Abstract
The environment disseminates antimicrobial-resistance genes; however, it remains challenging to distinguish whether human activities exacerbate antimicrobial resistance or what is natural. Here, we quantified ~300 resistance-related genes in 200+ Scottish soil samples. Location or land use does not explain gene differences, but nutrient levels reduce gene richness. Elevated levels of metals increased gene richness, and selenium increased transposase levels. Rainfall and persistent organic pollutants also increased transposase relative abundance, possibly promoting conditions conducive to the horizontal transfer of antimicrobial-resistance genes. Selenium and polychlorinated biphenyls were primary factors in gene abundance, while polychlorinated biphenyls, polycyclic aromatic hydrocarbons, and pH influenced gene diversity. Polychlorinated biphenyls are derived from anthropogenic activities, highlighting human activities’ potential impact on gene prevalence. This is the first national-scale, high spatial resolution dataset of antimicrobial-resistance genes in Scottish soils and provides a novel resource on which to build future studies.
Similar content being viewed by others
Introduction
Antimicrobial resistance (AMR) threatens the effectiveness of drug therapies to treat bacterial infections. It is predicted to become the leading cause of mortality in the future1, including economic consequences2. It was presumed that AMR was primarily acquired in the clinical environment for years. However, it is now accepted that the acquisition of AMR genes or mutations might also occur elsewhere, including soil, water, wastewater, food and gut3,4,5.
Analyses of AMR genes in pristine soils6,7 demonstrated that the natural environment has an innate level of AMR; however, this environmental reservoir of AMR can be altered through anthropogenic activities that exert environmental pressures on microbial communities. These pressures include biocides, heavy metals and organic pollutants3. However, the relative importance of different human activities—such as clinical and veterinary medicine, agriculture and environmental pollution—in increasing AMR is unknown due to the difficulty in linking direct AMR consequences to specific causative actions. This is because most experimental studies and sites have confounding factors, such as antibiotic residues, toxic organic pollution and potentially toxic elements (PTE; e.g., metals)3,8,9 making associated results less conclusive. Furthermore, many datasets cover a limited range of environmental conditions, have limited background information, and quantify a narrow range of AMR gene biomarkers; and examination of specific “human impacted” locations can create a myopic view of AMR and parochial bias. Moreover, there are no agreed international reporting standards or meta-data standards for AMR, meaning that it is usually impossible to produce composite datasets to test hypotheses. As a result, the debate lingers, and we must strive to delineate relative contributions and influences of different factors that correlate with the emergence and propagation of AMR.
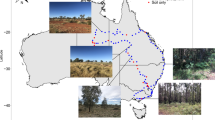
Extending the scale and depth of monitoring, including places with no apparent human impact, is critical to distinguish between natural and anthropogenically driven AMR. This will help us better understand background conditions and define factors that can alter the environmental resistome. Here, we quantified the abundance and distribution of AMR genes belonging to eleven antibiotic or mobile genetic element classes across Scotland, including clinically relevant genes associated with vancomycin and ESBL resistance, using a high throughput multi-array qPCR chip. Specifically, over 200 archived soil samples from the National Soils Inventory of Scotland (NSIS2 at James Hutton Institute) were characterised, ranging from low-impacted, relatively pristine soils (such as those in the Highlands) to high-impacted agricultural soils. Therefore, this study represents the first Scottish national-scale, high spatial resolution (20 km grid) assessment of AMR genes in soils. The results should form a foundation towards understanding the impact of human activities and future scenarios, such as climate change, on AMR gene prevalence and whether future interventions effectively reduce environmental AMR.
Results and discussion
AMR genes in Scottish soils
AMR genes were measured in soil samples collected between 2007 and 2009 as part of the re-sampling campaign for the second National Soil Inventory for Scotland (NSIS2). AMR genes were clustered into classes according to antibiotic or genetic element type to facilitate the analysis (5 < n < 54 genes in each class). AMR gene distributions and relative abundances found in the Scottish soils are summarised in Table 1, sorted according to antibiotic class. AMR genes conferring resistance to beta-lactam, multidrug resistance and vancomycin were ubiquitous in soils (i.e. 100% of the locations tested contained those resistances). Results for vancomycin are concerning, given that this is a last-resort antibiotic reserved for treating multidrug-resistant infections10. Conversely, the distribution of sulphonamide resistance was very variable, with only 46% of the locations containing detectable levels of those genes.
The mean relative abundance and richness of resistance genes belonging to each antibiotic class were determined, respectively. We defined AMR gene richness as the number of AMR genes detected per antibiotic class. Genes conferring multidrug resistance were the most abundant at each location, while sulphonamide resistance was the least abundant. The highest gene richness at each location conferred multidrug resistance (20 genes, 39%) and beta-lactam resistance (13 genes; 24%), while the lowest richness of genes conferred aminoglycoside resistance (6 genes, 13%) and sulphonamide resistance (1 gene, 14%).
The three most frequently detected resistance genes from each antibiotic class or MGE group were also identified (Table 1). In many cases, these were the most abundant resistance genes. Of note is the dominance of clinically important AMR genes such as blaTEM, blaCTX-M, and ampC and the potentially clinically relevant intl1.
