Abstract
Due to climate change the pathogenic bacterium Vibrio vulnificus proliferates along brackish coastlines, posing risks to public health, tourism, and aquaculture. Here we investigated previously suggested regulation measures to reduce the prevalence of V. vulnificus, locally through seagrass and regionally through the reduction of eutrophication and consequential formation of algal blooms. Field samples collected in the summer of 2021 covered the salinity and eutrophication gradients of the Baltic Sea, one of the largest brackish areas worldwide. Physico-, biological- and hydrochemical parameters were measured and variables explaining V. vulnificus occurrence were identified by machine learning. The best V. vulnificus predictors were eutrophication-related features, such as particulate organic carbon and nitrogen, as well as occurrence of potential phytoplankton blooms and associated species. V. vulnificus abundance did not vary significantly between vegetated and non-vegetated areas. Thus, reducing nutrient inputs could be an effective method to control V. vulnificus populations in eutrophied brackish coasts.
Similar content being viewed by others
Introduction
The Baltic Sea is a semi-enclosed marginal sea of the Atlantic located in northern Europe, with a coastline of approximately 8000 km and covering an area of 415,266 km2. Saline inflows through the North Sea produce a 2000 km long lateral surface salinity gradient throughout the entire Baltic Sea, ranging from high salinities (>25) in the transition zone of the Kattegat to low salinities (<5) in the Gulf of Bothnia1. The Baltic Sea is characterized by an estuarine-like circulation due to the positive freshwater budget. The drainage area of the Baltic Sea encompasses a population of approximately 85 million, and is consequently heavily influenced by eutrophication2. In addition, annual mean sea-surface temperatures are rising2 and the ecosystem is expected to be increasingly affected by warming in the coming decades3, and will be faced with extended heat wave durations4.
These changes favour the growth of pathogenic bacteria of the genus Vibrio and an increase in Vibrio spp. abundances, infection rates, and fatal cases along the Baltic Sea coastline has been reported5,6,7,8. The infections of predominantly immunodeficient humans can be associated with the consumption of raw or undercooked shellfish, but in the Baltic Sea, they frequently manifest as skin infections resulting from direct contact with coastal brackish water. Only a low number of infections are currently associated with Vibro vulnificus in the Baltic Sea9, but these are usually severe and often lethal6,10,11.
Temperature and salinity are widely accepted as the two primary regulators of V. vulnificus abundance and distribution12,13,14,15, but factors related to eutrophication, such as elevated dissolved organic carbon [DOC16] concentration or dinoflagellate blooms17, have been shown to stimulate V. vulnificus growth in laboratory settings. Due to its preference for intermediate salinities and proliferation at water temperatures >15 °C18, V. vulnificus experiences optimal growth conditions during the summer in the Baltic Sea. The expected spread of V. vulnificus does not only pose a significant threat to public health but also to the tourist, fishing, and aquaculture industries19,20,21. In consequence, the question arises, whether measures can be taken to regulate V. vulnificus abundances.
In a natural setting, macrophytes such as seagrass (Zostera marina) might reduce the abundance of pelagic and potentially pathogenic Vibrio spp. and other pathogens within the seagrass canopy22,23. The underlying mechanisms for this decrease are elusive but could include increased sedimentation rates due to hydrodynamic attenuation24,25,26, filter-feeding by benthic fauna27,28, or allopathic chemicals exuding directly from the seagrass plants29. Regardless of the specific causal mechanisms through which Z. marina beds potentially impact V. vulnificus abundance, they are putative nature-based solutions for reducing V. vulnificus23. Consequently, this study aims to elucidate important factors for mitigating V. vulnificus abundances along the Baltic coast, evaluating the potential of seagrass as a nature-based solution at the local scale and the reduction of eutrophication at the regional scale, respectively. We measured a large array of physical, biological, and hydrochemical parameters, and used three parallel methods (cultivation, amplicon sequencing, and droplet digital PCR; ddPCR) for the quantification of V. vulnificus within, adjacent and far from seagrass meadows. This enabled us to simultaneously assess the relationship between seagrass, environmental factors, such as temperature, salinity, and (in)organic nutrients, eukaryotic and prokaryotic microbial communities, and V. vulnificus over a vast salinity range, facilitating the detection of more generalizable patterns. These patterns are applicable to a wide range of environmental conditions found in estuaries and marginal seas worldwide. By combining a large number of parameters with a machine learning approach, complex and non-linear relationships are identified between the environment and V. vulnificus abundance.
V. vulnificus is likely stimulated by eutrophication-induced algal blooms; therefore, reducing eutrophication appears to be a promising strategy for mitigating the health risks associated this bacteria. Based on the results of this study, a regulatory effect of seagrass on V. vulnificus could not be determined.
Results
Environmental gradient and eutrophication index
A northeast-to-southwest salinity gradient (6–15.4) across the Baltic Sea was observed (sampling points: Fig. 1), consistent with the increase of average seagrass leaf length from 22 to 109 cm. The density of the meadows (125 to 1059 stalks per m²), depth (0.6−4.7 m), temperature (15.8–21.4 °C), dissolved oxygen (DO) (7.66 to 14.51 mg L−1), pH (7.96−8.98) and average grain size diameter (dx50) (98−1623 µm) were not structured along the salinity gradient.
Abundance of heterotrophic cells, ranging between 3.44 × 106 and 2.68 × 107 mL−1, varied across the Baltic Sea, with the highest and lowest abundances along the German (BV-06 to BV-11 and BV-24) and Estonian (BV-14 to BV-16 and BV-25) coasts, respectively. Among these cells, high nucleic acid (HNA) cell counts were very similar with 1.3−1.4 × 106 cells mL−1, but low nucleic acid (LNA) counts varied between 1.37 × 106 and 1.44 × 107 cells mL−1. Autofluorescent organisms were similarly distributed among stations, with Synechococcus abundances ranging from 4.09 × 104 to 1.12 × 106 cells mL−1, picoeukaryotes from 1.41 × 103 to 5.92 × 104 cells mL−1, and nanoeukaryotes from 2.17 × 102 to 4.95 × 104 cells mL−1.
Phosphate (PO43-) and nitrate (NO3−) concentrations ranged from <0.1 to 0.93 and <0.2 to 1.46 µM, respectively, with the highest concentrations at the Swedish coast (BV-20, BV-21). The highest concentrations of nitrite (NO2-) and ammonium (NH4+), reaching up to 0.15 µM and 6.94 µM, were observed on the Danish coast (BV-26). Silica (SiO2) concentrations ranged between <2 and 48.92 µM and did not show strong geographic structuring. Particulate organic carbon (POC) varied between 11.21 and 276.83 µM, particulate organic nitrogen (PON) between 1.64 and 41.76 µM, DOC between 180.10 and 681.30 µM, dissolved organic nitrogen (DON) between 8.30 and 30.60 µM and chlorophyll-a (chl-a) between 0.63 and 21.16 µg L−1, with the highest concentrations found along the German coast. The eutrophication index was highest on the German coast (BV-11) and lowest on the Danish coast (BV-26).
No significant differences in physicochemical parameters within the water column were observed between substations A (in the macrophyte meadow), B (15 m from the macrophyte meadow), and C (100 m from the macrophyte meadow), for both Z. marina and Fucus spp. (serving as controls, see Methods) stations (Supplementary Fig. 1).
