Abstract
Anthropogenic activities are causing detrimental changes to coastal plants– namely seagrass, mangrove, and tidal marshes. Looking beyond recent times to past vegetation dynamics is critical to assess the response and resilience of an environment to change. Here, we develop a high-resolution multi-proxy approach, providing a new evidence base to decipher long-term change in coastal plant communities. Combining targeted environmental DNA analysis with chemical analysis of soils, we reconstructed 4,000 years of change at a temperate wetland on Torrens Island South Australia and identified an ecosystem shift that occurred ~ 1000 years ago. What was once a subtidal seagrass system shifted to an intertidal mangrove environment that persists at this site today. We demonstrate that high-resolution historical changes in coastal vegetation can be attained using these proxies. This approach could be applied to other ecosystems to improve the way we protect, conserve, and restore vegetated ecosystems.
Similar content being viewed by others
Introduction
Life within the ocean relies on coastal marine plants such as seagrasses, mangroves and tidal marshes to provide habitat, protection and nurseries for a multitude of marine organisms1. These plant environments are also vitally important to modern society for their ability to store carbon, stabilise coastlines, and support food webs, yet they are in decline due to anthropogenic change2. Indeed, the Industrial Revolution era became a turning point for rapid degradation in coastal environments, but these habitats have experienced a history of change over geologic timescales (e.g., sea level rise and fall, climate, mass extinction, and plate tectonics). Uncovering historical change and identifying tipping points in ecosystem conditions are necessary for contextualising present-day changes, understanding natural resilience and successfully restoring degraded coastal habitats3.
Successful restoration relies on what we deem as the natural state of the system. This natural state is arbitrary and is often impacted by the shifting baseline syndrome—what we perceive as the natural state is only what has been measured in our lifetime4. This skewed perspective means restoration goals can often be unattainable or set up to fail. Incorrectly perceiving the natural state of an environment usually stems from a lack of data over meaningful timescales5. Traditional methods for assessing historical change in coastal plants include archival observations, which can be biased and only provide information at the scale of decades6. Information on longer time scales stems from the analysis of multiple proxies, like isotopes, fossils (in the form of plant fragment remains) and pollen trapped in natural archives7. For example, soil core archives can be dated, and historical reconstructions inferred. However, the fossil record is biased toward plants that deposit material readily, and the pollen record may not be reliable in coastal settings, owing to a lack of preservation of pollen from macrophytes such as seagrasses8. While isotopes, fossils and pollen do provide useful information at longer time scales, their use with emerging methods could increase the robustness and accuracy of change assessments in coastal vegetated environments.
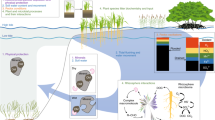
The recovery and analysis of environmental DNA (eDNA) from soil cores is an emerging tool for historical reconstructions of coastal plant communities9,10,11. When these data are coupled with information on the environmental conditions of a site, a detailed picture of environmental change can emerge12. Here, we analysed four soil cores, applying multiple proxies to look at environmental change in a temperate wetland on Torrens Island, South Australia (Fig. 1). This wetland is presently dominated by the grey mangrove (Avicennia marina), and cores were specifically taken here due to the high sedimentation rates and the dense root systems of mangroves that work to stabilise the sediment13. This stabilisation ensures a coherent stratigraphy, justifying their use as soil archives that can be dated. We documented 4000 years of historical change at this site and implemented targeted capture of eDNA, instead of the traditional metabarcoding approach, to capture multiple chloroplast gene regions and improve the ability to detect coastal plants. We combined this with the analysis of chemical proxies to build multiple lines of evidence for past changes. This proof-of-concept study successfully reconstructed millennial ecosystem changes in a temperate coastal wetland, demonstrating the potential for implementing these analyses in other environments to better understand and manage plant communities in the future.
Results
Radiocarbon dating
Radiocarbon (14C) dates were obtained from all four soil cores to assess the time scale of environmental change. The presence of a hiatus in the dating profiles of all cores, as indicated by the jump in 14C dates (Fig. 2a, Supplementary Table 2), precludes the construction of reliable age-depth models for the full core profiles. Instead, age-depth models were developed only for the pre-hiatus time period, where this was specified at 63 cm and were determined by changepoint analyses using principal component 1 scores of stable organic carbon isotope composition (δ13C), % organic carbon content (% Corg) and dry bulk density in g/mL, see below. The pre-hiatus age-depth models were generated for cores 1, 2 and 4 but not core 3 due to the limited number of 14C data pre-hiatus. These models (Supplementary Fig. 2), provide reliable ages up to ~ 1000 Calibrated Years Before the Present (Cal Year BP), thus, we define the pre-hiatus time period as <1000 Cal Year BP and the post-hiatus time period as >1000 Cal Year BP. For post-hiatus ages (and pre-hiatus ages for core 3) the absolute 14C calibrated dates were used to assign approximate age, with the oldest 14C date recorded at ~4200 Cal Year BP. Due to the lack of full age-depth models, decompressed depths (cm) were used to look at changes in plant communities and chemical proxies, where identified changes were matched to the approximate hiatus age of 1000 Cal Year BP.
a Radiocarbon dating results with a dashed line at 63 cm representing the approximate hiatus point. b Changes in high intertidal, intertidal and subtidal vegetation types plotted against decompressed depth using a gam smoothing term with 95% confidence intervals. The dashed line at 63 cm represents the approximate hiatus point. c Breakdown of plant community composition by relative abundance with Cal Year BP, colours indicate vegetation type; Red/orange = high intertidal, green = intertidal and blue = subtidal. Silhouettes (Integration and Application Network, ian.umces.edu/media-library) represent the species detected within each family; Acanthaceae = Avicennia marina, Aizoaceae = Disphyma crassifolium, Chenopodiaceae = Salicornia and Tecticornia (genus level resolution only), Convolvulaceae = Wilsonia humilis, Cymodoceaceae = Amphibolis antarctica, Posidoniaceae = Posidonia australis, Primulaceae = Samolus repens, Ruppiaceae = Ruppia maritima, Scrophulariaceae = Myoporum insulare, Zosteraceae = Zostera nigricaulis. Absolute radiocarbon dates are indicated in brackets with subscript numbers indicating which cores these dates are from. Cores 1, 2 and 4 are plotted up to the dashed line using age-depth models which is defined as the approximate pre-hiatus period and >1000 Cal Year BP as the post-hiatus period.