While it is impossible to know which of these resistance genes are expressed, this dataset shows the reservoir of resistance genes in soil that could be horizontally transferred or expressed by resistant microbes under certain conditions. The genes represent three broad mechanisms of resistance: efflux pumps (n = 88 genes), antibiotic deactivation (n = 115 genes) and cellular protection (n = 71 genes), while a fourth group includes mobile genetic elements and resistance genes with unknown function (n = 24 genes)11. The average number of genes per sample conferring these different resistance mechanisms was found across four broad categories of land use (arable, semi-natural, managed grassland and woodland/forestry) (Fig. 1a). This showed that more genes conferring efflux pumps were detected, followed by antibiotic deactivation, then cellular protection, with others being the least detected. There were no significant differences in the number of genes according to land use (p > 0.05).
Resistance mechanisms (efflux pumps, antibiotic ‘deactivation,’ cellular ‘protection’ and ‘other’ denoting mobile genetic elements and genes of unknown function) are displayed according to (a) the number of genes detected or (b) relative abundance of genes across four broad land use types (arable, semi-natural, managed grassland and woodland/forestry). Values of n refer to the number of genes belonging to that group of resistance mechanisms assayed in the soil samples. The error bars represent standard errors of the mean, and the letters above the bars show significant differences, where p < 0.05.
The mean relative abundance of genes conferring the different resistance mechanisms was also found across the four land use categories (Fig. 1b). This showed that efflux pumps were the most abundant, followed by antibiotic deactivation, then others, with cellular protection being the least abundant. There were significant differences according to land use, with arable land having significantly lower abundances of genes for efflux pumps and other genes than other land uses (p < 0.05). This is surprising given that genes with specific resistance mechanisms are enriched in farm environments.11.
AMR genes are ubiquitous but uneven
Mapping the total relative abundances and richness of AMR genes shows that AMR genes are ubiquitous in Scottish soils (see Fig. 2), which is unsurprising given that AMR is a natural phenomenon12. Furthermore, AMR genes have been found in all environmental matrices, including in relatively pristine environments6. Almost all gene classes displayed no significant relationships with geographical features regarding gene richness or abundance (i.e., northing, easting, latitude or longitude; Spearman correlations, p > 0.01). Furthermore, there were no statistical differences when split among Scottish councils (i.e., 32 local-authority regions; Supplementary Tables 1-2; K-W test, p > 0.01), consistent with Spearman’s rank results.
However, the data mapping showed localised hot spots of some AMR genes (and some not), influencing relative abundances and richness patterns (Figs. 3, 4, Supplementary Figs. 1–16). For example, the relative abundance and richness of vancomycin genes predominate in NE Scotland (Fig. 3a, b), though the correlations were weak (rs = 0.20, p < 0.01 for both). Conversely, other antibiotic resistance classes, such as beta-lactam genes, show no significant spatial variation in relative abundance and richness as this was not significant (p > 0.01) (Fig. 3c, d).
Despite spatial heterogeneity in some AMR gene classes, there were no significant differences among land-use patterns (e.g., arable, grassland, etc.; Supplementary Tables 3, 4; K-W, p > 0.01). Since the NSIS soil archive encompasses a range of soils from highly impacted to relatively pristine soils, it was surprising that land use did not significantly influence endemic AMR genes, especially since several studies have shown that impacted land (e.g. from agriculture13) can increase relative abundances of AMR genes.
Here, land-use data were limited at the time of sampling, i.e., no historical information was associated with the sites, even though past anthropogenic activities have been shown to leave legacy pollutants that continue influencing AMR genes in the environment14. Furthermore, some relatively pristine environments could be impacted by agriculture, e.g. peatlands can be used for grazing. These factors may confound detecting AMR gene patterns relative to land use.
Least squares regression analysis was undertaken to explore the potential influence of land use fully. Data were log-transformed, and categorical variables were dummy-coded as appropriate. The analysis showed no significant associations between AMR gene abundance or richness with any of the land use types recorded as part of the original soil survey (17 different categories, dummy coded) or when these were aggregated into four major classes (arable, managed grassland, semi-natural, woodland/forestry). This suggests that other factors not measured in the original survey might be more explanatory or that the categorisation used while suitable for the original purpose of the survey is less appropriate when it comes to explaining spatial variation in AMR gene richness/abundance (i.e., traditional land use categories might not adequately reflect impacts that promote AMR). This may be reflected in the fact that specific indicators of anthropogenic impact (carbon, nitrogen, pH, persistent organic pollutants, etc.) did show significant correlations with AMR gene richness/abundance (see below).
Rainfall increases transposon relative abundance
Climate change has been predicted to affect AMR gene prevalence, given that higher temperatures and moisture can accelerate bacterial growth rates and increase horizontal gene transfer, possibly promoting increased environmental transmission15. Therefore, we looked for potential correlations between temperature and rainfall. The temperature was not significantly correlated with either AMR gene relative abundance or richness, possibly because temperature ranges within Scotland are too limited to see a correlation. However, rainfall significantly correlated with transposon relative abundance (Table 2) but not transposon richness. While this is consistent with a previous study that showed that rainfall can be associated with AMR gene transmission and proliferation from air to soil16, it may also reflect the additional influence of non-point pollution in run-off from high rainfall. We cannot discern the difference from the archived soil samples.
