Abstract
Acid sulfate soils release metal laden, acidic waters that affect the environment, buildings, and human health. In this study, 16S rRNA gene amplicons, metagenomes, and metatranscriptomes all demonstrated distinct microbial communities and activities in the unoxidized potential acid sulfate soil, the overlying transition zone, and uppermost oxidized actual acid sulfate soil. Assembled genomes and mRNA transcripts also suggested abundant oxidized acid sulfate soil populations that aligned within the Gammaproteobacteria and Terracidiphilus. In contrast, potentially acid tolerant or moderately acidophilic iron oxidizing Gallionella and sulfur metabolizing Sulfuricella dominated the transition zone during catalysis of metal sulfide oxidation to form acid sulfate soil. Finally, anaerobic oxidation of methane coupled to nitrate, sulfate, and ferric reduction were suggested to occur in the reduced parent sediments. In conclusion, despite comparable metal sulfide dissolution processes e.g., biomining, Gallionella and Sulfuricella dominated the community and activities during conversion of potential to actual acid sulfate soils.
Similar content being viewed by others
Introduction
Natural deposits containing sulfidic sediments1 are widespread along coastal areas worldwide such as Baltic Sea, Asia, and Australia2. These sediments are predominantly stable if they remain anoxic, such as under the sea or the groundwater table3. However, coastal areas containing sulfidic sediments have been dredged for purposes such as agriculture and housing. This exposes the sulfides to atmospheric oxygen and initiates microbial-aided chemical reactions4 that turn potential acid sulfate soil materials (PASS) into actual acid sulfate soil materials (ASS; pH ≤ 4), which are described as the “nastiest soils on earth”5. This conversion to ASS releases large quantities of acidity that concomitantly increases the mobility of toxic metal(loid)s such as Al, As, Cd, and Zn that are transported to surrounding waters where they cause severe problems for the environment6,7, economy8, and human health5.
Pyrite (FeS2) oxidation is a complex biogeochemical process that involves a series of chemical reactions that are catalyzed by microbial re-oxidation of ferrous iron9 and metabolism of intermediate sulfur compounds formed during mineral breakdown10. Pyrite dissolution is extensively studied in the context of the industrial process of “biomining”11 and acid mine drainage12 and the geochemical reactions that occur when PASS are oxidized to ASS have also been elucidated2,13. Culture based studies of ASS identify sulfur-oxidizing microbes14 and acidophiles such as Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans15. In addition, 16S rRNA gene amplicon-based studies of Australian coastal PASS are dominated by sequences aligning with Proteobacteria16 while sub-tropical ASS contain iron-oxidizers such as Alicyclobacillus tolerans, Leptospirillium sp., Sideroxydans lithotrophicus, and S. paludicola17. Metagenomic analysis also demonstrates a shift in microbial populations when a Chinese sub-tropical PASS was exposed to oxygen18. Finally, Australian coastal ASS split into “high organic,” “surface tidal,” and “sulfuric” zones shows a disproportionally higher number of active microbial populations from the “rare biosphere” (defined as the great number of low abundance microorganisms found in an environment) that were involved in iron- and sulfur cycling19. Despite the importance of ASS in the release of metals to the environment, the microbial communities and activities in the underlying sediments and during the process of metal sulfide oxidation have been largely neglected.
The Risöfladan experimental field is situated on the Ostrobothnia coast in Western Finland. The Risöfladan parent marine sediments contain approximately 50% FeS2 and 50% metastable iron sulfides (FeSn; n = 1.0–1.3) that were drained over 60 years ago and have since developed into a typical clay-type ASS20,21. The experimental field is utilized to investigate the mitigation of metal and acid release22,23,24,25,26,27 and the remediation of ASS. Microbial communities in the Risöfladan ASS include populations with 16S rRNA gene sequences most similar to acidophiles such as Acidocella sp. and Acidithiobacillus ferrivorans as well as populations similar to species identified from low temperature and metal contaminated environments13,26. Finally, 16S rRNA gene amplicons assigned to sulfur and/or iron cycling populations including Acidithiobacillus, Gallionella, Sulfuricurvum, and Sulfurimonas were identified from metal sulfide containing river sediments from the vicinity of Risöfladan when they were exposed to air28. However, most previous ASS studies only used 16S rRNA gene sequencing to identify relative abundances of populations during the generation of boreal ASS and the metabolic potential and active processes of the populations during this transition remain uncertain.