Environmental DNA
Plant families detected using eDNA for all soil cores were divided into subtidal (Cymodoceaceae, Posidoniaceae, Ruppiaceae and Zosteraceae), intertidal (Acanthaceae and Chenopodiaceae) and high intertidal (Scrophulariaceae, Aizoaceae, Primulaceae and Convolvulaceae). Intertidal and high intertidal plants were present throughout all soil core profiles (Fig. 2b), but as intertidal plants decreased in relative abundance with depth, subtidal plants increased. Subtidal vegetation increased exponentially (edf = 2.3, P < 0.05) but intertidal vegetation decreased linearly (edf = 1.0, P < 0.05) and high intertidal vegetation did not change (edf = 1.988, P = 0.3) in all four soil cores (non-significant (P = 1) random effect of core) which illustrates a changing community structure in the soil profile (Fig. 2b). The community structure in older sections of the cores (>1000 years) comprised subtidal plants, particularly the seagrass Zostera nigricaulis (Zosteraceae), which was only observed once in core 1 but multiple times in cores 2, 3 and 4 with maximum relative abundances of 32%, 40%, 66% and 32% respectively (Fig. 2c). Additional subtidal plants detected in older sections of the cores (all <10% of the total community) were Posidonia australis (Posidoniaceae) and Ruppia maritima (Ruppiaceae) in cores 2 and 4, and Amphibolis antarctica (Cymodoceaceae) in core 2, Fig. 2c. The plant community in younger sections of the cores (<1000 years) was dominated by the intertidal grey mangrove species Avicennia marina, comprising 100% of the community in some instances, Fig. 2c. Additional detections of intertidal species were only obtained at a single point in core 1, which were of the family Chenopodiaceae. High intertidal plant communities were detected in the soil profile of all four cores but never exceeded 35% of the total community. These high intertidal plants consisted of Myoporum insulare (Scrophulariaceae) and Wilsonia humilis (Convolvulaceae), with additional plant species detected at a single point in core 1, Disphyma crassifolium (Aizoaceae) and Samolus repens (Primulaceae) (Fig. 2c).
%Corg, δ13C, dry bulk density
The measured variables δ13C and dry bulk density were higher in the upper sections of all four cores, and %Corg was lower (Supplementary Fig. 3), suggesting a shift in environmental conditions along the soil profiles corresponding to ~1000 Cal Year BP (Fig. 3a). These variables were analysed with a Principal Component Analysis where principal component 1 explained 73.9% of the variance. All variables contributed similarly to principal component 1 (PC1), with δ13C contributing the greatest to the observed change (37%), followed closely by dry bulk density (33%) and %Corg (30%) (Supplementary Table 3). All measured variables were strongly correlated with PC1 (R2 > 0.6), supporting the use of this principal component for identifying significant change points, with one identified at 63 cm (cores 1–4), defining a shift in environmental conditions.
a Dry bulk density, δ13C and %Corg and b elements; Moly ratio, Ti/Ca, Br/Cl, Sr (normalised) and Ca/Fe for soil Cores 1–4 and Core 3, respectively. Principal component 1 explained 73.9% of the variance in a and 76.4% in (b). How well each variable correlated to PC1 is shown by the R2 values. The dashed lines are placed at the depths of significant change points identified by change point analyses, 63 cm in a. and 60 cm in (b). Colours indicate the dominant vegetation type found <1000 Cal Year BP, green = intertidal and >1000 Cal Year BP, blue = subtidal with silhouettes depicting the dominant species here, Avicennia marina and Zostera nigricaulis, respectively (Integration and Application Network, ian.umces.edu/media-library).
X-ray fluorescence
Chemical elements measured using X-ray fluorescence (XRF) confirmed changing environmental conditions down the soil profile of core 3 (Fig. 3b). Calcium/Iron (Ca/Fe), a proxy for carbonate productivity14 and Strontium (Sr) (normalised to Molybdenum incoherent + coherent scattering), a marine indicator15, increased with depth. The chemical proxies Molybdenum Compton (incoherent) and Rayleigh (coherent) scattering (Moly ratio) and Bromine/Chlorine (Br/Cl), which are indicators of organic matter content16,17, and Titanium/Calcium (Ti/Ca), a proxy for terrigenous sediment supply15,17, all decreased with depth (Supplementary Fig. 4). When these variables were analysed using principal components, principal component 1 (PC1) explained 76.4% of the variance. Sr contributed the most to this variation (24%, Supplementary Table 3), followed by Br/Cl, Moly ratio, and Ca/Fe while Ti/Ca was the lowest contributor (13%). All variables strongly correlated with PC1 (R2 > 0.7) except Ti/Ca, which was only moderately correlated with PC1 (R2 = 0.5). This justified using PC1 to identify significant change points, with one of them identified at 60 cm (core 3), supporting a change in elemental composition of the soil profile where this is estimated to have occurred at ~1000 Cal Year BP.
Discussion
Uncovering the dynamics of coastal plant communities over hundreds to thousands of years is important to understand their responses and resilience to current and forecasted changing environmental conditions18. In this study, we successfully coupled targeted capture of eDNA with soil chemical analyses to obtain high-resolution reconstructions of coastal plant changes through time. We identified a historical ecosystem shift ~1000 years ago from a subtidal seagrass environment to the present-day intertidal mangrove habitat. Using the combined approach of targeted eDNA and chemical analyses, we built multiple lines of evidence for the 4000-year history of this temperate coastal wetland (Fig. 4).
The oldest 14C date is shown on the x-axis to indicate the timescale of change. Indicative water level is shown on the y-axis. Wedges on the bottom of the figure indicate the measured variables that increased and decreased over time. Changes in the dominant plant community type are indicated by coloured bars at the top of the image. Grey mangrove (Avicennia marina) was detected in the full soil profiles of each core, but this is likely not a true signal in older time periods when a subtidal seagrass community was detected. This signal likely stems from mangrove roots growing deep into the sediment (per observation), which is represented in the diagram by mangrove soil/roots extending into the seagrass habitat.