Factors affecting bacterial richness also affect AMR genes
Here, the biggest contributor to AMR gene richness was bacterial abundance. Significant positive correlations existed between bacterial abundance and the resistance gene richness for aminoglycoside, beta-lactam, am-/chlor-/fluoro-quinolones, multidrug resistance, sulphonamide, tetracycline and transposase genes. We also measured bacterial richness using the Chao Index. There were significant positive correlations between bacterial richness and the richness of resistance genes for beta-lactam and sulfonamide (0.20 < rS < 0.31, p < 0.01). In other words, the greater the bacterial abundance and richness, the greater the AMR gene richness.
Therefore, the environmental factors often correlated with bacterial communities (Supplementary Table 5) were also examined with AMR genes (Tables 2 and 3). There were no consistent correlations between carbon, nitrogen and pH with AMR gene abundance (Table 2); however, these factors often correlate with AMR gene richness (Table 3). Carbon was negatively correlated with bacterial abundance and richness (Supplementary Table 5; rs = −0.34 and rs = −0.78, respectively, both p < 0.01) and, therefore, negatively correlated with the richness of the AMR genes for beta-lactam, multidrug resistance, sulfonamide, tetracycline and vancomycin.
Similar correlations were observed with LOI (loss on ignition; rs = −0.36 and rs = −0.75, p < 0.01), which is unsurprising since LOI also indicates the amount of organic carbon in soils. Nitrogen was also negatively correlated with bacterial abundance and richness (Supplementary Table 5; rs = −0.19 and rs = −0.58, respectively, p < 0.01) and sulfonamide gene richness. On the other hand, soil pH was positively correlated with bacterial abundance and richness (Supplementary Table 5; rs = 0.33 and rs = 0.84, respectively, p < 0.01) and positively correlated with the richness of the AMR genes associated with beta-lactams and sulphonamides.
While we cannot explicitly assign how much carbon and nitrogen came from human activity versus natural sources, our results suggest that human activities that alter concentrations of crucial nutrients in the environment, such as those in fertilisers, can influence AMR gene richness. Nutrient availability impacts bacterial abundance and richness, so it is reasonable to expect that these factors also affect AMR genes. Nitrogen application to soil can decrease microbial biomass17, reducing the pool of potential hosts for AMR genes. Increased assimilable carbon can diminish soil biodiversity18, whereas in water, it can decrease microbial richness as eutrophication tends to select for fewer dominant, typically opportunistic species19. This selection process potentially explains why AMR richness declines with increased nutrient loadings in our study.
Our results are consistent with other studies that found soil carbon significantly influenced microbial community composition and AMR gene prevalence. For example, Li et al.20 showed that higher total organic carbon led to lower AMR gene abundance in Tibetan soils, which was attributed to higher autochthonous microbial abundances that out-compete and prevent invasions of antibiotic-resistant bacteria. In a different context, higher carbon concentrations in a biofilm from a drinking water sand filter also decreased bacterial diversity and AMR gene richness, attributed to a few species’ dominance at higher nutrient concentrations19. An alternative explanation is the reversion of resistant bacteria to susceptible forms (i.e. plasmid curing) without stress conditions, especially when the carriage of resistance genes comes at a fitness cost21.
Our results also showed that higher soil pH is associated with increased bacterial abundance and richness (Supplementary Table 5) and AMR gene richness (Table 3). Many studies have shown soil pH to be the overarching factor influencing microbial community composition22,23,24,25, so it is reasonable that it might also affect AMR gene prevalence. For example, one study on rice paddy soils showed that changes in microbial community structure were strongly influenced by pH, which consequently influenced local AMR gene spread26. The positive correlation between soil pH and AMR gene richness found in this study is contrary to other studies on soil AMR27,28; however, soils in those studies encompassed a broader pH range, whereas Scottish soils are mostly acidic (the average was pH 4.7 and the range was pH 3.2–7.9 within our sample set).
Metals influence AMR gene richness
Metals can influence AMR prevalence and selection; however, their bioavailability remains the critical factor in the relationship29. Despite this, we looked for correlations between total metal concentrations and AMR genes in the soils studied here. Surprisingly, Spearman’s rank correlation showed that metal concentrations were largely negatively correlated with AMR gene relative abundances. Specifically, negative correlations were observed between copper, strontium, mercury, nickel, zinc, boron, magnesium, manganese and chromium and the relative abundance of resistance genes conferring multidrug resistance, vancomycin and transposons. Only selenium was positively correlated with transposase relative abundance (Table 2). The predominantly negative correlations with metals may result from antimicrobial activity and fewer hosts for AMR genes30. Moreover, metal bioavailability and the abundance and types of anions consequentially alter the influence of heavy metals on microbial communities31, including relative AMR gene abundances29, which might also explain some of the negative correlations.
Conversely, elevated total metal concentrations were associated with increased AMR gene richness. Specifically, the richness of resistance genes for beta-lactam, sulphonamide, tetracycline, and vancomycin increased with increasing concentrations of manganese, barium and aluminium (Table 3). Therefore, higher metal concentrations for the soils in this study increased AMR gene richness but did not necessarily increase overall AMR gene abundance (apart from the case with selenium).