In this study, soil samples were collected from a Risöfladan depth profile that were used to investigate the microbial community structure by high throughput amplicon sequencing of the 16S rRNA marker gene; metabolic processes coded within reconstructed genomes by community DNA (metagenomics) sequencing; and community RNA (metatranscriptomics) based activity of the reconstructed genomes that shows the processes being carried in the oxidized, transition, and unoxidized soil zones. The tested hypotheses were that (i) the community structure of microbes is altered during PASS conversion to ASS and (ii) in contrast to extreme acidophiles found in biomining or acid mine drainage environments29,30, the conversion of PASS to ASS selects for moderate acidophiles.
Results
Sampling was performed when the microbes were most active i.e., when the cracks in the ASS were depleted of water and filled with air. Triplicate samples were taken from each soil zone (Fig. 1); namely, the ASS oxidized zone (defined as “OX”; depth 75–140 cm), the transition zone (“TR”; 140–190 cm) that has a steep pH gradient from acidic to near neutral, and a pH neutral unoxidized PASS zone (“UN”; >190 cm).
The figure shows the sampling site near Vaasa, Finland marked with a red dot (a), a photograph of the plough layer at the top, oxidized ASS, transition zone, and unoxidized PASS at the deepest depth (b), and basic geochemical characteristics (c) (data are averages ± SD; n = 3; complete data in Supplementary Table S1). The maps were generated by © OpenStreetMap under an Open Database License.
Geochemical characteristics of the soil zones
The oxidized zone was characterized as a typical ASS, with a pH <4 and high redox potential (Fig. 1 and Supplementary Table S1). The pH and conductivity significantly increased with soil depth, respectively (Spearman’s rank correlation; pH-depth R2 = 0.97 and p < 0.001, conductivity-depth R2 = 0.84 and p = 0.004). The concentration of total leachable S, most likely as sulfate (SO42−), was significantly different (S-depth R2 = 0.90 and p = 0.001) in the soil zones with the highest concentrations in the transition zone and the lowest in the unoxidized zone. Acid volatile sulfide (AVS; comprising of mainly metastable iron sulfides) showed significantly highest values in the unoxidized zone (AVS-depth R2 = 0.80 and p = 0.01). Although chromium reducible sulfur (CRS; mainly pyrite) showed highest values in the transition zone, no significant differences were found between the soils (CRS-depth R2 = 0.48 and p = 0.193). Furthermore, no significant differences were found between the soils regarding the total reduced sulfur (TRS; all reduced sulfur species; TRS-depth R2 = 0.47 and p = 0.197) although the TRS in the oxidized zone was lower than in the other two zones. All total leachable Fe was identified as ferrous iron in all soil zones with the highest concentrations in the transition zone, although not found statistically significant (FeII-depth R2 = –0.369 and p = 0.33; Fe-depth R2 = 0.16 and p = 0.68). This was likely due to ferric attack on the iron sulfide generating ferrous iron in the bioleaching process along with ferric precipitation at pH values >1.811 that gave the highest total Fe value in the transition zone followed by lower values in the oxidized and unoxidized zones. Seasonal contributions to the Fe value in the transition zone may come from the oxidized zone via percolating water and from the unoxidized zone through capillary movement. The concentrations of leachable Al (Al-depth R2 = 0.74 and p = 0.023), Cu (Cu-depth R2 = 0.69 and p = 0.042), and Zn (Zn-depth R2 = 0.95 and p < 0.001) were significantly higher in the oxidized zone compared to the transition and unoxidized zones. Furthermore, Ca (Ca-depth R2 = –0.85 and p = 0.004) and Ni (Ni-depth R2 = 0.84 and p = 0.004) were significantly higher in both the oxidized and the transition zones compared to the unoxidized zone. Finally, the concentrations of leachable As (As-depth R2 = 0.48 and p = 0.195), Cr (Cr-depth R2 = 0.05 and p = 0.893), Co (Co-depth R2 = –0.47 and p = 0.197), and Mn (Mn-depth R2 = –0.47 and p = 0.197) did not differ significantly between the three soil zones.