The present-day habitat of our study site is a temperate wetland dominated by the grey mangrove (Avicennia marina), which was captured in the eDNA results through the full profiles of all four cores. Given that chemical analyses indicate this site was a subtidal system over 1000 years ago, mangrove presence is unlikely at that time. It is more likely that this signal is an artefact of more recent mangrove roots growing deep into the sediment19 and shedding DNA into the older sediment layers. As a matter of fact, laboratory observations confirmed the presence of roots in deeper sections of the cores. While mangrove roots can help to stabilise sediments, leading to a clear stratigraphy, this may have led to a false positive eDNA signal for mangroves. Analysing additional proxies, such as pollen, could support the hypothesis that mangroves were not present at this site over 1000 years ago during a time when subtidal conditions would have impeded their establishment. Indeed, eDNA data confirm the presence of multiple subtidal seagrass species, Z. nigricaulis, R. maritima, P. australis and A. antarctica at >1000 Cal Year BP. The first two species can be found in shallow water, and the latter two are typically found in deeper waters (>1 m)20. These results indicate that sea-level was likely higher at that time and is supported by previous evidence21 that this was the case ~6000 years ago, and it is hypothesised the sea level slowly dropped to present-day conditions.
The sea level at the site today supports intertidal communities with some high intertidal plants occurring in nearby adjacent areas. This could explain the eDNA signals from high intertidal plant families detected through the full profiles of all four soil cores, where this is the result of detrital material being brought into the system, rather than these plants growing at the specific site the cores were taken. Plant material is known to be imported and exported within coastal landscapes and can end up sequestered in the soils of adjacent habitats22. We may have detected eDNA from adjacent habitats due the targeted eDNA approach designed for multiple chloroplast gene regions, which increased our sensitivity beyond traditional single gene approaches23,24. These more traditional (metabarcoding) approaches rely on the amplification of target gene regions using primers. This can cause amplification bias as the corresponding primer binding sites may not be intact in degraded DNA from soil samples and primers can be biased towards amplifying specific taxa25. Targeted capture of eDNA does not rely on this initial amplification step and improves the likelihood of recovering all taxa present in a sample because multiple genes are targeted24,26,27. This could make it difficult to know whether an eDNA signal is from a plant present at the study site or an adjacent one. Fortunately, pairing the eDNA results with chemical analyses identified conditions likely to support mangroves in younger sections of the cores (<1000 years) and seagrasses in older sections (>1000 years).
Chemical analyses characterised the older sections of the soil cores as a marine environment and younger sections as having a marine and terrestrial influence. This intertidal environment was characterised by high levels of organic matter as evidenced by the %Corg results and supported by higher values for the Moly ratio16. Higher organic matter here was also observed in the greater Br/Cl values, signifying marine organic matter15, and a decrease in this ratio downcore corresponds to an increase in salinity17, indicating a more marine environment in older sediment layers. Increased values of Sr validate this15, as well as a higher ratio of Ca/Fe—both are indicators of marine productivity14,15. Seagrass presence is supported here by higher values of δ13C, and while we could not use these values to reliably infer plant community presence (as δ13C values can be influenced by mangrove roots28 and organic matter diagenesis29) we can conclude that older sections of the core (>1000 Cal Year BP) were more marine (seagrass) in nature than younger sections (<1000 Cal Year BP) which had a more terrestrial influence. The terrestrial signal stems from the higher Ti/Ca, which is a proxy for terrigenous sediment supply15,17. However, we observed a spike in this ratio at ~25 cm (Supplementary Fig. 4), explaining the low correlation to PC1. This proxy may be driven by a specific event or a changing sediment regime that occurred at this time21. Regardless, the higher values of Ti/Ca in the younger sections of the core support an intertidal environment. These characteristic environmental conditions validate that this site was a marine environment that supported a subtidal seagrass community up until ~1000 years ago when it shifted to an intertidal system as the sea level dropped and mangroves colonised the area.
The environmental conditions of the four soil core profiles provide a framework to interpret the eDNA results and by combining these proxies, a holistic picture of change has emerged. This picture describes a previous community of subtidal plants and a higher sea level than the present-day that then shifted to an environment with a lower sea level where mangroves could thrive. There is evidence to suggest this shift occurred due to tectonic uplift30, dramatically reducing sea level and paving the way for mangroves to colonise the area, which would have exposed the soil surface to erosion and could explain the gap (hiatus) in the dating profile. This did not necessarily need to be a sudden event, as sea level could have dropped gradually due to climate changes, leading to erosion of the soil from exposure or the exposed soil could have been removed during a major storm. The possible causes of the ecosystem shift are difficult to decipher, but the synergies between targeted capture of eDNA and chemical proxies of the soil, enabled this shift to be detected and a clear colonisation of mangroves into seagrass habitat was documented. These multiple lines of evidence for historical dynamics in coastal plants can be employed in other vegetated environments prone to change i.e., coastal sites at the interface of sea level rise or permafrost areas in regions that experienced climatic warming, to develop high-resolution historical reconstructions. This will lead to an improved understanding of long-term change dynamics facilitating the protection, conservation, and restoration of vegetated ecosystems.
Methods
Study site
Four biological replicate soil cores were collected from Torrens Island, South Australia (−34.7929, 138.5265). This site is characterised by a Mediterranean climate with hot, dry summers and mild, wet winters. Historically, Torrens Island was occupied by the Kaurna people31, the Traditional Owners of the land. The island is currently unoccupied, and most of the site was declared a Conservation Park in 1972 to protect the diverse animal and plant life found there, including mangroves, temperate saltmarshes and seagrasses. Common species include Atriplex paludosa, Wilsonia humilis, Disphyma crassifolium, Samolus repens, Suaeda australis, see32 for a complete list); samphire shrublands dominated by Sarcocornia, Salicornia and Tecticornia species; and one mangrove species (Avicennia marina). The seagrass communities surrounding the island include Posidonia spp., Zostera spp., Amphibolis spp. and Halophila spp.33
Soil core collection and processing
Four soil cores (~ 1 m long and 7.5 cm diameter) were collected using PVC pipes by manual percussion and rotation in 2017 (TI2.2017; Core 1) and 2018 (TIN1.2018, Core 2; TIN2.2018, Core 3; TIN3.2018, Core 4). Compression during coring was assessed by measuring the length of the core protruding above the soil’s surface both inside and outside the PVC pipe before retrieving the core. These measurements were used to calculate a compression rate that enabled back-calculation of decompressed sample depths after the core was sectioned34. Following retrieval, each core was sealed at both ends, transported vertically, and stored at 4 °C until it was processed in the laboratory. Core 3 was sliced lengthways, with an intact half retained for scanning X-ray fluorescence (details below) and the remaining half treated as per the following description for all other cores. Throughout the soil sample preparation, steps were taken to reduce eDNA contamination, including wiping all laboratory benches with bleach, water and ethanol and cleaning all equipment with ethanol between each sample35.