Several studies have shown that metals enhance AMR genes and mobile genetic elements through co-resistance (inheritance of different resistance determinants on the same genetic element) and cross-resistance (the same resistance determinant confers resistance to metal and antibiotic)8,32. For example, resistance to both barium and aluminium was prevalent in antibiotic-resistant bacterial isolates from a polluted river in Turkey33. Furthermore, Robins et al.29 discovered that mobile genetic elements in NE England were associated with AMR genes in rural settings, whereas in urban samples, the AMR genes directly correlated with metal pollutants.
In agricultural soils, the metal composition tends to be more closely governed by the parent material34, i.e. from natural sources. Analysis of historical samples from the National Soils Archive of Scotland (1940–1970, representing the early antibiotic era) showed a baseline level of AMR gene relative abundance that correlated with a combination of baseline levels of copper, chromium, nickel, lead and iron35. Since this new study focussed on rural locations, probable anthropogenic sources of metals in these soils include fossil fuel combustion, organic material application, fertilisers and atmospheric deposition34. Although it is impossible to determine how many specific metals are from natural or anthropogenic sources here, the results suggest that local metals influence AMR gene richness; therefore, reducing anthropogenic metals to Scottish landscapes will help decrease AMR gene richness in the soils.
Persistent organic pollutants drive transposase abundance
There has been increasing interest in the potential for persistent organic pollutants (POPs) to drive AMR9,36,37. The concentrations of several POPs, including polychlorinated biphenyls (PCBs), polycyclic aromatic hydrocarbons (PAHs) and polybrominated diphenyl ethers (PBDEs) were previously measured in the NSIS soils38 so these were used to determine any possible relationships with AMR genes. There are no known natural sources of PCBs or PBDEs, and they are an explicit measure of environmental anthropogenic pollution.
Spearman’s rank showed that there were significant positive correlations between the relative abundance (but not richness) of transposase genes and the PCBs, 2,3′,4,4′,5-pentachlorobiphenyl, 2,2′,3,4,4′,5′-hexachlorobiphenyl, 2,2′,4,4′,5,5′-hexachlorobiphenyl and 2,2′,3,4,4′,5,5′-heptachlorobiphenyl, and the PAH fluorene (Tables 2 and 3). On the other hand, significant negative correlations were observed between the richness of resistance genes for beta-lactam, sulfonamide, tetracycline and vancomycin with the PCBs 2,2′,4,5,5′-pentachlorobiphenyl, 2,3′,4,4′,5-pentachlorobiphenyl, 2,2′,3,4,4′,5′-hexachlorobiphenyl, 2,2′,4,4′,5,5′-hexachlorobiphenyl and 2,2′,3,4,4′,5,5′-heptachlorobiphenyl and the PAH chrysene (Tables 2 and 3). Only one correlation with PBDEs was observed with AMR genes, where the relative abundance of tetracycline was negatively correlated with 2,2′,4,4′, and 6-pentabromo-diphenyl ether.
Studies have shown that catabolic genes responsible for the degradation of organic compounds are closely associated with mobile genetic elements, such as transposons, which promote their spread and de novo construction of catabolic pathways, allowing bacteria to adapt to such pollutants39. Our results corroborate transposons’ possible role in the bioremediation of organic compounds in the environment40. The increase in transposase abundance driven by PCBs would be expected to promote AMR transmission. However, the apparent decrease in AMR gene richness with PCBs and PAHs possibly results from toxicity to host cells41. These confounding effects have unclear consequences for AMR in the environment. As noted by others9, there is a gap in the literature on the possible enrichment of AMR genes in PCB-degrading bacteria in soil. As such, further work is needed to help elucidate the effects of PCBs on AMR genes.
Transposon-mediated co-selection of PAH-degrading genes has been noted previously42, which may explain higher transposon relative abundances with increased fluorene concentrations. Field studies have shown that PAH-contaminated environments can select for AMR genes37,43, possibly mediated by horizontal gene transfer36, while in vitro studies have shown that PAHs can block plasmid-mediated transfer of AMR genes44. Again, these confounding effects have unclear consequences for AMR in the environment, so further studies are required to determine the effects of PAHs on AMR genes.
A principal components analysis (PCA) determined the relative associations between environmental parameters (climate, pH, carbon/LOI, nitrogen, metals/metalloids and POPs) and AMR gene mean relative abundance and richness. Overall, the plots show no clustering according to land use (Fig. 5), which agrees with our correlation and regression analysis. There was also no clustering according to location from the correlation analysis. For AMR relative abundance, the axes explain 71.6% of the variation and are most influenced by selenium, followed by PCBs. For AMR richness, the axes explain 79.5% of the variation and are most influenced by PCBs and PAHs, followed by soil pH. This indicates that human activities are one of the strongest drivers of AMR abundance and richness since PCBs can only be derived from human activity.
Integrons and transposons drive AMR gene richness and abundance
Since integrons and transposons can be vectors for the AMR gene to spread45,46, they were used as explanatory variables to determine whether they correlated with AMR gene richness and abundance (Tables 2, 3). Interestingly, the relative spatial distribution of transposons versus integrons is quite different across Scottish soils (Fig. 4a, b). Transposon relative abundances are much higher in west Scotland than east, whereas integron relative abundances are more evenly distributed across the country.