Macrophytes had no significant effect on V. vulnificus abundance
V. vulnificus was detected in 47% of the samples using ddPCR, in 33% through 16S rRNA gene sequencing, and both methods identified it in 20% of the cases. Across all sampled stations, local effects of Z. marina or Fucus spp. on V. vulnificus absolute or relative abundances were insignificant. V. vulnificus was equally distributed between substations A, B, and C (Wilcoxon rank sum test (Fig. 2)). This was consistent for vvhA gene copy numbers and 16S rRNA gene relative abundances, green colony forming units (CFUs), which are presumed, but not unequivocally identified, V. vulnificus colonies, and for heterotrophic bacterial cell counts (p-values > 0.05). Likewise, there was no consistent pattern between substations A, B, and C in vvhA gene copy number nor relative 16S rRNA gene abundances when observing the individual stations (Supplementary Figs. 2 & 3). V. vulnificus in sediments was also evenly distributed among substations (Supplementary Fig. 4). Based on the vvhA genes and 16 S rRNA gene sequencing, V. vulnificus was not present in Z. marina biofilms. However, one V. vulnificus strain was isolated from seagrass leaves at station BV-12. In addition, other potential pathogenic Vibrio spp. found in the Baltic Sea were also distributed equally between substations on a Baltic Sea-wide level (Supplementary Fig. 5).
a V. vulnificus abundances (vvhA gene copies mL−1), b Relative V. vulnificus abundance based on 16 S rRNA genes, c Presumptive V. vulnificus based on green CFUs mL−1 on TCBS agar, d Number of heterotrophic bacteria mL−1. Station A is located within a macrophyte meadow, while station B is located 15 m, and station C is located 100 m from the meadow edge. Boxes represent the 25th and 75th percentiles.
Eutrophication impacted V. vulnificus abundance
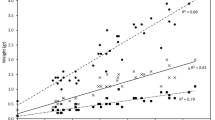
A positive relationship was observed between temperature and V. vulnificus abundance (approximated by vvhA gene copies mL−1, Fig. 3). Within the temperature range of 19.5–20.5 °C, however, strongly eutrophied samples exhibited higher vvhA gene numbers compared to other samples in the same, as well as other, temperature ranges, coinciding with higher cell abundances of heterotrophic bacteria in general (Fig. 3). The average concentration of the vvhA gene in this temperature bin was higher than that at higher temperatures, and, on average, the bin exhibited higher eutrophication. The eutrophication index captured more of the variance in V. vulnificus abundance (36%) than temperature (12%) (Supplementary Fig. 6). The combination of both explained 43% while the more comprehensive and non-linear random forest (RF) including all parameters explained 59% of the variance.
The x-axis consists of 1-degree temperature bins for the bars and is continuous for the points, the y-axis is the log10 transformed vvhA gene copy numbers. Colours represent the eutrophication status per sample and are averaged per bin for the bars. The point shape indicates heterotrophic bacterial cell abundance, grouped into five bins, ranging from 3.4 × 106 to 2.7 × 107 cells mL−1. Error bars represent standard deviation.
Prevalence of V. vulnificus was correlated with indicators of blooming situations
Both RF models exhibited moderate performance, explaining 0.59 ± 0.32 and 0.59 ± 0.45 of the variance in the derived total V. vulnificus abundances and of the variation of vvhA gene copies mL−1, respectively. Root Mean Square Error (RMSE) was 697.16 ± 597.50 and 6.33 ± 4.66 of the target values 8,130.20 and 37.93, and the mean absolute error (MAE) was 519.76 ± 307 and 4.56 ± 2.55.
Prokaryotes identified as the most important predictors for V. vulnificus abundance belonged, among others, to the phylum Cyanobacteria, with representatives of the genera Cyanobium (ASV219, ASV379) and Nodularia (ASV2690), as well as Actinobacteria, with Candidatus Aquiluna(ASV236), ML602J-51 (ASV654) and Nocardioides (ASV1070) (Fig. 4; Supplementary Table 1). The most important eukaryotic predictor taxa for V. vulnificus, which could be classified at the genus level, were the rodophyte Ceramium sp. (ASV71), the Cercozoa Thaumatomastix and Aplanochytrium (ASV320, ASV430), the mixotrophic organism Teleaulax (ASV6) and fungi Rhizophydium spp. (ASV312) (Fig. 4; Supplementary Table 2). The relative abundances of these taxa, except Teleaulax and Nodularia, were notably higher at elevated vvhA gene concentrations and derived V. vulnificus cell numbers (Fig. 4). Cyanobium spp. encompassed up to 18.4%, and Ceramium up to 9.2%, of the ASV counts, respectively. The high relative abundances of these pro-and eukaryotic taxa coincided with high POC and PON, high Synechococcus abundance, and high LNA cell concentration (Fig. 4).
Predictors are ordered from high (left) to low (right) importance for the respective models. Observations are ordered according to decreasing vvhA gene copy numbers. Values of each predictor are scaled from 0 to 1 for color representation in the heatmap, while the original values are printed in each cell.
Discussion
We present a comparative study of 19 coastal Baltic Sea sampling stations in summer/early fall 2021, exploring the potential of seagrass as a local solution and the reduction of eutrophication as a more regional strategy to mitigate the proliferation of potentially pathogenic V. vulnificus. For these stations, physical-, biological- and hydrochemical parameters were measured and variables explaining V. vulnificus occurrence were identified. We found that eutrophication-related parameters such as POC/PON and high Cyanobium sp. and Synechococcus sp. abundance predicted high V. vulnificus abundances, while the occurrence and density of Z. marina showed no predictive value. This implies that reducing eutrophication on a regional level could be a promising strategy for constraining further proliferation of V. vulnificus along the Baltic Sea coast.
The distribution and abundance of V. vulnificus has been extensively studied, with temperature and salinity being consistently identified as key factors12,13,14,15. Our study observed temperatures exceeding 15 °C and salinities ranging from 6 to 16, which are generally considered favorable conditions for V. vulnificus5,30, yet abundances varied between stations. Notably, stations with a high eutrophication index tended to exhibit higher V. vulnificus abundances compared to those with similar or even higher temperatures but a lower eutrophication index (Fig. 3). Furthermore, the eutrophication index explained a greater proportion of the variance in the V. vulnificus abundance than temperature (Supplementary Fig. 6). This finding is consistent with earlier studies, which also related elevated concentrations of DOC and chl-a to high V. vulnificus abundances16,30. Chl-a was, besides being part of the eutrophication index, also closely associated with the predictors for V. vulnificus identified by RFE, namely POC, PON, Cyanobium sp. and Synechococcus (relative) abundance in this study (Supplementary Fig. 7). Additionally, the effectiveness of heterotrophic cell counts (LNA cells in Fig. 4) in the prediction underscored the link to eutrophication. These results align with observations from the Neuse River estuary, which faces similar problems with eutrophication as the Baltic Sea, where elevated levels of both total Vibrio spp. and V. vulnificus corresponded with high phytoplankton biomass31 and an investigation after Hurricane Ian (October 2022), where high phytoplankton mass, approximated by chl-a concentration, and associated zooplankton abundance, was found to potentially stimulate V. vulnificus proliferation32.
