anisotropic confocal images along the y axis.
- Spectral unmixing
-
Data processing procedure in which the spectral signatures of voxels are divided into collections of constituent spectra.
- Fourier transform
-
A mathematical operation that transforms a real intensity-based image into a spatial frequency-based array. These arrays are used to perform mathematical operations in spatial frequency space and inverse Fourier transforms are used to calculate real images.
- Fluorescence correlation spectroscopy
-
A time correlation-based statistical imaging method for the quantification of fluorescence fluctuations that is used to measure diffusion coefficients or characterize reaction kinetics.
- Fluorescence lifetime imaging
-
The imaging of the spatial distribution of the lifetimes of fluorophores in a specimen; fluorescence lifetimes are sensitive to their close environment.
- Hot pixels
-
Dysfunctional pixels that appear white in every image independent of the actual signal.
- Dead pixels
-
Dysfunctional pixels that appear black in every image independent of the actual signal.
- Stitching
-
In fluorescence microscopy image processing, the combination of multiple partially overlapping stacks recorded along the same direction to generate larger three-dimensional images.
- Multi-view fusion
-
In fluorescence microscopy data processing, the calculation of a single three-dimensional image based on multiple image stacks recorded along multiple directions of an opaque specimen.
- Deconvolution
-
A mathematical approach to take advantage of a priori information regarding the properties of an optical system to emphasize certain spatial frequencies and, in consequence, improve the resolution of an image.
- Block-wise
-
Describes parallelized image processing using blocks such as fractions of a line, area or volume to process data on multiple computation and graphic processing units.
- Hierarchical data formats
-
File formats that utilize a file directory-like structure to organize the data within the file in a structured fashion.
- Lossy image compression
-
A data size-reduction approach based on approximation and/or partial data discard; the original data cannot be completely reconstructed.
- Bit-shuffling
-
The rearrangement of bits to, for example, allow or simplify image processing.
- Segmentation
-
Detection and highlighting of morphological features, for example cell membranes or nuclei, in a two-dimensional or three-dimensional image.
- Spheroids
-
Multicellular, usually densely packed, three-dimensional tissue-like cell culture models.
- Organoids
-
A miniaturized and functionally simplified cell culture version of an organ.
- Trichoblasts
-
Cells at the surface of plant roots that are responsible for the formation of root hairs.
- Actinic
-
Describes light that is able to induce photochemical reactions.
- Beam divergence
-
In Gaussian beams, the increase in beam radius as a function of distance from the beam waist.
- Point spread function engineering
-
The modification of the plane wave usually encountered in the entrance aperture of a lens to change the properties of the point spread functions. For example, Bessel beams are generated by point spread function engineering.
Rights and permissions
About this article
Cite this article
Stelzer, E.H.K., Strobl, F., Chang, BJ. et al. Light sheet fluorescence microscopy. Nat Rev Methods Primers 1, 73 (2021). https://doi.org/10.1038/s43586-021-00069-4
Accepted:
Published:
DOI: https://doi.org/10.1038/s43586-021-00069-4
This article is cited by
-
Fluorescence-based multifunctional light sheet imaging flow cytometry for high-throughput optical interrogation of live cells
Communications Physics (2024)
-
Brillouin microscopy
Nature Reviews Methods Primers (2024)
-
Fluorescent microscopy evaluation of diode laser effect on the penetration depth of turmeric (Curcuma longa) extract cream on skin tissues of Wistar rats
Lasers in Medical Science (2024)
-
Geometric deep learning of particle motion by MAGIK
Nature Machine Intelligence (2023)
-
Light-sheets and smart microscopy, an exciting future is dawning
Communications Biology (2023)