Relative integron and transposon abundances were positively correlated with the relative abundances of most AMR gene classes (Table 2). The exception was for AMR genes conferring resistance to beta-lactams and am-/chlor-/fluoro-quinolones, where correlations with relative transposon levels were not significant (p > 0.01). Overall, integron relative abundance had the strongest positive correlation with AMR gene relative abundance compared to the correlation between transposon relative abundance and AMR gene relative abundance (rs = 0.34–0.66 for integrons vs rs = 0.19–0.34 for transposons), indicating that they may have a greater influence on possible AMR gene transfer.
For AMR gene richness (Table 3), integron and transposon richness appeared closely associated with the richness of most AMR gene classes. Only the richness of genes conferring resistance to am-/chlor-/fluoro-quinolones was not correlated with either integrons or transposons. Numerous studies have demonstrated that the abundance of the intl1 gene increases with pollution and human impact47; therefore, such associations might be expected. Equally, transposons can increase with pollution29,32. Overall, such associations between mobile genetic elements (MGEs) and AMR genes suggest an elevated potential risk of AMR gene transmission across Scottish soils via horizontal and vertical gene transfer.
Practical implications
This study shows that “intrinsic” soil resistomes in Scotland are significantly influenced by environmental factors, including climate (rainfall), soil pH, nutrients (carbon, nitrogen), metals (selenium, manganese, barium, and aluminium) and POPs (PAHs, PCBs, PBDEs), and that increased MGEs strongly associate with relative AMR gene abundances and diversity. The strongest associations with relative AMR gene abundances were with ambient selenium and PCB levels. By contrast, PCB, PAH, and soil pH conditions substantially influenced relative AMR gene richness in soil. This suggests that in situ resistance potential has been altered by pollutants from anthropogenic activities, as PCBs can only be derived from human activity. Rainfall, selenium, fluorene and PCBs positively correlated with transposase relative abundance, which may increase the potential for AMR gene transfer. Increased AMR gene richness potentially increases the number of different mechanisms bacteria can use to evade the antimicrobial effects of antibiotics. On the other hand, increased AMR gene relative abundances increase potential human exposure via different possible soil- and water-related exposure pathways. However, it remains to be seen whether increased AMR gene richness or abundance indeed presents a greater risk to human health, either immediate or latent.
The uneven distribution of genes belonging to each antibiotic class across Scotland raises questions of when and how the AMR genes emerged in local hot spots, how the AMR genes were propagated, and why the AMR genes persisted more in some soils than others. Care needs to be taken over the interpretation of our data. Since the soil is highly heterogeneous, the results cannot necessarily be extrapolated to soils near our sampling points. Nevertheless, the extensive soil sample series shows that some geographical regions in Scotland may have a greater opportunity for environmental AMR genes to move into strains of greater health importance. This work further confirms that targeted studies using historical soil samples can help elucidate mechanisms that impact the emergence and evolution of environmental AMR. This is very important given the greater emphasis at the United Nations on broader integrated surveillance as an essential element in understanding AMR prevalence and spread5. This valuable national-scale dataset describes background conditions across the country, which allows the identification of locations for additional surveillance and, in turn, permits the development of mitigation strategies to control the spread of AMR.
Materials and methods
Soil sampling
Soil samples were collected as part of a re-sampling campaign for the second National Soil Inventory of Scotland (NSIS2, 2007–9)48, based on standardised methods developed for the first National Soil Inventory (NSIS1). This survey recorded the sampling sites’ location (northing, easting, longitude, latitude) and altitude48. The collection contains representative and unbiased soil samples from 195 sites across Scotland, based on a 20 km grid sampling pattern, plus 21 rare soils off the 20 km grid pattern, giving a complete archive of 216 soil samples. Briefly, soil pits were excavated at each site, and the soil horizons were identified by visual inspection. Approximately 1.5 to 2 kg of stone-free soil was collected from the middle of each horizon, placed in bags, and sealed immediately to avoid contamination. Meteorological data (temperature and rainfall) was recorded from the Met Office49.
Once returned to the James Hutton Institute, a sub-sample was taken for microbiological analysis; only the top horizon was considered for this study. A second sub-sample was frozen at −80 °C until required, and the remaining soil was air-dried and sieved (2 mm) for chemical analyses and subsequent storage in the National Soils Archive (www.hutton.ac.uk/about/facilities/national-soils-archive). Persistent Organic Pollutants (POPs), including polychlorinated biphenyls (PCBs), polycyclic aromatic hydrocarbons (PAHs) and polybrominated diphenyl ethers (PBDEs), were determined as described in Zhang et al.28 and Cui et al.38. Soil pH was determined on the supernatant of a 3:1 water to the soil mix.