Cyanobium sp., which thrives under eutrophic conditions and is known to form blooms and correlate positively with the decay phases of other blooms33, was found to be an important V. vulnificus predictor. The identified ASVs for Cyanobium sp. comprised up to 18.7% of the total prokaryotic sequence reads at some of the highest vvhA gene copy numbers. Associations between pathogenic Vibrio spp. and (harmful) algal blooms have been observed before34 and our results substantiate this relationship. It is plausible that eutrophication, by stimulating blooms, served as an important indirect driver of the distribution and abundance of V. vulnificus via the release of DOC/DON and POC/PON. The idea that the increase in DOC/DON triggered by the bloom contributed to the proliferation of V. vulnificus prior to sampling is supported by the predictive power of the actinobacteria Candidatus Aquiluna (ASV236), ML602J-51 (ASV654), and Nocardioides (ASV1070). Actinobacteria are commonly associated with the demise of phytoplankton blooms and the generation of cyanobacterially sourced DOC35,36. The stimulation of V. vulnificus abundance by algal blooms was also inferred from the high V. vulnificus abundances coinciding with high filamentous Ceramium sp. abundances, peaking at 9.2% of the sequence reads during both the highest vvhA gene concentrations and derived V. vulnificus cell counts at station BV-08-A (Fig. 4). Ceramium sp. is an opportunistic fast-growing algae, with high nutrient uptake rates37. Interestingly enough, the eutrophication index, POC and PON are all low at this station. This could be an indication that the blooming (in terms of relative abundance) of specific plankton, for example, Ceramium sp. and Cyanobium sp. can already convey some benefit. POC/PON are still good predictors of V. vulnificus in that scenario, as they generally increase with phytoplankton blooms, including all the blooms that provide species-specific benefits. Regardless of the specific combination of factors that promote V. vulnificus proliferation, reducing eutrophication would limit the extent of algal blooms in the Baltic Sea, likely reducing infection risk. Among the top five instances of highest vvhA gene copy abundances and derived V. vulnificus cell numbers, either Ceramium or Cyanobium sp. was consistent, except at station BV-24-B, found at high relative abundance.
The high relative abundance of Cyanobium sp. was concurrent with other species expected to either infect or co-occur with a bloom. The parasite Aplanochytrium sp., which is known to bloom38 and to correlate strongly with phototrophs39, and the fungi Rhizophydium sp. (ASV 312)40, reached their highest relative abundance (max 0.4% and 1%, respectively) at stations with high Cyanobium sp. and V. vulnificus (relative) abundance. Under conditions conducive to extensive bacterial growth, such as during blooms, Thaumatomastix (ASV320) might be able to temporarily compete with other organisms41. Thaumatomastix exceeded 0.1% of the eukaryotic community only in samples where Cyanobium sp., Synechococcus, and their potential parasites were relatively abundant, further pointing to a highly productive system. This scenario coincided with elevated V. vulnificus abundance.
We speculate that when grazing pressures on the algal bloom reached critical levels, the bloom ceased, leading to a depletion of the organic nutrients crucial for the proliferation of V. vulnificus. In instances where we observed high relative abundances of the potential predator Teleaulax (>1%), there was a concurrent decrease in Synechococcus and Cyanobium sp. (relative) abundances and a decrease in the concentrations of PON and POC (e.g. BV-18 & BV-20). This phenomenon was consistently associated with the absence or low relative abundance of V. vulnificus. This may be attributed to the potential role of Teleaulax in depleting the Synechoccocus population42,43 and thereby reducing the food sources for V. vulnificus.
While organic nutrients were good predictors of V. vulnificus abundance, inorganic nutrients were not. Additionally V. vulnificus was generally less correlated with inorganic nutrients (e.g. PO43-) than heterotrophic bacteria overall (Supplementary Fig. 8). This suggests that potential bloom-forming organisms may play a pivotal role in the chain leading from inorganic nutrient input to increased V. vulnificus abundance by converting inorganic nutrients into organic forms.
In addition to the increase in organic constituents present during a phytoplankton bloom, the higher abundance of phytoplankton and the associated zooplankton community could potentially offer protection against grazing by bacterivorous protozoa, as previously demonstrated for V. cholerae44. V. cholerae population growth is normally balanced by protozoan predation, but during declining blooms, V. cholerae has been shown to multiply at an increased rate due to the higher availability of DOC and DON and/or change to a particle-attached lifestyle for protection from predation45. A similar protective effect has been shown for other species of Vibrio, which significantly benefitted from association to phytoplankton during periods of intense grazing46. We hypothesize that V. vulnificus might also benefit from a similar interaction.
The discussed microorganisms and eutrophication index exhibited greater explanatory power than temperature for V. vulnificus abundance. This was likely due to their sensitivity to historical and current temperatures and nutrient availability. As a result, they served as a record of the water mass that was more predictive than temperature alone. The suggested pathway through which eutrophication could stimulate V. vulnificus abundance is summarized in Fig. 5. The identity of the blooming organism and its predators might not be as critical as the fact that the bloom and its associated predators stimulate organic nutrient availability and might provide a degree of protection.
P and N represent the total dissolved inorganic phosphorus and nitrogen. Inorganic nutrient inflow from the land induces algal blooms, providing the organic material required for V. vulnificus proliferation and potentially protection from bacterivorous protozoa. The figure also displays other organisms identified as important predictors and their potential connections to blooms. Notably, Zostera marina could not be shown to significantly affect abundance in our study.
While eutrophication is often a regional problem, with regional solutions, such as the Baltic Sea action plan47. Intraregional differences exist between bays due to differences in nutrient sensitivity48 and nutrient input from point sources49. Engineering solutions to eutrophication in semi-enclosed Baltic Sea bays have been tested successfully50, potentially providing a solution for some Baltic Sea coasts. Besides allowing for a potential (multi)-national approach to reduce the V. vulnificus hazard, the possible impact of eutrophication can be used to improve risk assessment and early warning by integrating the existing HELCOM Eutrophication Assessment Tool48 into the currently available ECDC vibrio viewer, which relies solely on temperature and salinity, for the Baltic Sea51. In other estuaries or marginal seas, the integration of chl-a data from remote sensing into early warning systems may provide an improved prediction of V. vulnificus abundance.
The assessment of V. vulnificus predictors was hampered by the observed discrepancies between the quantification methods employed. They are likely due to multiple biases associated with each methodology. These are, in no particular order: CFUs by nature of the method, underestimate the real abundance, as they are unable to quantify viable but not culturable cells, resulting in the lowest abundances measured by any of the methods (Fig. 2). Differences between vvhA gene copy numbers and derived cell counts can be partly attributed to multiple copies of the 16S rRNA gene52, which range between 8 and 12 for V. vulnificus53. We did not correct for these, according to the advice of Louca et al.54. Additionally, not all V. vulnificus cells contain the vvhA gene, which further complicates the comparison. The discrepancy between vvhA and derived V. vulnificus abundance could be an indication that a sizeable part of the sampled V. vulnificus community lacks this pathogenicity gene. Further differences might arise from primer bias, PCR biases and biases in quantification of bacterial counts. When assessing the vvhA gene in our study, fewer instances of V. vulnificus are identified per station; however, V. vulnificus is detected at more stations. A prime example is station BV-11-C (Fig. 4), which has the second highest vvhA gene copy numbers, and where presumable V. vulnificus colonies were cultured, but no V. vulnificus was detected with the 16S rRNA amplicon sequencing, while at stations BV-11-A and BV-11-B V. vulnificus is detected by all methods. In this case, the discrepancy arose because the 16S rRNA gene sequences amplified at station BV-11-C could not unambigiously be identified as V. vulnificus. A similar degree of similarity was observed with a related Vibrio species. Additional discrepancies might arise from V. vulnificus variants not accounted for in the database used for comparison. This complicates the comparison between the methods, but overall, the association between algal blooms and V. vulnificus could still be shown across all measured parameters.