The soil cores were sliced into 0.5 cm thick increments in the top 20 cm and at 1 cm thick increments for the remainder of each core. Each slice was weighed, then a 0.5 g sub-sample was taken for DNA analyses and the remainder of the soil was dried at 60 °C until constant dry weight to estimate dry bulk density (DBD). One sub-sample of the dried bulk soil was used for 14C (age-dating) analysis. Another sub-sample was ground in a ball mill grinder and analysed for soil organic carbon content (%Corg), with some of these sub-samples also being analysed for organic carbon stable isotopes (δ13C). A summary of the analyses done on each core is provided in Table 1, with further details on the sample preparation associated with each analysis described in the sections below.
Environmental DNA extraction
A 0.5 g sub-sample of soil was collected from the middle of each soil slice and was stored at 4 °C in Powerbead solution buffer (QIAGEN) until DNA extraction. Collecting from the middle of the slice helped prevent contamination (as the outside edges had been in contact with the PVC pipe). Each 0.5 g soil sample was divided into two equal-weight (250 mg) replicates (A and B). DNA was extracted from each replicate sample using the DNeasy Powerlyzer Soil Kit (QIAGEN) with zirconia beads and using strict handling protocols to prevent contamination. We chose this extraction kit based on in-house trials and previous research24,36 and used zirconia beads instead of the standard glass beads to ensure plant cells could be properly lyzed (personal observation). Post-extraction is usually followed by sonication in targeted capture analyses for non-degraded samples37, however, due to the degraded nature of DNA in soils38, the impact of sonication on plant community recovery was tested before proceeding. A subset of samples from core 1 was sonicated using a Diagenode Bioruptor® Pico to a size distribution peaking around 400-600 base pairs (bp) (cycle of 15 s on, 90 s off, and repeat 7 times). Sonication reduced the number of unique taxa recovered in older sediment samples due to fragmenting already degraded DNA, whereas in recent samples, sonication did not impact the number of unique taxa recovered (Supplementary Fig. 1). Based on this, all samples were processed without sonication, along with extraction blank controls (2 per 20 samples) to monitor contamination35.
Library preparation and bait capture
These methods are described in detail in24 briefly, library preparation was completed using the NEBNext Ultra II Library preparation kit (New England Biolabs®) with the following modifications: 1/3 the recommended reaction volume (16.7 µL) was used, and custom-made stubby (incomplete, P5 and P7 indexes missing) Y-adaptors (25uM)39 were used at the ligation step. Following adaptor ligation, libraries were amplified using the Q5 Master Mix at the original reaction volume of 50 µL with in-house primers P7 preCap Long and P5 preCap Long (Cycling conditions: [98 °C 10 s, 65 °C 30 s, 72 °C 30 s] × 17 cycles, 72 °C 120 s, 4 °C hold). From here, 2 µL of each uniquely indexed library was examined with gel electrophoresis and pooled into batches of eight samples. These batches were then purified using AMPure XP (at 0.8 × volume concentration).
Bait design was conducted using the RefSeq release of plastid sequences (https://ftp.ncbi.nlm.nih.gov/refseq/release/plastid/: accessed Oct 2017) across ~160 taxa to design baits targeting a set of 20 plastid gene regions for angiosperms (Supplementary Table 1) including standard plant barcoding regions40. The plastid region trnL was not included in this bait set despite its utility in ancient DNA studies41 because the chosen plastid gene regions were believed to offer greater species discrimination. Using Arabidopsis lyrata (Genbank reference NC_034379) as a reference, target regions were extracted from the RefSeq data using Blast (blastn, e value < 1e-50) and were clustered using CD-HIT42 with a 95% identity cut-off, retaining the longest sequence per cluster for probe design. A total of c. 2800 representative sequences, ranging in length from 180–900 bp (mean 370 bp) were used to design c. 15,000 120-mer probe sequences with 2X tiling (i.e., each probe overlaps half its length). Further information can be found in43. Baits were ordered through myBaits®, and hybridisation was performed following their instructions. The hybridisation temperature/time was 65 °C for 48 h to maximise hybridisation of degraded DNA. The hybridised product was amplified using custom P7 and P5 indexed primers designed in-house using cycling conditions: 98 °C 120 s, [98 °C 20 s, 60 °C 30 s, 72 °C 45 s] × 17 cycles, 72 °C 30 s, 4 °C hold. Molarity was calculated between 300 and 800 bp on a 2100 Bioanalyzer (Agilent) and then pooled in equimolar concentration and purified using AMPure XP (New England Biolabs) at 0.7 × concentration. The final library underwent further size selection using a Pippin Prep (Sage Science) with a 1.5% agarose gel cassette set to select between 300 and 600 bp and was quantified using a QuantStudio 6 Flex Real-Time PCR (Thermo Fisher Scientific. The final library was sent to the Garvan Institute of Medical Research (Sydney, Australia) to be sequenced on one lane of an Illumina HiSeq X Ten using 2 × 150 bp chemistry. Further details of library preparation and targeted capture can be found in43.
Bioinformatics
Full details for bioinformatic processing of sequences can be found in24, and all scripts, including the reference sequence database used in this study are available on Github (https://github.com/NicoleRFoster/Targeted-capture-of-eDNA-in-sediment-cores).
For read processing and mapping, raw sequences were demultiplexed based on indexes using Illumina Bcl2fastq v2.18.0. The output Read 1 and Read 2 fastq.gz files were then demultiplexed based on the Y-adaptor internal barcodes using AdapterRemoval v244. PALEOMIX45 was used to trim adaptors (using AdapterRemoval), discard sequences less than 25 bp and trim for ambiguous nucleotides and low-quality base calls. BWA-MEM aligner46 was selected within PALEOMIX as the mapping tool with a mapping quality of 30 and discarding unmapped reads. A specified reference database was made by combining the temperate coastal plant reference database from47 with a curated database from the National Centre for Biotechnology Information48 that contained all plants (both terrestrial and coastal) within the local sampling location. Following mapping, Picard Mark Duplicates (Version 4.0.10.1) was used to remove clonality (duplication in read alignment) and the resulting BAM files were then used to generate VCF files using SAMtools mpileup49, specifying ploidy as 1 (as haplotypic organellar DNA) and filtering for base quality <30, mapping quality <30 and depth <50, outputting N’s if read coverage was not met. Variant calls were normalised with BCFtools norm49 and the consensus caller in BCFtools was then used to call final consensus FASTA files, outputting variants with N’s. All blanks failed to pass filtering and so were discarded.