Total carbon and nitrogen concentrations (C%, N%) were determined using a Flash EA 1112 Series Elemental Analyser (Thermo Fisher, Germany). Loss on Ignition (LOI) was measured gravimetricly. The soil was dried at 105 °C until a constant mass was achieved, then weighed into pre-weighed silica or porcelain crucibles. The sample was ignited at 450 °C and 900 °C for a minimum of 2 h. For metals and metalloids (arsenic, barium, cadmium, cobalt, chromium, copper, mercury, molybdenum, nickel, lead, selenium, strontium, zinc, aluminium, boron, calcium, iron, magnesium and manganese), soils were milled to a fine powder and digested in a mixture of hydrochloric acid and nitric acid (aqua regia) under reflux for two hours. The digest was cooled, filtered and made up to a known volume with 0.5 M nitric acid before being analysed by inductively coupled plasma optical emission spectroscopy (ICP-OES) or Inductively Coupled plasma-mass spectrometry (ICP-MS) to determine element concentration.
DNA extraction and quantification of AMR genes
DNA was extracted from triplicate samples of 250 mg of soil using a modified CTAB method, which were pooled prior to analysis50. Modifications to the CTAB method included carrying all steps on ice rather than at room temperature (including the lysis step), using higher centrifugation speeds for more extended periods, using MaXtract High-density Tubes (Qiagen) to separate the DNA from the phenol-chloroform, and carrying out two ethanol wash steps to remove impurities. The purity and concentrations of the extracts were verified by UV spectrometry (Nanodrop and QuBit, respectively; both ThermoFisher). The DNA were normalised to 10 ng/µL and stored long-term at −80 °C. 100 µL (1 µg) of DNA was Analysed using the Applied Biosystems OpenArray platform.
All high-throughput quantitative PCR (HT-qPCR) reactions were performed in triplicate using the Applied Biosystems OpenArray system according to Looft et al.51, with threshold cycles (CT) set at 27 cycles as the minimum detection limit. The ΔΔCT method52 was used to calculate the relative abundances of the genes, and 16S rRNA genes were quantified to normalise the AMR gene abundances11.
Primer sets for the 295 AMR and AMR-related assays were previously developed and validated11. Average CT values were calculated from triplicates; a sample without amplification was considered a “false negative” and discarded.
Individual genes were grouped according to their antibiotic class or AMR-related activity: aminoglycosides (36 genetic determinants), beta-lactam (54), am-/chor-/fluoro-quinolones (FCA, 9), macrolide-lincosamide-streptogramin B (MLSB, 46), sulfonamide (7), tetracycline (39), vancomycin (32), other antibiotic resistances (i.e. to nitroimidazole, pyrazinamide, streptothricin, bacitracin and fosfomycin, 11), multidrug mechanisms (51, here we refer to non-specific resistance mechanisms, such for efflux pumps), integrases (mobile genetic elements, 5) and transposases (mobile genetic elements, 8). The relative abundance of the AMR and AMR-related genes were expressed as the abundance of genes relative to the abundance of bacteria (i.e. the 16S rRNA gene). The total relative abundances per antibiotic class or AMR-related activity were used for statistical analysis. In addition, the total number of genes (referred to as AMR gene richness) for each antibiotic class or AMR-related activity was used for statistical analysis.
Statistical analysis
Statistical analyses were conducted in R version 4.2.1 and SPSS v26. Spearman’s rank determined correlations with environmental variables53. Kruskal–Wallis (K–W) test was used to compare multiple populations (a non-parametric equivalent of ANOVA), i.e. Scottish council areas and land use types including arable, bog/wetland, dunes, grassland, improved grassland, rough grassland, heath, larch plantation, moorland, scrub, broadleaf woodland and coniferous woodland. For all correlations, those with p < 0.01 were considered statistically significant. Least squares regression analysis was undertaken to explore the potential influence of land use fully. Data were log-transformed, and categorical variables were dummy coded, as appropriate.
A t test was performed in Excel using a two-tailed distribution and a two-sample unequal variance to determine significant differences between the distribution of resistance mechanisms (antibiotic deactivation, cellular protection, efflux pumps and ‘other’ denoting mobile genetic elements and resistance genes of unknown function) amongst different land uses (arable, semi-natural, managed grassland and woodland/forestry).
Sequenced 16S rRNA genes were used to determine bacterial richness (Chao Index) using Primer754. Principal Components Analysis was done to determine the relative strength of environmental variables to explain the variation in the AMR gene abundance and richness55. Data were square-root transformed, and the Bray-Curtis Index determined similarities. The PCA plot was drawn using Primer7, and the environmental variables with the strongest correlations (rS > 0.1) were overlaid on the plot.
Data mapping
AMR gene relative abundance and richness distributions across Scotland were plotted using the ArcGIS Pro GIS software package v 3.056. Each dataset was summarised by quintiles (except in cases where numbers were too low, e.g. integron richness) using graduated symbols of increasing size and blue-red colour ramp to indicate increasing relative abundance and richness.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data has been archived at the UK Environmental Information Data Centre57 and the James Hutton Institute. Bacterial 16S-rRNA gene sequences have been deposited to NCBI, with BioProject ID number PRJNA997939.
References
O’Neill 2016. Tackling drug-resistant infections globally: final report and recommendations. Review on Antimicrobial Resistance/Wellcome Trust. London, UK. 80pp.
WEF-World Economic Forum 2021. Antimicrobial Resistance and Water: The risks and costs for economies and societies. Briefing Paper, https://www3.weforum.org/docs/WEF_Antimicrobial_Resistance_and_Water_2021.pdf.