V. vulnificus was found to be absent from seagrass leaves by molecular methods and only one strain could be isolated from them, even though V. vulnificus is known to form biofilms55 under a variety of conditions56,57,58. This is surprising given the release of labile DOC from seagrass leaves59 but could be due to allelopathic chemicals exuded from the seagrass leaves hampering the growth of V. vulnificus, as observed for a general bacterial community60. An alternative explanation is that there is a strong association between the seagrass and their associated microbiome and that V. vulnificus was consistently outcompeted61,62. V. vulnificus was, however, found in the benthic and pelagic environment, in similar abundances at substations inside and outside of the meadows (Fig. 2, Supplementary Figs. 1, 2), implying that the impact of seagrass meadows on V. vulnificus abundance, relative abundance, and culturable potential within the surrounding environment was minor in the Baltic Sea. Additionally, no effect on the relative abundance of other potential Vibrio pathogens could be detected within the sampled salinity range (Supplementary Fig. 5). Similar to the findings in San Diego coastal waters, our results contrast the reduction in pathogenic bacteria abundance observed by Lamb et al. in 201763. Future findings may differ for other conditions, e.g., water temperatures, salinity ranges, or less eutrophied systems, which were outside our study framework. The discrepancy between our study and earlier work on the influence of seagrass on potential pathogens may also be ascribed to a difference in methodology or study area. Reusch et al. (2021) used solely chrom-agar based blue colony counts at salinities between 14.3 and 22.123, while we employed methods more specific to V. vulnificus over a different salinity gradient. The salinity of our study areas was mostly lower and may have affected the physiology of seagrass and its possible effect on V. vulnificus. Based on our results, it could not be shown that seagrass meadows are a direct effective nature-based solution for reducing V. vulnificus abundance and associated infections in the Baltic Sea. However, the current high nutrient concentrations in the Baltic Sea may have masked possible direct effects of seagrass, and probably also Fucus spp., on V. vulnificus, and indirect effects through the reduction of the nutrient load.
Our study suggests that a reduced anthropogenic nutrient input could mitigate the cascade of events that stimulate algal blooms and through that, organic nutrient availability. This in turn, would make conditions less conducive for V. vulnificus proliferation. This could reduce public health risks and be a critical management tool for V. vulnificus in the Baltic Sea, as well as in other coastal brackish water systems worldwide.
Materials and Methods
Sampling stations and SCUBA sampling strategy
Samples from fifteen Z. marina and, for comparison, four Fucus spp. stations were collected from July 25th to September 2nd, 2021, covering the German, Estonian, Finnish, Polish, Swedish, and Danish coasts (Fig. 1). Water and sediment samples were collected inside the macrophyte meadows (substation A), as well as the leaves/fronds of the macrophyte. Analogously, water and sediment samples were collected at control stations without macrophytes, located 15 m (substation B) and 100 m (substation C) from the edge of the meadow (Fig. 1). Sampling was carried out by SCUBA or snorkel divers at water depths ranging from 0.6 to 4.7 m. Sampling was only performed up to a moderate breeze of 4 on the Beaufort scale with wave heights <0.5 m to prevent heavy mixing between substations. The prevailing current direction was visually determined at the sampling depth by releasing food colourant (Dr. Oetker 4 Back & Speisefarbe). Divers approached the sampling site against the current and maintained buoyancy to prevent sediment resuspension.
Water samples were collected following two methods. First, for DNA extraction, flow cytometry, and plating, 100 mL of water was collected in 9 replicates using rinsed syringes. The samples were collected ca. 5 cm from a macrophyte and ca. 20 cm above the sediment at substation A, and ca. 20 cm above the sediment at substations B and C. Second, sterile 5 L plastic bags, used for chl-a, inorganic nutrients, POC and PON, DOC and DON determination, were filled by swimming against the current while keeping the bag open 20 cm above the sediment. This water was split between 1 L plastic and glass bottles after triplicate rinsing. Sediment was collected in nine sterile 50 mL Falcon tubes by scraping the top 1 cm of sediment. At substation A, Z. marina/Fucus spp. samples were collected in nine replicates by pinching the connection between the root and stalk. Subsequently, the leaves/fronds were separated by hand for further processing. All samples were transferred to a 4 °C cooler immediately and stored (maximally 8 h) until processing.
Macrophyte characteristics
Z. marina plant densities were determined by counting 20 × 20 cm squares in triplicate. The length of 30 leaves was measured per meadow. Fucus spp. densities were not determined due to the large heterogeneity within the individual beds.
Environmental parameters
Salinity, temperature, and depth were measured using a CTD48M (Sea & Sun Technology, Trappenkamp, Germany) attached to a diver. DO and pH were measured using a handheld multimeter (HQ40D Portables 2-Channel Multimeter) (HACH, Iowa, USA) with Hach Intellical LDO101 and Intellical PHC101 probes, respectively.
Water from the plastic bottles was filtered through 25Ø GF/F filters (Whatman plc, Maidstone, UK), with the filters stored for chl-a determination at −80 °C (250–500 mL) and the filtrate for nutrient analysis stored at −20 °C. Water from the glass bottles was filtered through pre-combusted (450 °C, 4 h) 25Ø GF/F filters for POC and PON determination and the filtrate for DOC and DON analysis, all stored at −20 °C. PO43−, NO3−, NO2−, ammonium NH4+, and SiO2 concentrations were measured using a Seal Analytical QuAAtro automated constant flow analyzer (SEAL Analytical Ltd, Nordestedt, Germany), with detection limits of 0.1, 0.2, 0.05, 0.5 and 2 µM, respectively64. DOC and DON were analyzed with a TOCL-CPH/TOC-VCPH TOC-Analyzer (Shimadzu, Germany)65, and POC/PON measurements were conducted with a varioMICRO cube element analyzer (Elementar Analysensysteme, Germany). Chl-a was measured fluorometrically using a 10- AU-005-CE fluorometer (Turner, San Jose, USA) according to the HELCOM66 guidelines and corrected for phaeopigment.
Sediment grain size
The grain size was analysed after 5 d of lyophilization with a Delta 1-24 LSCplus (Martin Christ Gefriertrocknungsanlagen, Osterode am Harz, Germany). Sediment samples were sieved through a 3.5 mm sieve and visible biological elements were removed. The remaining sediment chunks were broken by grinding for a minimum of 90 s with mortar and pestle. Samples were measured using the dry cell of a Mastersizer 3000 with a range from 0.01 to 3.500 μm (Malvern Panalytical, Malvern, UK).