Despite stringent mapping, some of the gene regions may be unable to distinguish among closely related species (i.e. a gene region may not be able to resolve to species level in certain groups50) or the correct species in the sample is not represented in our database. For example, a specific gene region (i.e., psbA47) may only be capable of separating certain taxa at the family level and therefore, reads may not map to this region for the specific species in the sample, instead mapping to this region for a species not in the sample but from the same family. To overcome this issue, we clustered that data by assigning rankings to consensus FASTA sequences based on the ability of the sequence to distinguish taxa at the Order, Family, Genus or species level. To do this, FASTA files were filtered for length <100 bp and all ambiguity and missing data codes were removed prior to clustering the reads with the reference database using CD-HIT-EST42. The threshold for sequence identity was set to 95% with clustering set to the most similar and alignment was set to cover at least 10% of the representative sequence and 100% of the shorter sequence (-aL 0.1 -aS 1). If the sample consensus sequence clustered broadly with other sequences, it meant either, the gene in question did not have high enough variation to resolve taxonomy below a certain level or the sample consensus sequence contained a high number of ambiguous base calls that confounded species identification, i.e., read depth did not meet the assigned threshold to call a base (depth <50 bp) and instead missing data values were inserted (N’s).
For data processing, a custom script was written in R version 3.5.151 to unpack the output.clstr file from CD-HIT-EST and generate upstream taxonomic assignment for each sample in each cluster. The script identified the highest common taxonomic ranking as an output for each cluster, denoting this as ‘rank’. The final dataset contained assigned sample taxonomy generated from the mapping of reads and a ranking for the level of taxonomic clustering this sequence provided. This script was written using the R packages taxize52, TAI53, dplyr54, stringi55, stringr56 and tidyr57.
Replicate samples A and B were then combined to assess overall gene recovery for each section of the soil cores. Taxa with less than four gene regions recovered were not included in the final community evaluation (informed by recommendations in24). All samples were then combined for each core such that there was a single file documenting species recovery and taxonomic ranking for all samples analysed along the length of each core. These were then converted into separate phyloseq58 objects and data were subset to only include taxonomic rankings at the family level or below (for visualisation purposes—species and genus level assignments were included in the interpretation of results). Taxonomic assignments were then summarised by families to calculate relative abundances as a percentage of the total community composition at each position along each core.
To summarise changes in coastal plant communities over time, we designated three community types to the detected plant families. These were subtidal (seagrasses), intertidal (mangrove and samphire) and high intertidal (other coastal plants). To assess changes in relative abundances of communities with depth across all four cores we fitted a generalised additive model using the gam function in the mgcv package59 in R51. We tested several models and used the lowest AIC and most variance explained by the model to select a final model candidate. This model included relative abundance as the response variable with community type as an explanatory variable, a smoothing term of depth by community type and core as a random factor.
Relative abundance ~ community type + s(depth, by = community type) + s(core, bs = “re”)
The degree of smoothing was estimated using restricted maximum likelihood (REML).
Radiocarbon dating
Radiocarbon (14C) analyses were conducted on three to five bulk soil sub-samples taken from below 22 cm in each core. The sampling depths from each core were selected based on taking one sample from just below the top section of the core (that was thought to be mixed based on previous studies); one sample from near the bottom of the core (the oldest soil); and at least one (and up to three) samples from in between these upper and lower sample positions. Radiocarbon dating was performed on bulk soil samples from Core 1 at the AMS Direct Laboratory (USA) and at the Centre for Accelerator Science, Australian Nuclear Science and Technology Organisation (ANSTO) for the remaining three cores. Carbonates and humic acids were removed by treating the sample with 2 M HCl (at 60 °C) for carbonate removal, a series of sodium hydroxide (NaOH) treatments (at 60 °C) for the removal of humic and a final treatment of 2 M HCl (at room temperature). After drying, the samples were combusted to carbon dioxide (CO2) and reduced to graphite using H2/Fe at 600 °C. The graphite targets were analysed by accelerator mass spectrometry (AMS) using the Vega 1MV NEC accelerator.
Radiocarbon ages were calibrated in OxCal 4.4.460 using a P-Sequence deposition model. Calibrations were performed against the SHCal20 calibration curve61 or a mixed SHCal20/Marine2062 curve where a marine influence is identified. Marine influence was corrected using a local ΔR = −150 ± 59 taken from the Australian ΔR calculator for the Torrens Island region (https://delta-r-calc.jcu.io/)63.
The degree of marine influence was assessed based on the organic δ13C stable isotope ratio (‰) for which lower values are more indicative of terrestrial vegetation and higher values marine64. Some of our samples had more depleted δ13C values than would be expected in a 100% marine environment, indicating a mixed terrestrial/marine carbon source (as expected in estuarine settings, such as Torrens Island). For these samples, we calibrated the dates using mixed curves, based on a value of −25.0‰ being 100% terrestrial, and a value of −10.0‰ being 100% marine. These thresholds assume that the marine organic inputs at our site would be primarily seagrass, and seagrass δ13C stable isotope values typically range from −13.3‰ to −7.3‰65.
Due to the presence of a hiatus in the dating profile of all cores, age-depth models could not be reliably constructed. Thus, we generated only pre-hiatus age-depth models for cores 1, 2 and 4 using the R routine “Bacon” for Bayesian chronology building66 and specifying the end depth of the model at 63 cm, which is the depth identified in the change point analyses using principal component 1 scores for %Corg, δ13C and dry bulk density variables (see below). Owing to the limited number of 14C data pre-hiatus (core 3), it was not reliable to report age-depth models for core 3, nor the post-hiatus periods in cores 1, 2 and 4, and only the absolute 14C calibrated ages are reported to indicate the approximate ages in these cases.
Dry bulk density, organic carbon and stable isotope analyses
Dry bulk density (DBD; g cm−3) was calculated for all depth increments from all four cores (see Table 1 for a number of analyses) using the dry weight data and sample volume. All samples were analysed for organic carbon (%) and stable isotope composition (δ13C).