Singer, A. C., Shaw, H., Rhodes, V. & Hart, A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7. https://doi.org/10.3389/fmicb.2016.01728 (2016).
Anthony, W. E., Burnham, C. A. D., Dantas, G. & Kwon, J. H. The gut microbiome as a reservoir for antimicrobial resistance. J. Infectious Dis. 223, S209–S213 (2021).
UNEP - United Nations Environment Programme. Bracing for Superbugs: Strengthening environmental action in the One Health response to antimicrobial resistance. (UNEP, Geneva, 2023).
Kim, H., Kim, M., Kim, S., Lee, Y. M. & Shin, S. C. Characterisation of antimicrobial resistance genes and virulence factor genes in an Arctic permafrost region revealed by metagenomics. Environ. Pollut. 294, 118634 (2022).
Knapp, C. W. et al. Relationship between antibiotic resistance genes and metals in residential soil samples from western Australia. Environ. Sci. Pollut. Res. 24, 2484 (2017).
Baker-Austin, C., Wright, M. S., Stepanauskas, R. & McArthur, J. V. Co-selection of antibiotic and metal resistance. Trends. Microbiol. 14, 176–182 (2006).
Gorovtsov, A. V., Sazykin, I. S. & Sazykina, M. A. The influence of heavy metals, polyaromatic hydrocarbons, and polychlorinated biphenyls pollution on the development of antibiotic resistance in soils. Environ. Sci. Pollut. Res. 25, 9283–9292 (2018).
Walsh, C. Microbiology—deconstructing vancomycin. Science 284, 442–443 (1999).
Zhu, Y. G. et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl Acad. Sci. USA 110, 3435–3440 (2013).
D’costa, V. M. et al. Antibiotic resistance is ancient. Nature 477, 457–461 (2011).
Thanner, S., Drissner, D. & Walsh, F. Antimicrobial resistance in agriculture. Mbio. 7 (2016).
Rodgers, K. et al. Can the legacy of industrial pollution influence antimicrobial resistance in estuarine sediments? Environ. Chem. Lett. 17, 595–607 (2019).
Lio, R. A. S., Favara, G., Maugeri, A., Barchitta, M. & Agodi, A. How antimicrobial resistance is linked to climate change: an overview of two intertwined global challenges. Int. J. Environ. Res. Public Health 20, 1681 (2023).
Wang, Q. et al. Rainfall facilitates the transmission and proliferation of antibiotic resistance genes from ambient air to soil. Sci. Total Environ. 799, 149260 (2021).
Zhang, T. A., Chen, H. Y. H. & Ruan, H. H. Global negative effects of nitrogen deposition on soil microbes. ISME J. 12, 1817–1825 (2018).
Drenovsky, R. E., Vo, D., Graham, K. J. & Scow, K. M. Soil water content and organic carbon availability are major determinants of soil microbioal community composition. Microbial Ecol. 48, 424–430 (2004).
Wan, K. et al. Organic carbon: An overlooked factor that determines the antibiotic resistome in drinking water sand filter biofilm. Environ. Int. 125, 117–124 (2019).
Li, B., Chen, Z., Zhang, F., Liu, Y. Q. & Yan, T. Abundance, diversity and mobility potential of antibiotic resistance genes in pristine Tibetan Plateau soil as revealed by soil metagenomics. FEMS Microbiol. Ecol. 96. https://doi.org/10.1093/femsec/fiaa172 (2020).
Millan, A. S. & MacLean, R. C. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spect. 5 https://doi.org/10.1128/microbiolspec.mtbp-0016-2017 (2017).
Fierer, N. & Jackson, R. B. The diversity and biogeography of soil bacterial communities. Proc. Natl Acad. USA 103, 626–631 (2006).
Rousk, J. et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351 (2010).
Lauber, C. L., Hamady, M., Knight, R. & Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 (2009).
Zhalnina, K. et al. Soil pH determines microbial diversity and composition in the park grass experiment. Microbial Ecol. 69, 395–406 (2015).
Xiao, K. Q. et al. 2016. Metagenomic profiles of antibiotic resistance genes in paddy soils from South China. FEMS Microbiol. Ecol. 92.
McCann, C. M. et al. Understanding drivers of antibiotic resistance genes in High Arctic soil ecosystems. Environ. Int. 125, 497–504 (2019).
Zhang, Z. L. et al. Long term temporal and spatial changes in the distribution of polychlorinated biphenyls and polybrominated diphenyl ethers in Scottish soils. Sci. Total Environ. 468-469, 158–164 (2014).
Robins, K. et al. Bioavailability of potentially toxic elements influences antibiotic resistance gene and mobile genetic element abundances in urban and rural soils. Sci. Total Environ. 847, 157512 (2022).
Lemire, J. A., Harrison, J. J. & Turner, R. J. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 11, 371–384 (2013).
Gupta, S., Graham, D. W., Sreekrishnan, T. R. & Ahammad, S. Z. Heavy metal and antibiotic resistance in four Indian and UK rivers with different levels and types of water pollution. Sci. Total Environ. 857, 159059 (2023).
Zhao, Y. et al. Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci. Total Environ. 656, 512–520 (2019).