Microbial analyses
Vibrio spp. CFUs and V. vulnificus isolates were obtained from water, sediment, and macrophytes from six independent replicate samples. For this, water aliquots of 50, 100 or 200 μL were (a) plated directly onto Vibrio selective thiosulfate citrate bile sucrose (TCBS) agar (Merck, Darmstadt, Germany) and (b) aliquots of 2, 5, 10 or 25 mL were filtered onto 0.2 μm PC-filters (Merck-Millipore, Burlington, USA) and placed onto TCBS agar. The sediment samples were homogenized after the removal of the overlying water, a subsample of approximately 10 g (dry-weight determined accurately after lyophilization) was transferred from six sediment samples to sterile 50 mL falcon tubes, where 40 mL of double 0.2 μm- filtered station water was added. To detach bacteria, five ultrasonic pulses of 10 s at 25% capacity at 5 s intervals using the Bandelin SONOPULS HD 2200.2 (Bandelin, Berlin, Germany) were applied. After subsequent vortexing and settling of the sediment, water aliquots of 50, 100, or 200 μL were plated on TCBS agar in six biological replicates. The same method was applied to the macrophytes. The only modification was that 5 − 20 mL of the supernatant was filtered over a PC-filter to obtain colonies. After 24 h incubation at 37 °C, CFUs of green colonies were determined for all plates.
To isolate V. vulnificus, green colonies from stations BV-05, BV-06, BV-09, BV−10, BV−12, BV−15, BV-20, and BV-21 were further cultivated on CHROMagar_vibrio™ (Chromagar Ltd. Paris, France) for 24 h at 37 °C and blue-colored colonies were restreaked on TCBS agar, CHROMagar, and Columbia sheep blood agar (Oxoid, Basingstoke, UK). DNA of isolates which were green on TCBS, blue on CHROMagar, and confirmed to be pure cultures on blood agar, was extracted (DNeasy Blood and Tissue Kit, Qiagen, Hilden, Germany), and purity and concentration were determined using a NanoDrop Spectrophotometer (Thermo Fisher, Waltham, USA). To confirm V. vulnificus, the species-specific vvhA gene sequence was targeted using multiplex real-time PCR (5´ nuclease assay). In the same assay, an internal amplification control (KOMA) was detected67. Primers for the detection of the vvhA gene45 are provided (Supplementary Table 3).
Enumeration of heterotrophic bacterial cells and autofluorescent phytoplankton
Four mL of water from three replicate 100 mL syringes, which were also used for the DNA extraction, was pipetted in duplicate into 5 mL sterile cryovials (VWR, Radnor, USA), and 200 μL formaldehyde (37%) was added. After homogenization and 1 h incubation at 8 °C, the samples were shock frozen and stored at −80 °C until flow cytometry analysis. Autofluorescent and heterotrophic cells were counted separately using a CytoFLEX S flow cytometer (Beckman coulter, Brea, USA) and unstained and SYBR Green I (Invitrogen, Waltham, USA) stained samples, respectively68. Autofluorescent cells were analysed with CytExpert software (Beckman coulter, Brea, USA) and grouped into Synechococcus, picoeukaryotes, and nanoeukaryotes. Stained samples were gated and clustered using FlowClust69 after quality control by FlowAI70 based on green and red fluorescence and side scatter68. The minimum Bayesian Information Criterion was reached for three clusters, showing beads, HNA, and LNA cells. The flow rate was monitored by adding beads to each sample (NFPPS-52-4K, Spherotech, Lake Forest, USA).
Molecular prokaryotic and eukaryotic community composition
Water (92 mL) from three syringes was filtered through 0.22 μm polyvinylidene fluoride membrane filters (Merck, Darmstadt, Germany), which were shock-frozen in liquid nitrogen and stored at −80 °C for downstream molecular analysis. Three falcon tubes with sediment from each substation and three falcon tubes with macrophytes from substation A were frozen at −80 °C. DNA extractions were performed using the DNeasy PowerSoil Pro Kit (Qiagen, Hilden, Germany). The sediment and macrophyte samples were thawed and homogenized. A subsample of 500 mg of sediment or 4-6 pieces of a macrophyte of 2–3 cm lengths were transferred into bead-beating tubes. These were placed on ice and sonicated twice for 7 min and bead-beaten for 30 s at 4 m/s. Subsequently, the manufacturer’s instructions were followed and DNA yield was quantified using Picogreen (Thermo Fisher, Waltham, USA).
16S rRNA genes were amplified using primers, covering the prokaryotic V3–V4 hypervariable region71, and 18S rRNA genes by targetting the eukaryotic V4 region (Supplementary Table 3)72, with a PCR protocol derived from Latz et al.73. (Supplementary Table 4). Illumina sequencing adapters were included in the 5’ ends of the 16S and 18S primers (Supplementary Table 3). Phased primers74 were used to increase the complexity of the sequencing libraries. For 16S rRNA genes, forward and reverse primers were phased (CTAGAGT, TAGAGT, etc. for the forward and ACTACTG, CTACTG, etc. for the reverse primer), for 18S rRNA only the forward primer (ATG, TG, G, or no base) was phased. PCR thermal conditions and master mix details are provided (Supplementary Tables 4 & 5). Leftover adapters were removed using the MagSi-NGS PREP Plus Kit (MDKT00010075, magtivio BV., Nuth, the Netherlands). The purified product was indexed through a second PCR (Supplementary Table 4) following the Adapterama indexing scheme75, pooled in equimolar ratios, and sequenced on MiSeq for 16S and 18S rRNA gene metabarcoding by SciLifeLab/NGI (Solna, Sweden). In addition to the MiSeq sequencing, the pelagic 16S rRNA gene libraries were deep-sequenced on NovaSeq 6000 (Illumina Inc, San Diego, CA, US).
Phased primer sequences were removed from the reads using a snakemake pipeline76,77. The pipeline (https://github.com/biodiversitydata-se/amplicon-multi-cutadapt) encompassed: the removal of read-pairs that contain Illumina adapters, exclusion of read-pairs lacking the expected primer sequences located at the 5’ ends of the reads, removal of the primer sequences from the remaining reads and elimination of read-pairs that have misplaced primer sequences. DADA278 was used for denoising, concatenating paired-end reads, and chimera removal. The resulting amplicon sequence variants (ASVs) were taxonomically assigned with DADA2 using PR279 (V. 4.14.0) as a training set for 18S rRNA gene amplicons and Silva 138.180 for 16 S rRNA gene amplicons.
Species-level classification of Vibrio ASVs was achieved by sequence comparison (BLAST, V. 2.13.081) to a custom database. This database includes 16S rRNA gene sequences of complete Vibrio genomes from RefSeq82 (51 species, 317 strains—including 22 V. vulnificus strains), 41 draft V. vulnificus genomes from clinical isolates from the Baltic Sea region11,83, and 84 draft V. vulnificus genomes from environmental Baltic Sea isolates. In order for an ASV to obtain a species-level assignment to a Vibrio spp., we required perfect (100% identity) alignment of the full ASV to 16 S rRNA genes of a single species in the custom database. None of the ASVs that perfectly matched to V. vulnificus 16S rRNA genes also matched perfectly to 16S genes of other species.
In order to obtain derived V. vulnificus cells mL−1, the integrated relative abundace of V. vulnificus ASVs was multiplied by the flow-cytometry measured heterotrophic cells per mL−1.