For organic carbon (%), each dried bulk soil sample was acid-treated to remove inorganic carbon by adding 4% HCl. The sample was then centrifuged at 3400 rpm for 5 min and the supernatant (including acid residues) was carefully removed with a pipette, avoiding resuspension. The pelleted soil sample was then washed with ultrapure water, centrifuged again, and the supernatant carefully removed. Post acid treatment, the remaining soil sample was re-dried at 60 °C until a constant weight was achieved and then placed into a tin capsule for analysis. The % organic carbon was determined on the acid-treated samples using a Costech Elemental Analyser at the University of Hawaii Hilo Analytical Laboratory, USA (Core 1 samples) or a Perkin Elmer EA2400 Series II at the MASS Facility, in the SET Faculty at the University of Adelaide, Australia (samples from Cores 2–4), with calibration against an Acetanilide certified reference standard.
The acid-pre-treated dried bulk soil samples were also used for organic carbon stable isotope analyses (δ13C). A mass of sample equivalent to 100 µg of organic carbon was accurately weighed into a tin capsule based on the organic carbon content determined by the elemental analyses (as described above). The weighed samples from Cores 2–4 were analysed for δ13C using a continuous flow isotope ratio mass spectrometer (Nu Horizon, Wrexham, UK) equipped with an elemental analyser (EA3000, EuroVector, Pavia, Italy) at the MASS Facility in the SET Faculty at the University of Adelaide, Australia, whereas samples from Core 1 were analysed using a Thermo-Finnegan Delta V Isotope Ratio Mass Spectrometer at the University of Hawaii Hilo Analytical Laboratory, USA.
For all samples, the stable isotope ratios were expressed in δ notation as deviations from a standard in parts per mil (‰):
Where Rsample is the ratio of abundance of 13C /12C in the sample, and Rstandard is this ratio in the standard. δ13C was reported relative to the standard Vienna Pee Dee Belemnite (VPDB). All samples were corrected for instrument drift and normalised according to reference values using in-house standards (n = 19 per 60 samples); δ13C = glycine −31.2‰, glutamic acid −16.7‰ & triphenylamine (TPA) −29.2‰ calibrated against USGS and IAEA certified reference materials (USGS40, USGS 41, IAEA-2, NIST 8704).
X-ray fluorescence (XRF)
A full-length ½ longwise section of Core 3 was analysed using an Itrax XRF core scanner at the Australian Nuclear Science and Technology Organisation (ANSTO). This is a non-destructive method for rapidly generating high-resolution elemental profiles using XRF spectrometry. The Itrax XRF scans were performed using a molybdenum tube set at 55 kV and 30 mA with a dwell time of 10 s. A step size of 200 μm was selected to capture elemental variations occurring in laminations observed in the radiographic image. The Itrax XRF analysis produced data for a standard suite of 36 elements and Molybdenum Compton (incoherent) and Rayleigh (coherent) scattering (Moly ratio). Incoherent scattering occurs when the energy of the X-ray photons is greater than the energy binding electrons to the atom nucleus, often occurring for elements H, C, N and O and resulting in a loss of energy. Rayleigh scattering occurs when there is no loss of energy, and therefore, a higher Moly ratio can be an indication of the presence of these elements and a proxy for organic matter content16. From these, we selected five elements/ elemental ratios that were most relevant to our research question; these were Moly ratio, Ti/Ca, Br/Cl, Sr and Ca/Fe where Sr was normalised against Molybdenum incoherent + coherent scattering to reduce the impact of water content and density changes in the soil core67.
Assessing change in DBD, %Corg, δ13C and XRF
To reduce dimensionality and assess consistent downcore trends across DBD, %Corg, and δ13C, we applied Principal Component Analysis (PCA)68 to all four cores. PCA analysis was run in R using the ‘stats’ package51 and the function ‘princomp’ with data centred and scaled. We similarly applied a PCA to the five-element dataset produced by the XRF analysis on core 3, to generate a single data point for each depth increment that represents the variance in all five element ratios: Moly ratio, Ti/Ca, Br/Cl, Sr (normalised) and Ca/Fe.
We subsequently used the outputs from both the PCA analyses (component 1 values for each depth increment) in a changepoint analysis, run in R using the package ‘changepoint’69 and the function ‘cpt.meanvar’. Changepoint analyses statistically assess whether any significant, sustained changes in the mean and variance of a combined dataset have occurred (not assuming constant variance). The test identifies the number of changes and their position in the data series and provides confidence estimates (0–1, with 1 being full confidence) for any changes detected. This approach allowed us to test for any clear inflection or change points in soil biogeochemistry through the core depth profiles.
All plots and figures presented in the results were created in R using the package ggplot70. The conceptual model of the system, which is based on the suite of data produced by our multiple analyses, was generated using the software Adobe Illustrator.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Environmental DNA sequence data that support the findings of this study have been deposited in Genbank (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA749388/) with the accession codes SAMN26810607- SAMN26810698; NCBI SRA; and TLS: PRJNA749388. All chemical data that support the findings of this study and the radiocarbon dating results have been deposited on Github https://github.com/NicoleRFoster/Targeted-capture-of-eDNA-in-sediment-cores.
Code availability
All scripts for processing the environmental DNA sequence data and reference libraries used for mapping sequences are available on Github https://github.com/NicoleRFoster/Targeted-capture-of-eDNA-in-sediment-cores.
References
Barbier, E. B. et al. The value of estuarine and coastal ecosystem services. Ecol. Monogr. 81, 169–193 (2011).
Halpern, B. S. et al. Recent pace of change in human impact on the world’s ocean. Sci Rep 9, 11609 (2019).
Bálint, M. et al. Environmental DNA time series in ecology. Trends Ecol. Evol. 33, 945–957 (2018).
Pauly, D. Anecdotes and the shifting baseline syndrome of fisheries. Trends Ecol. Evol. 10, 430 (1995).
McClenachan, L., Ferretti, F. & Baum, J. K. From archives to conservation: why historical data are needed to set baselines for marine animals and ecosystems. Conserv. Lett. 5, 349–359 (2012).
Alleway, H. K. et al. The shifting baseline syndrome as a connective concept for more informed and just responses to global environmental change. People Nat. 5, 885–896 (2023).
Powell, T. M. & Steele, J. H. Ecological Time Series. (Springer Science & Business Media, 2012).
López-Sáez, J. A. et al. Palaeoecological potential of the marine organic deposits of Posidonia oceanica: a case study in the NE Iberian Peninsula. Palaeogeogr. Palaeoclimatol. Palaeoecol. 271, 215–224 (2009).