Icgen, B. & Yilmaz, F. Co-occurrence of antibiotic and heavy metal resistance in Kizilirmak river isolates. Bull. Environ. Contamination. Toxicol. 93, 735–743 (2014).
Alloway, B. J. S in Heavy Metals in Soils (ed. Alloway, B. E.) (Springer, 2013).
Knapp, C. W. et al. Antibiotic resistance gene abundances correlate with metal and geochemical conditions in archived Scottish soils. PLoS ONE 6, e27300 (2011).
Azhogina, T. et al. Bioaccessible PAH influence on distribution of antibiotic resistance genes and soil toxicity of different types of land use. Environ. Sci. Pollut. Res. 30,12695–12713 (2022).
Chen, B. W. et al. Polycyclic aromatic hydrocarbons (PAHs) enriching antibiotic resistance genes (ARGs) in the soils. Environ. Pollut. 220, 1005–1013 (2017).
Cui, S. et al. Long-term spatial and temporal patterns of polycyclic aromatic hydrocarbons (PAHs) in Scottish soils over 20 years (1990-2009): a national picture. Geoderma 361, 114135 (2020).
Brenner, V., Arensdorf, J. J. & Focht, D. D. Genetic construction of PCB degraders. Biodegradation 5, 359–377 (1994).
Top, E. M., Springael, D. & Boon, N. Catabolic mobile genetic elements and their potential use in bioaugmentation of polluted soils and waters. FEMS Microbiol. Ecol. 42, 199–208 (2002).
Chavez, F. P., Gordillo, F. & Jerez, C. A. Adaptive responses and cellular behaviour of biphenyl-degrading bacteria toward polychlorinated biphenyls. Biotechnol. Adv. 24, 309–320 (2006).
Obayori, O. S. & Salam, L. B. Degradation of polycyclic aromatic hydrocarbons: role of plasmids. Sci. Res. Essays 5, 4093–4106 (2010).
Wang, J. et al. PAHs accelerate the propagation of antibiotic resistance genes in coastal water microbial community. Environ. Pollut. 231, 1145–1152 (2017).
Kang, F. X., Hu, X. J., Liu, J. & Gao, Y. Z. Noncovalent binding of polycyclic aromatic hydrocarbons with genetic bases reducing the in vitro lateral transfer of antibiotic resistant genes. Environ. Sci. Technol. 49, 10340–10348 (2015).
Babakhani, S. & Oloomi, M. Transposons: the agents of antibiotic resistance in bacteria. J. Basic Microbiol. 58, 905–917 (2018).
Zhang, S. et al. Dissemination of antibiotic resistance genes (ARGs) via integrons in Escherichia coli: A risk to human health. Environ. Pollut. 266, 115260 (2020).
Gillings, M. R. et al. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279 (2015).
Lilly, A., Bell, J. S., Hudson, G., Nolan, A. J. & Towers, W. National Soil Inventory of Scotland 2007-2009: Profile description and soil sampling protocols. (NSIS_2). Technical Bulletin, James Hutton Institute. https://doi.org/10.5281/zenodo.7688040 (2011).
Perry, M. & Hollis, D. The generation of monthly gridded datasets for a range of climatic variables over the UK. Int. J. Climatol. 25, 1041–1054 (2005).
Griffiths, R. I., Whiteley, A. S., O’Donnell, A. G. & Bailey, M. J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491 (2000).
Looft, T. et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl Acad. Sci. USA 109, 1691–1696 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408 (2001).
Gauthier, T. D. Detecting trends using Spearman’s rank correlation coefficient. Environ. Forensics 2, 359–362 (2001).
Chao, A. & Chiu, C. H. Species richness: estimation and comparison. Wiley StatsRef: Statistics Reference Online. 22. https://doi.org/10.1002/9781118445112.stat03432.pub2 (2016).
Groth, D., Hartmann, S., Klie, S. & Selbig, J. In Computational Toxicology. Methods in Molecular Biology Vol. 930 (eds. Reisfeld, B., Mayeno, A.) (Humana Press, 2013).
ESRI. ArcGIS Pro: v3.0. Redlands, CA, USA https://www.esri.com/ (2022).
Knapp, C. W. et al. Antibiotic Resistance Genes Found in Soils Across the Entire Scottish Landscape (2007-10) (NERC Environmental Information Data Centre, 2019).
Acknowledgements
The project was funded by UKRI Natural Environment Research Council (NE/019776/1). EP, RH, LA, LR and TF are supported by the Scottish Government’s Rural and Environmental Science and Analytical Services (RESAS) Division.
Author information
Authors and Affiliations
Contributions
E.P. and C.W.K. wrote the manuscript; R.H. and M.C. generated data and provided statistics support; L.R., X.Y.Z., J.Q.S., and T.P. provided laboratory support and generated data; L.A., T.F., Y.G.Z., D.W.G., and C.W.K. conceived the project and managed elements of the project. C.W.K. was the principal investigator of the project. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth & Environment thanks Guo-Yu Yin, Nwadiuto Esiobu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Fiona Tang, Clare Davis. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pagaling, E., Hough, R., Avery, L. et al. Antibiotic resistance patterns in soils across the Scottish landscape. Commun Earth Environ 4, 403 (2023). https://doi.org/10.1038/s43247-023-01057-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-023-01057-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.