Quantification of V. vulnificus
V. vulnificus was quantified using ddPCR (QX200™ Droplet Digital PCR System, Bio-Rad, München, Germany) targeting the vvhA gene84 (Supplementary Table 3). PCR thermal conditions and master mix details are provided (Supplementary Tables 4 & 5). Droplets were generated using the QX100 droplet generator (Bio-Rad, Hercules, USA). Emulsified samples were transferred to a 96-well plate and sealed by a pierceable foil hot seal (BioRad, 181–4040) using PX1 PCR Plate Sealer™ (Bio-Rad, Hercules, USA) (5 s at 180 °C). The PCR was performed using a Bio-Rad C1000 Touch™ thermal cycler (Supplementary Table 4). Subsequently, the plate was analyzed with the QX100 droplet reader using the Quantasoft 1.74.09.17 software. Positive and negative controls were 50 ng of DNA of a V. vulnificus isolate and of a V. harveyi isolate, respectively. In the contamination control, template DNA was substituted with DEPC water.
Statistical analyses and machine learning
To assess the Baltic Sea-wide differences in V. vulnificus abundance between substations A (N = 42), B (N = 45), and C (N = 42) of seagrass stations, a two-sided Wilcoxon rank sum test, assuming independence and equal variance, using the rstatix (V. 0.7.2) package in R (V. 4.3.0)85 was performed and holm correction for multiple testing was applied. Additionally, heterotrophic bacterial cell abundances and green CFUs, inside and outside of the seagrass meadows, were compared. Sample sizes for comparison between substations are consistent for the water column and sediment. No repeat measurements were performed, every data point is a distinct sample.
Three separate linear models for V. vulnificus were used to compare the explanatory power (R2) of the traditional predictor temperature with the more integrated predictor eutrophication index and their combination, using averaged vvhA gene copies mL−1 (log10 transformed) per substation as a response.
To identify the environmental conditions associated with high V. vulnificus abundance, two random forests (RF) in combination with a recursive feature elimination (RFE) algorithm were employed in caret (V. 6.0.94)86 predicting both the vvhA gene copies mL−1 and the derived V. vulnificus cells mL−1. These models used sequencing and environmental data as predictors. Combining RF and RFE addressed correlated variables in this high-dimensional dataset and was able to detect complex and non-linear relationships between measured variables and V. vulnificus87,88.
Prior to model training, the 16S and 18S rRNA relative gene proportions were pre-processed: Rare (absent in at least 25 samples) for both datasets were removed. All ASVs taxonomically classified as Vibrio were removed from the predictors. After pre-processing, the data set included 964 predictors, namely 314 eukaryotic, 627 prokaryotic, 14 physico-chemical, 7 biological ones, substation and macrophyte type. Samples collected from the same station were grouped, to avoid data leakage between test and train data.
Both RF models consisted of 2000 trees with 30 randomly sampled variables as candidates at each split. The model was trained and evaluated using 10-fold cross-validation on a dataset of 52 observations, each representing the average of three biological replicates. Performance metrics used for evaluation included the mean absolute error (MAE), average absolute difference between predicted and actual values, root mean squared error (RMSE), a measure of the differences between the values predicted by the model and the actual values, and coefficient of determination (R2) of the 10-fold cross-validation. The top 10 predictors of both models are discussed.
Eutrophication index
A eutrophication index, defined as the organic matter availability in an ecosystem89, was derived by performing a principal component analysis (PCA), using the “FactoMineR (V 2.8)” R package90, on the environmental parameters DOC, POC, PON, DN, chl-a, NO3−, and PO43−. All organic nutrients strongly aligned with principal component one, explaining 63%, which was chosen as the eutrophication index (Supplementary Fig. 9).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
16S and 18S rRNA gene data were archived in the European Nucleotide Archive under the accession number PRJEB6822291 in compliance with the Minimal Information about any (X) Sequence (MIxS) standard92 through the brokerage service GFBio93. Environmental data are available at IOWMeta (doi.io-warnemuende.de/10.12754/data-2023-0010)94. The reference V. vulnificus database is available under https://doi.org/10.5281/zenodo.1087510895.
Code availability
All custom code used for the bioinformatics processing,the machine learning and analysis of the flow cytometry data in this study is available at https://github.com/lfdelzam/ASV_dada2_chunck/ and https://git.io-warnemuende.de/riedinge/Baltvib_RF_RFE.
References
Reissmann, J. H. et al. Vertical mixing in the Baltic Sea and consequences for eutrophication—A review. Prog. Oceanogr. 82, 47–80 (2009).
Team, B. I. A. et al. Second Assessment of Climate Change for the Baltic Sea Basin. 291-292 (Springer, 2015).
Meier, H. E. M. et al. Climate change in the Baltic Sea region: A summary. Earth Syst. Dyn. 13, 457–593 (2022).
Rutgersson, A. et al. Natural hazards and extreme events in the Baltic Sea region. Earth Syst. Dyn. 13, 251–301 (2022).
Baker-Austin, C. et al. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Chang. 3, 73–77 (2013).
Baker-Austin, C., Trinanes, J., Gonzalez-Escalona, N. & Martinez-Urtaza, J. Non-cholera vibrios: the microbial barometer of climate change. Trends Microbiol. 25, 76–84 (2017).
Frank, C., Littman, M., Alpers, K. & Hallauer, J. Vibrio vulnificus wound infections after contact with the Baltic Sea, Germany. Wkly. releases 11, 3024 (2006).
Ruppert, J. et al. Two cases of severe sepsis due to Vibrio vulnificus wound infection acquired in the Baltic Sea. Eur. J. Clin. Microbiol. Infect. Dis. 23, 912–915 (2004).
Brehm, T. T. et al. Non-cholera Vibrio species — currently still rare but growing danger of infection in the North Sea and the Baltic Sea. Internist 62, 876–886 (2021).
Linkous, D. A. & Oliver, J. D. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174, 207–214 (1999).
Amato, E. et al. Epidemiological and microbiological investigation of a large increase in vibriosis, northern Europe, 2018. Eurosurveillance 27, 2101088 (2022).
Blackwell, K. D. & Oliver, J. D. The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina Estuaries. J. Microbiol. 46, 146–153 (2008).
Banakar, V. et al. Temporal and spatial variability in the distribution of Vibrio vulnificus in the Chesapeake Bay: a hindcast study. Ecohealth 8, 456–467 (2011).
Johnson, C. N. et al. Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl. Environ. Microbiol. 78, 7249–7257 (2012).
Randa, M. A., Polz, M. F. & Lim, E. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl. Environ. Microbiol. 70, 5469–5476 (2004).
Eiler, A., Gonzalez-Rey, C., Allen, S. & Bertilsson, S. Growth response of Vibrio cholerae and other Vibrio spp. to cyanobacterial dissolved organic matter and temperature in brackish water. FEMS Microbiol. Ecol. 60, 411–418 (2007).
Eiler, A., Johansson, M. & Bertilsson, S. Environmental influences on Vibrio populations in northern temperate and boreal coastal waters (Baltic and Skagerrak Seas). Appl. Environ. Microbiol. 72, 6004–6011 (2006).
Rippey, S. R. Infectious diseases associated with molluscan shellfish consumption. Clin. Microbiol. Rev. 7, 419–425 (1994).
Yun, N. R. & Kim, D. M. Vibrio vulnificus infection: A persistent threat to public health. Korean J. Intern. Med. 33, 1070–1078 (2018).