Nguyen, N.-L. et al. Sedimentary ancient DNA: a new paleogenomic tool for reconstructing the history of marine ecosystems. Front. Mar. Sci. 10, 1185435 (2023).
Pedersen, M. W. et al. A comparative study of ancient environmental DNA to pollen and macrofossils from lake sediments reveals taxonomic overlap and additional plant taxa. Quat. Sci. Rev. 75, 161–168 (2013).
Parducci, L. et al. Shotgun environmental DNA, pollen, and macrofossil analysis of lateglacial lake sediments from Southern Sweden. Front. Ecol. Evol. 7, 189 (2019).
Barrenechea Angeles, I. et al. Encapsulated in sediments: eDNA deciphers the ecosystem history of one of the most polluted European marine sites. Environ. Int. 172, 107738 (2023).
Pérez, A., Libardoni, B. G. & Sanders, C. J. Factors influencing organic carbon accumulation in mangrove ecosystems. Biol. Lett. 14, 20180237 (2018).
Becker, L. W. M. et al. Palaeo-productivity record from Norwegian Sea enables North Atlantic Oscillation (NAO) reconstruction for the last 8000 years. npj Clim. Atmos. Sci. 3, 1–12 (2020).
Rothwell, R. G. & Croudace, I. w. Twenty years of XRF core scanning marine sediments: what do geochemical proxies tell us? in Micro-XRF Studies of Sediment Cores: Applications of a non-destructive tool for the environmental sciences (eds. Croudace, I. W. & Rothwell, R. G.) 25–102 (Springer Netherlands, 2015). https://doi.org/10.1007/978-94-017-9849-5_2.
Woodward, C. A. & Gadd, P. S. The potential power and pitfalls of using the X-ray fluorescence molybdenum incoherent: coherent scattering ratio as a proxy for sediment organic content. Quat. Int. 514, 30–43 (2019).
McCloskey, T. A., Smith, C. G., Liu, K., Marot, M. & Haller, C. How could a freshwater swamp produce a chemical signature characteristic of a saltmarsh? ACS Earth Space Chem. 2, 9–20 (2018).
Foster, N. R., Gillanders, B. M., Jones, A. R., Young, J. M. & Waycott, M. A muddy time capsule: using sediment environmental DNA for the long-term monitoring of coastal vegetated ecosystems. Mar. Freshwater Res. 71, 869 (2020).
Andersen, F. Ø. & Kristensen, E. Oxygen microgradients in the rhizosphere of the mangrove Avicennia marina. Mar. Ecol. Prog. Ser. 44, 201–204 (1988).
Duarte, C. M. Seagrass depth limits. Aquat. Bot. 40, 363–377 (1991).
Bowman, G. & Harvey, N. Geomorphic evolution of a holocene beach-ridge complex, LeFevre Peninsula, South Australia. J. Coast. Res. 2, 345–362 (1986).
Duarte, C. M., Middelburg, J. J. & Caraco, N. Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences 2, 1–8 (2005).
Fahner, N. A., Shokralla, S., Baird, D. J. & Hajibabaei, M. Large-scale monitoring of plants through environmental DNA metabarcoding of soil: recovery, resolution, and annotation of four DNA Markers. PLoS ONE 11, e0157505 (2016).
Foster, N. R. et al. A multi-gene region targeted capture approach to detect plant DNA in environmental samples: a case study from coastal environments. Front. Ecol. Evol. 9 (2021).
Deiner, K. et al. Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895 (2017).
Murchie, T. J. et al. Collapse of the mammoth-steppe in central Yukon as revealed by ancient environmental DNA. Nat Commun 12, 7120 (2021).
Lentz, D. L. et al. Environmental DNA reveals arboreal cityscapes at the Ancient Maya Center of Tikal. Sci. Rep. 11, 12725 (2021).
Saintilan, N., Rogers, K., Mazumder, D. & Woodroffe, C. Allochthonous and autochthonous contributions to carbon accumulation and carbon store in southeastern Australian coastal wetlands. Estuar. Coast. Shelf Sci. 128, 84–92 (2013).
Kaal, J., Martínez Cortizas, A., Mateo, M.-Á. & Serrano, O. Deciphering organic matter sources and ecological shifts in blue carbon ecosystems based on molecular fingerprinting. Sci. Total Environ. 742, 140554 (2020).
Belperio, A. P., Hails, J. R., Gostin, V. A. & Polach, H. A. The stratigraphy of coastal carbonate banks and Holocene sea levels of northern Spencer Gulf, South Australia. Mar. Geol. 61, 297–313 (1984).
Telfer, K. W. & Malone, G. A People’s Living Cultural Landscape. (City of Charles Sturt, 2012)
Telfer, S. Torrens Island Biodiversity Action Plan. (Department for Environment and Water, 2013).
Edyvane, K. S. Coastal and marine wetlands in Gulf St Vincent, South Australia: understanding their loss and degradation. Wetl. Ecol. Manag. 7, 83–104 (1999).
Glew, J. R., Smol, J. P. & Last, W. M. Sediment core collection and extrusion. in Tracking Environmental Change Using Lake Sediments: Basin Analysis, Coring, and Chronological Techniques (eds Last, W. M. & Smol, J. P.) 73–105 (Springer Netherlands, 2001). https://doi.org/10.1007/0-306-47669-X_5.
Ficetola, G. F., Taberlet, P. & Coissac, E. How to limit false positives in environmental DNA and metabarcoding? Mol. Ecol. Resour. 16, 604–607 (2016).
Hermans, S. M., Buckley, H. L. & Lear, G. Optimal extraction methods for the simultaneous analysis of DNA from diverse organisms and sample types. Mol. Ecol. Resour. 18, 557–569 (2018).
Lemmon, A. R., Emme, S. A. & Lemmon, E. M. Anchored hybrid enrichment for massively high-throughput phylogenomics. Syst. Biol 61, 727–744 (2012).
Corinaldesi, C., Beolchini, F. & Dell’anno, A. Damage and degradation rates of extracellular DNA in marine sediments: implications for the preservation of gene sequences. Mol. Ecol. 17, 3939–3951 (2008).
Glenn, T. C. et al. Adapterama I: Universal Stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext). PeerJ 7, e7755 (2019).
Hollingsworth, P. M., Graham, S. W. & Little, D. P. Choosing and Using a Plant DNA Barcode. PLoS ONE 6, e19254 (2011).