16S rRNA gene-based microbial diversity in the soil zones
The 16S rRNA gene amplicon sequencing generated an average of 369,027 reads per soil zone and were assigned to a total of 4380 different individual gene sequences that are termed “amplicon sequence variants” or ASVs31. The rarefaction curves showing the number of individual ASVs versus the sequencing depth (Supplementary Fig. S1) plateaued for all samples indicating sufficient sequencing was performed to identify the majority of the populations. A significantly higher Shannon alpha diversity (Supplementary Fig. S2) was found in the unoxidized zone compared to the oxidized (one-way ANOVA with Tukey’s post hoc test; OX-UN p < 0.001) and transition zones (TR-UN p < 0.001), while no difference in diversity was found between the oxidized and transition zones (OX-TR p = 0.103).
All of the 16S rRNA gene ASVs were classified as Bacteria of which eleven phyla had a total relative abundance >1% (Fig. 2 and Supplementary Fig. S3). The reason why no archaeal 16S rRNA genes were amplified may have been due to primer bias as the amplicons were generated using polymerase chain reaction (PCR) primers designed for bacterial sequences (although they have been demonstrated to amplify some Archaea32) in combination with the low abundance of archaea (described below). A significant portion of the microbial communities in the three soil zones were represented by unassigned Bacteria (21–27, 1–6, and 37–39% relative abundance in the oxidized, transition, and unoxidized zones, respectively). The bacterial community composition and proportions of the identified phyla varied between the soil zones. Proteobacteria (46–51%), Chloroflexota (11–16%), Actinobacteriota (6–9%), and Acidobacteriota (3%) dominated in the oxidized zone. Of these four phyla, unassigned ASVs within the Gammaproteobacteria (43–47%), the Ktedonobacteraceae family (10–15%), and unassigned Actinobacteria (3%) accounted for the majority of the relative abundance. In the transition zone, the Proteobacteria (49–84%) comprised of ASVs aligning to the Gallionellaceae (8–69%), Sulfuricellaceae (1–47%), and Rhodanobacteraceae (3–9%) families. In the unoxidized zone, the community was dominated by Chloroflexota (22–23%), Planctomycetota (9–11%), Acidobacteriota (7–9%), Desulfobacterota (5%), and Caldatribacteriota (2–4%). Of these phyla, dominating ASVs belonged to the envOPS12 (10–11%), Dehalococcoidaceae (4–5%), unassigned Planctomycetota (4–5%), Aminicenantaceae (6–7%), unassigned BSN033 (1–2%), and the 34–128 family (2–4%).
Correlation between geochemical factors and microbial populations
The compositional principle component analysis biplot (Fig. 3; explaining 92.2% of the variation) showed that the biogeochemical environments in the three zones were distinct. The oxidized zone microbial populations were linked with high redox and high concentrations of Al, Cu, Ni, and Zn. In contrast to the analysis of the geochemical data alone (described above) and when ratios between all factors were taken into account; S, Fe2+, Mn, As, Cd, and Pb were all grouped with the samples from the oxidized zone. Although the ASVs from the Burkholderiaceae, Acidobacteraceae, and Acidimicrobiaceae families showed low total relative abundances (1, 1–2, and 1%, respectively) in the 16S rRNA gene amplicon data, the short links between metals and these ASVs suggested they have a substantial oxidizing role in this zone. The transition zone samples had some of the longest links from the center (e.g., ASVs from the Williamwhitmaniaceae, Gallionellaceae, and Treponemataceae B families), which indicated that their relative abundance were the most dissimilar to the other zones. The negative correlation between ASVs belonging to the Sulfuricellaceae and Nap2-2B families and TRS indicated that these ASVs might be sulfur oxidizers. Finally, the unoxidized zone ASVs also had some of the longest links from the center, i.e., ASVs from the Aminicenantaceae and 34–128 families that were not identified in the other zones.
Metagenome and metatranscriptome data
The triplicate metagenomes per soil type had an average mapping percentage of 84% between DNA reads and metagenomic assemblies31 (Supplementary Table S2) suggesting a good coverage of the three soil communities. Bioinformatic assignment of the community DNA reads (binning) generated 882 metagenome assembled genomes (MAGs) with >75% completeness and <10% contamination that were de-replicated to give 293 unique populations (Supplementary Data S1). Based upon the minimum information for publication33, the data included 140 “high-quality draft” MAGs with >90% completeness and <5% contamination. The three metatranscriptomes per soil type had an average read mapping percentage of 85% to the assemblies31 (Supplementary Table S2). Non-metric multidimensional scaling analysis of the reads and transcripts separated the soils on the first axis while the second axis separated the metaomics data for the transition zone community (Supplementary Fig. S4). This indicated that MAG presence was proportional to RNA transcript activities in the unoxidized PASS and oxidized ASS zones.