Baker‐Austin, C., Stockley, L., Rangdale, R. & Martinez‐Urtaza, J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ. Microbiol. Rep. 2, 7–18 (2010).
Haenen, O. L. M. et al. Vibrio vulnificus outbreaks in Dutch eel farms since 1996: Strain diversity and impact. Dis. Aquat. Organ. 108, 201–209 (2014).
Lamb, J. B. et al. Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science. 355, 731–733 (2017).
Reusch, T. B. H. et al. Lower Vibrio spp. abundances in Zostera marina leaf canopies suggest a novel ecosystem function for temperate seagrass beds. Front. Mar. Sci. 168, 1–6 (2021).
Fonseca, M. S., Fisher, J. S., Zieman, J. C. & Thayer, G. W. Influence of the seagrass, Zostera marina L., on current flow. Estuar. Coast. Shelf Sci. 15, 351–364 (1982).
Nowell, A. R. M. & Jumars, P. A. Flow environments of aquatic benthos. Annu. Rev. Ecol. Syst. 15, 303–328 (1984).
Worcester, S. E. Effects of eelgrass beds on advection and turbulent mixing in low current and low shoot density environments. Mar. Ecol. Prog. Ser. 126, 223–232 (1995).
Peterson, B. J. & Heck, K. L. Jr Positive interactions between suspension-feeding bivalves and seagrass a facultative mutualism. Mar. Ecol. Prog. Ser. 213, 143–155 (2001).
Gonzalez-Ortiz, V. et al. Interactions between seagrass complexity, hydrodynamic flow and biomixing alter food availability for associated filter-feeding organisms. PLoS One 9, e104949 (2014).
Bodhaguru, M. et al. Screening, partial purification of antivibriosis metabolite sterol-glycosides from Rhodococcus sp. against aquaculture associated pathogens. Microb. Pathog. 134, 103597 (2019).
Brumfield, K. D. et al. Environmental Factors Influencing Occurrence of Vibrio parahaemolyticus and Vibrio vulnificus. Appl. Environ. Microbiol. 89, e00307–e00323 (2023).
Hsieh, J. L., Fries, J. S. & Noble, R. T. Vibrio and phytoplankton dynamics during the summer of 2004 in a eutrophying estuary. Ecol. Appl. 17, S102–S109 (2007).
Brumfield, K. D. et al. Genomic diversity of Vibrio spp. and metagenomic analysis of pathogens in Florida Gulf coastal waters following Hurricane Ian. MBio 14, e01476–23 (2023).
Matcher, G., Lemley, D. & Adams, J. Bacterial community dynamics during a harmful algal bloom of Heterosigma akashiwo. Aquat. Microb. Ecol. 86, 153–167 (2021).
Greenfield, D. I. et al. Temporal and environmental factors driving Vibrio Vulnificus and V. Parahaemolyticus populations and their associations with harmful algal blooms in South Carolina detention ponds and receiving tidal creeks. GeoHealth 1, 306–317 (2017).
Hugerth, L. W. et al. Metagenome-assembled genomes uncover a global brackish microbiome. Genome Biol. 16, 1–18 (2015).
Bunse, C. et al. Spatio-temporal interdependence of bacteria and phytoplankton during a Baltic Sea spring bloom. Front. Microbiol. 7, 517 (2016).
Wallentinus, I. Comparisons of nutrient uptake rates for Baltic macroalgae with different thallus morphologies. Mar. Biol. 80, 215–225 (1984).
El-Hadary, M. H., Elsaied, H. E., Khalil, N. M. & Mikhail, S. K. Molecular taxonomical identification and phylogenetic relationships of some marine dominant algal species during red tide and harmful algal blooms along Egyptian coasts in the Alexandria region. Environ. Sci. Pollut. Res. 29, 53403–53419 (2022).
Hamamoto, Y. & Honda, D. Nutritional intake of aplanochytrium (labyrinthulea, stramenopiles) from living diatoms revealed by culture experiments suggesting the new prey–predator interactions in the grazing food web of the marine ecosystem. PLoS One 14, 1–23 (2019).
Reñé, A. et al. The new chytridiomycete Paradinomyces triforaminorum gen. et sp. nov. co-occurs with other parasitoids during a Kryptoperidinium foliaceum (Dinophyceae) bloom in the Baltic Sea. Harmful Algae 120, 102352 (2022).
Thomsen, H. A., Hällfors, G., Hällfors, S. & Ikävalko, J. New observations on the heterotrophic protists genus Thaumatomastix (Thaumatomastigaceae, Protista incertae sedis) with particular emphasis on material from the Baltic Sea. Ann. Bot. Fenn. 30, 87–108 (1993).
Anschütz, A. A., Flynn, K. J. & Mitra, A. Acquired phototrophy and its implications for bloom dynamics of the Teleaulax-Mesodinium-Dinophysis-complex. Front. Mar. Sci. 8, 1–18 (2022).
Altenburger, A. et al. Dimorphism in cryptophytes—The case of Teleaulax amphioxeia/Plagioselmis prolonga and its ecological implications. Sci. Adv. 6, 1–9 (2020).
Matz, C. et al. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. USA. 102, 16819–16824 (2005).
Worden, A. Z. et al. Trophic regulation of Vibrio cholerae in coastal marine waters. Environ. Microbiol. 8, 21–29 (2006).
Main, C. R., Salvitti, L. R., Whereat, E. B. & Coyne, K. J. Community-Level and species-specific associations between phytoplankton and particle-associated Vibrio species in delaware’s inland bays. Appl. Environ. Microbiol. 81, 5703–5713 (2015).
HELCOM. HELCOM Baltic Sea Action Plan (adopted by the HELCOM Ministerial meeting, Krakow, Poland 15th November 2007).
Ranft, S. et al. Eutrophication assessment of the Baltic Sea Protected Areas by available data and GIS technologies. Mar. Pollut. Bull. 63, 209–214 (2011).
Voss, M. et al. History and scenarios of future development of Baltic Sea eutrophication. Estuar. Coast. Shelf Sci. 92, 307–322 (2011).
Rydin, E., Kumblad, L., Wulff, F. & Larsson, P. Remediation of a eutrophic bay in the Baltic Sea. Environ. Sci. Technol. 51, 4559–4566 (2017).
Semenza, J. C. et al. Environmental suitability of Vibrio infections in a warming climate: an early warning system. Environ. Health Perspect. 125, 107004 (2017).
Klappenbach, J. A., Saxman, P. R., Cole, J. R. & Schmidt, T. M. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29, 181–184 (2001).
Stoddard, S. F., Smith, B. J., Hein, R., Roller, B. R. K. & Schmidt, T. M. rrn DB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43, D593–D598 (2015).
Louca, S., Doebeli, M. & Parfrey, L. W. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome 6, 1–12 (2018).
Yildiz, F. H. & Visick, K. L. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17, 109–118 (2009).
Joseph, L. A. & Wright, A. C. Expression of Vibrio vulnificus capsular polysaccharide inhibits biofilm formation. J. Bacteriol. 186, 889–893 (2004).
Marco-Noales, E., Milán, M., Fouz, B., Sanjuán, E. & Amaro, C. Transmission to eels, portals of entry, and putative reservoirs of Vibrio vulnificus serovar E (Biotype 2). Appl. Environ. Microbiol. 67, 4717–4725 (2001).