Revéret, A. et al. Environmental DNA of aquatic macrophytes: the potential for reconstructing past and present vegetation and environments. Freshw. Biol. 68, 1929–1950 (2023).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Waycott, M., Van Dijk, K. & Biffin, E. A Hybrid Capture RNA Bait Set for Resolving Genetic and Evolutionary Relationships in Angiosperms from Deep Phylogeny to Intraspecific Lineage Hybridization. http://biorxiv.org/lookup/doi/10.1101/2021.09.06.456727 (2021).
Schubert, M., Lindgreen, S. & Orlando, L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9, 88 (2016).
Schubert, M. et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 9, 1056–1082 (2014).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv http://arxiv.org/abs/1303.3997 (2013).
Foster, N. R. et al. A targeted capture approach to generating reference sequence databases for chloroplast gene regions. Ecol. Evol. 12, e8816 (2022).
NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46, D8–D13 (2018).
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011).
Hollingsworth, M. L. et al. Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol. Ecol. Resour. 9, 439–457 (2009).
R Core Team, R. R: A Language and Environment for Statistical Computing. (2018).
Chamberlain, S. A. & Szöcs, E. taxize: taxonomic search and retrieval in R. F1000Research 2 (2013).
Drost, H.-G., Gabel, A., Liu, J., Quint, M. & Grosse, I. myTAI: evolutionary transcriptomics with R. Bioinformatics 34, 1589–1590 (2018).
Wickham, H., François, R., Henry, L., Müller, K. & Wickham, M. H. A Grammar of Data Manipulation. R package version 1.0.2. 8 (2019).
Gagolewski, M. stringi: Fast and Portable Character String Processing in R. J. Stat. Softw. 103, 1–59 (2022).
Wickham, H. & Wickham, M. H. Simple, Consistent Wrappers for Common String Operations. R package version 1.4.0 (2019).
Wickham, H. & Henry, L. tidyr: Tidy Messy Data. R package version 1.0.0. (2019).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE 8, e61217 (2013).
Wood, S. & Wood, M. S. Package ‘mgcv’. R package version 1, 729 (2015).
Ramsey, C. B. Deposition models for chronological records. Quat. Sci. Rev. 27, 42–60 (2008).
Hogg, A. G. et al. SHCal20 Southern Hemisphere Calibration, 0–55,000 Years cal BP. Radiocarbon 62, 759–778 (2020).
Heaton, T. J. et al. Marine20—the marine radiocarbon age calibration curve (0–55,000 cal BP). Radiocarbon 62, 779–820 (2020).
Ulm, S. et al. Australian marine radiocarbon reservoir effects: ΔR atlas and ΔR calculator for Australian mainland coasts and near-shore islands. Radiocarbon 65, 1139–1159 (2023).
Kennedy, H. et al. Seagrass sediments as a global carbon sink: Isotopic constraints. Glob. Biogeochem. Cycles 24, GB4026 (2010).
Hemminga, M. & Mateo, M. Stable carbon isotopes in seagrasses:variability in ratios and use in ecological studies. Mar. Ecol. Prog. Ser. 140, 285–298 (1996).
Blaauw, M. & Christen, J. A. Flexible paleoclimate age-depth models using an autoregressive gamma process. Bayesian Anal. 6, 457–474 (2011).
Evans, G., Augustinus, P., Gadd, P., Zawadzki, A. & Ditchfield, A. A multi-proxy μ-XRF inferred lake sediment record of environmental change spanning the last ca. 2230 years from Lake Kanono, Northland, New Zealand. Quat. Sci. Rev. 225, 106000 (2019).
Jolliffe, I. T. Principal Component Analysis for Special Types of Data. (Springer, 2002).
Killick, R. & Eckley, I. A. changepoint: An R Package for Changepoint Analysis. J. Stat. Soft. 58, 1–19 (2014).
Wickham, H. An introduction to ggplot: An implementation of the grammar of graphics in R. (2006).
Acknowledgements
We acknowledge and respect the Kaurna people, the Traditional Owners of the lands on which this study was undertaken. We pay respects to Kaurna Elders past and present. Access to ANSTO facilities (Centre for Accelerator Science, ITRAX XRF and 14C dating) was provided under ANSTO Award AP11907. We acknowledge the financial support from the Australian Government for the Centre for Accelerator Science at ANSTO through the National Collaborative Research Infrastructure Strategy (NCRIS). This work was supported by the Goyder Institute for Water Research (project reference: CA-16-04), with co-funding received from SA Water, the SA-EPA, ECU and the University of Adelaide’s Environment Institute, a Herman Slade Foundation grant (HSF 18-2) awarded to A.J., a Max Day Environmental Fellowship awarded by the Australian Academy of Science to NF and a Research Training Programme Scholarship awarded to NF. OS was supported by I + D + i projects RYC2019-027073-I and PIE HOLOCENO 20213AT014 funded by MCIN/AEI/10.13039/501100011033 and FEDER. The IAEA is grateful for the support provided to its environmental laboratories by the Government of the Principality of Monaco. Collection of soil samples was conducted under the State Herbarium of South Australia permit G25787-3. We would like to thank Luis Williamson for his assistance in the field.
Author information
Authors and Affiliations
Contributions
N.R.F. carried out the field and laboratory work, undertook the analyses and wrote the paper. A.R.J. provided supervision and mentorship, assisted with fieldwork, data analysis and writing, and edited the paper. O.S., A.L., P.M. and P.S.L. assisted with collecting and dating of core 1, interpretation of dating results and edited the paper. Kv.D. and E.B. developed the bait set and lab methodology and edited the paper. B.M.G. and J.Y. provided supervision and mentorship and edited the paper. P.S.G. and A.Z. assisted with XRF scanning and interpretation and edited the paper. GEJ assisted with carbon dating cores 2–4 and calibrated the raw radiocarbon ages for all cores and assisted with interpretation and method writing for this section. AG produced the graphical illustration, provided support, and edited the paper. MW provided supervision and mentorship, conceived the idea, and edited the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth & Environment thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Aliénor Lavergne. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Foster, N.R., Jones, A.R., Serrano, O. et al. Environmental DNA identifies coastal plant community shift 1,000 years ago in Torrens Island, South Australia. Commun Earth Environ 5, 115 (2024). https://doi.org/10.1038/s43247-024-01277-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-024-01277-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.