In many cases, phylogenetic assignment of the MAGs within the GTDB taxonomy was not achieved beyond, e.g., the class (Supplementary Data S1). For instance, the Planctomycetota MAGs UN2.108, UN2.11, and UN3.86 were not assigned to a class and four unclassified Gammaproteobacteria MAGs (OX1.71, OX3.35, OX3.60, and OX3.95) clustered with the Woesiales and Steroidobacterales orders (Supplementary Fig. S5).
MAG based microbial diversity and activity in the soil zones
All MAGs with a relative abundance or RNA transcript-based activity >1.0% in any one of the three soil zones were categorized into taxonomic groups (Supplementary Data S2 and S3). The phylum with the largest DNA and RNA based relative abundance in the oxidized zone was Proteobacteria with 34.7% and 21.5%, respectively (Fig. 4 and Supplementary Data S4). Of these abundances and activities, 31.2 and 15.1% were comprised of MAG group Ox_Gammaproteobacteria compared to <0.1% abundance and activity in the transition and unoxidized zones. Four MAG groups assigned to the Acidobacteriae soil bacteria Acidobacteriota34 made a further 18.4 and 34.8% of the relative abundance and activity, respectively. OxTr_Terracidiphilus genus MAGs had a low relative abundance of 1.2% but a high activity of 19.3% of populations typically growing at pH 3 to 6 that matches the acidic conditions in the oxidized ASS35. MAGs grouped as Ox_UBA7541 (13.9 and 10.7%) mediate iron cycling in tropical soils and are suggested to be named Candidatus Acidoferrales36. Actinobacteriota MAG groups constituted 11.7 and 10.1% of the oxidized zone community and activity, respectively and were dominated by MAGs assigned to the Acidimicrobiales that are typically associated with metal contaminated and acidic environments with the RAAP-2 family being identified in acid mine drainage37. In addition, five MAGs in two groups were assigned to the Streptosporangiaceae family found in soils and marine sediments38 named Ox_UBA9676 (3.4 and 3.9%) and OxTr_UBA8262 (2.1 and 2.8%). Groups Ox_Dormibacteraceae and Ox_UBA8260 assigned to the Dormibacteria class that has been identified from acid mine drainage39 accounted for 8.1 and 2.6% of the relative abundance and activity, respectively that was significantly higher than the deeper soil zones (Kruskal–Wallis test with Bonferroni correction; all p < 0.05; Supplementary Table S3). Finally, one Archaeal MAG group, Ox_Nitrososphaerales (3.4 and 8.0%) contained two MAGs with one further aligning with the autotrophic (i.e., carbon dioxide fixing) ammonia oxidizing Candidatus Nitrosotalea genus of phylum Thermoproteota40.
Numbers in brackets refer to the number of representative MAGs assigned to each phylum in the GTDB and the group taxonomy define selected abundant MAG groups for Archaeal (a) and Bacterial (b) domains. Bars indicate the relative abundance as percentages of the total reads mapped to the MAGs in the oxidized (OX), transition (TR), and unoxidized (UN) soil zones (metagenome, MG) and protein coding RNA transcripts mapped to the MAGs in the three soil zones (metatranscriptome, MT).
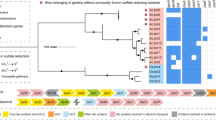
Proteobacteria also dominated the transition zone community with Gammaproteobacteria MAG groups constituting 54.4 and 59.3% of the relative abundance and activity, respectively (Fig. 4 and Supplementary Data S3). Proteobacteria groups included the Tr_Gallionella genus (29.1 and 33.2%) that are characterized as iron-oxidizers41 and the Tr_Sulfuricella genus (17.0 and 20.8%) that are autotrophic sulfur-oxidizers42. Both these MAG groups had significantly higher relative abundance and activity in the transition zone compared to both the oxidized and unoxidized zones (all p < 0.05). MAG group Tr_Rhodanobacter (4.9 and 4.2%) was significantly more abundant and active compared to the unoxidized zone (p < 0.05) with members of the genus identified as acid-tolerant denitrifiers43. The sulfur oxidizing Thiobacillus genus44 MAG group Tr_Thiobacillus (1.8 and 0.2%) was also only present in the transition zone. Finally, two MAG groups assigned to the Spirochaetota phylum were split into the mostly free living anaerobes in the Spirochaetales order45.