McDougald, D., Lin, W., Rice, S. & Kjelleberg, S. The role of quorum sensing and the effect of environmental conditions on biofilm formation by strains of Vibrio vulnificus. Biofouling 22, 161–172 (2006).
Ugarelli, K., Chakrabarti, S., Laas, P. & Stingl, U. The seagrass holobiont and its microbiome. Microorganisms 5, 1–28 (2017).
Guan, C. et al. Identification of rosmarinic acid and sulfated flavonoids as inhibitors of microfouling on the surface of eelgrass Zostera marina. Biofouling 33, 867–880 (2017).
Cúcio, C., Engelen, A. H., Costa, R. & Muyzer, G. Rhizosphere microbiomes of European seagrasses are selected by the plant, but are not species specific. Front. Microbiol. 7, 1–15 (2016).
Möller, L., Kreikemeyer, B., Luo, Z.-H., Jost, G. & Labrenz, M. Impact of coastal aquaculture operation systems in Hainan island (China) on the relative abundance and community structure of Vibrio in adjacent coastal systems. Estuar. Coast. Shelf Sci. 233, 106542 (2020).
Webb, S. J., Rabsatt, T., Erazo, N. & Bowman, J. S. Impacts of Zostera eelgrasses on microbial community structure in San Diego coastal waters. Elem Sci Anth. 7, 11 (2019).
Grasshoff, K., Kremling, K., Ehrhardt, M. (eds.), Methods of Seawater Analysis − 3rd edition. Wiley-VCH, 159-228 (1999)
Lysiak-Pastuszak, E. & Krysell, M. Chemical measurements in the Baltic Sea: guidelines on quality assurance. 35, 146 - 149 (2004).
2019). HELCOM. Guidelines for monitoring of chlorophyll a.
Kirchner, S. et al. Pentaplexed quantitative real-time PCR assay for the simultaneous detection and quantification of botulinum neurotoxin-producing clostridia in food and clinical samples. Appl. Environ. Microbiol. 76, 4387–4395 (2010).
Gasol, J. M. & Del Giorgio, P. A. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64, 197–224 (2000).
Lo, K., Hahne, F., Brinkman, R. R. & Gottardo, R. flowClust: a Bioconductor package for automated gating of flow cytometry data. BMC Bioinformatics 10, 1–8 (2009).
Monaco, G. et al. flowAI: automatic and interactive anomaly discerning tools for flow cytometry data. Bioinformatics 32, 2473–2480 (2016).
Herlemann, D. P. R. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579 (2011).
Balzano, S., Abs, E. & Leterme, S. C. Protist diversity along a salinity gradient in a coastal lagoon. Aquat. Microb. Ecol. 74, 263–277 (2015).
Latz, M. A. C. et al. Short‐and long‐read metabarcoding of the eukaryotic rRNA operon: evaluation of primers and comparison to shotgun metagenomics sequencing. Mol. Ecol. Resour. 22, 2304–2318 (2022).
Fadrosh, D. W. et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2, 1–7 (2014).
Glenn, T. C. et al. Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext). PeerJ 7, e7755 (2019).
Köster, J. & Rahmann, S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522 (2012).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Guillou, L. et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2012).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Brehm, T. T. et al. Heatwave-associated Vibrio infections in Germany, 2018 and 2019. Eurosurveillance 26, 2002041 (2021).
Panicker, G., Vickery, M. C. L. & Bej, A. K. Multiplex PCR detection of clinical and environmental strains of Vibrio vulnificus in shellfish. Can. J. Microbiol. 50, 911–922 (2004).
Kassambara, A. rstatix: Pipe-friendly framework for basic statistical tests. R package version 0.7.2, https://rpkgs.datanovia.com/rstatix/ (2023).
Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 28, 1–26 (2008).
Breiman, L. E. O. Random Forests. Mach. Learn. 45, 5–32 (2001).
Gregorutti, B., Michel, B. & Saint-Pierre, P. Correlation and variable importance in random forests. Stat. Comput. 27, 659–678 (2017).
Nixon, S. W. Coastal marine eutrophication: A definition, social causes, and future concerns. Ophelia 41, 199–219 (1995).
Lê, S., Josse, J. & Husson, F. FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25, 1–18 (2008).
Labrenz M. et al. 16S and 18S rRNA amplicon sequencing of coastal microbial communities across the salinity gradient of the Baltic Sea in pelagic, benthic, and biofilm environments. https://www.ebi.ac.uk/ena/data/view/PRJEB68222 (2024).
Yilmaz, P. et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420 (2011).
Diepenbroek, M. et al. Towards an integrated biodiversity and ecological research data management and archiving platform: the German federation for the curation of biological data (GFBio). Inform. 232, 1711–1724 (2014).
Labrenz M. et al. The occurrence of Vibrio spp. in the salinity gradient of shallow coastal waters of the Baltic Sea—data set including environmental and microbiological data. (https://doi.org/10.12754/data-2023-0010) (2023).
Delgado, L. F., Labrenz, M., & Andersson, A. F. Vibrio 16S rRNA gene sequences (v1.0). Zenodo. https://doi.org/10.5281/zenodo.10875108 (2024).
Acknowledgements
This work resulted from the BiodivERsA project “Pathogenic Vibrio bacteria in the current and future Baltic Sea waters: mitigating the problem” (BaltVib), funded by the European Union and the Federal Ministry of Education and Research, Germany (grant 16LC2022A), the Innovation Fund Denmark (grant 0156-00001B), the Estonian Research Council (grant T210076PKKH / P200028PKKH), the Research Council of Lithuania (grant S-BIODIVERSA-21−1), the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning FORMAS (grant 2020-02366), the Polish National Science Centre (grant 2020/02/Y/NZ8/00009) and the Academy of Finland (grant 344743). The authors acknowledge support from the National Genomics Infrastructure in Genomics Application Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. We are grateful to the crew and captain of the R/V Elisabeth Mann Borgese (EMB283), the research divers of the Leibniz Institute for Baltic Sea Research, the University of Rostock, and the Estonian University of Life Sciences, Jonas Nilsson for assisting with sampling in Sweden, and to Adam Woźniczka and Jarone Pinhassi for providing lab space. Lastly, the authors would like to thank Rita R. Colwell (University of Maryland, USA) for constructive criticism of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
M.L. initiated and supervised the project, M.L., L.R., A.F.A., D.P.R.H., C.P., T. B. H. R., M.R., G.G., D.J.R. and M.K. designed the project; M.L., D.J.R., V.F.J., D.P.R.H., K.P., S.K., A.S., G.G. and H.B.B. performed the sampling and conducted the laboratory work; L.F.D. and A.F.A. performed the bioinformatics processing; D.J.R., T.S. and C.H. analyzed the data and performed the machine learning; F.B. and A.A. carried out the ddPCR; D.S.B. analyzed the organic nutrients; S.D. performed the multiplex-PCR. The manuscript was written by D.J.R. and M.L. and reviewed by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth and Environment thanks Orr Shapiro and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: José Luis Iriarte Machuca, Joe Aslin, and Clare Davis. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Riedinger, D.J., Fernández-Juárez, V., Delgado, L.F. et al. Control of Vibrio vulnificus proliferation in the Baltic Sea through eutrophication and algal bloom management. Commun Earth Environ 5, 246 (2024). https://doi.org/10.1038/s43247-024-01410-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-024-01410-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.