Planctomycetota with 18.1 and 13.6% relative abundance and activity was most abundant in the unoxidized zone (Fig. 4 and Supplementary Data S3) that was further divided into MAG groups attributed to three Phycisphaerae, one Brocadiae, and one Planctomycetes class. The partitioning of these MAG groups differed in the other two zones with UnTr_Phycisphaerae constituting a minor portion of the transition zone but was all but absent in the oxidized zone (all p < 0.05 except for OxUn_Thermoguttaceae). Characterized Phycisphaerae are aquatic bacteria associated with marine algae46 while Brocadiae have been detected in marine sediments47 and therefore, may have been present from before the PASS materials were drained. The second most abundant and active phylum group was attributed to the Chloroflexota group UnTr_GIF9 from the Dehalococcoidia that constituted 7.5 and 9.2% of the relative abundance and RNA transcript activity, respectively that was equally abundant in the unoxidized and transition zones, but significantly more active in the unoxidized zone (p < 0.05). Characterized Dehalococcoidia are obligate anaerobes identified in e.g., soils and aquatic environments48,49. Two additional Chloroflexota MAG groups were UnTr_Anaerolineae and Un_Thermoflexales that were significantly more abundant and active (p < 0.05) in the unoxidized zone compared to oxidized zone. The next most abundant bacterial phylum was the Caldatribacteriota (group UnTr_34-128) with 10.9 and 7.9% of the total relative abundance and activity in the unoxidized zone, respectively that had significantly higher relative abundance (p < 0.05) compared to the oxidized zone and was most similar to candidate bacteria 34–128 that lacks a detailed description50. Further abundant and active populations included UnTr_Desulfobacteria (5.7 and 6.1%) that had significantly higher abundance and activity compared to the oxidized zone (all p < 0.05). MAG groups that aligned with the Archaea included Un_TCS64 from the Thermoproteota class Bathyarchaeia (6.7 and 21.7%) that have been identified in e.g., marine environments, soils, and sediments where they may have an ecological role in symbiosis with Methanomicrobia51. A further unoxidized zone archaeal group was Un_ANME-1ex4572 (0.6 and 7.1%) that aligned with the Halobacterota class Syntropharchaeia that are characterized as anaerobic methane producers52. A third archaeal MAG group was UnTr_Heimdallarchaeia (2.3 and 0.5%) that aligned with the Asgardarchaeota class Heimdallarchaeia that partitioned in the soil types with the highest abundance and activity in the unoxidized zone compared to the oxidized zone (all p < 0.05).
mRNA transcript based metabolic processes in the three soil zones
Key genes were investigated in the RNA transcripts (Figs. 5–7, Supplementary Data S4, and Supplementary Fig. S6) to create a model of the major metabolic processes in the three zones (Fig. 8). While some MAG groups were identified in more than one zone, key groups were predominantly found in a single zone suggesting a clear distinction between the active populations. For instance, the average number of RNA transcripts (n = 3 ± SD) were considerably higher in the transition versus unoxidized zone encoding ferrous iron (7177 ± 2198 and 680 ± 25 TPMs) and sulfur compounds (1244 ± 302 and 1 ± 1 TPMs) oxidation that likely catalyzed the conversion of PASS to actual ASS. In contrast, transcripts for methanogenesis (i.e., methane production) were predominantly identified in the unoxidized compared to the transition zone (4575 ± 533 and 582 ± 106 TPMs). Furthermore, transcripts for both ferrous oxidation and ferric reduction genes suggested separate populations in aerobic and anaerobic soil niches or alternatively, multiple populations with different metabolic strategies were included within some MAG groups.
Bars indicate protein coding RNA transcripts mapped to the MAGs in the soil zone metatranscriptome. Maxima for each band was set to: 881 (dissimilatory iron reduction), 29 (dissimilatory nitrate reduction), 1929 (dissimilatory sulfur reduction), 5277 (Fe oxidation), 5612 (low pH), 1654 (metals resistance), 2481 (methanogenesis), 6249 (oxidative stress), and 1259 (sulfur oxidation). Numbers in brackets refer to the number of representative MAGs assigned to each phylum in the GTDB and the group taxonomy define selected RNA transcript based active MAG groups.
Bars indicate protein-coding RNA transcripts mapped to the MAGs in the soil zone metatranscriptome. RNA transcript maxima for each band and numbers in brackets are as described for Fig. 5.
Bars indicate protein-coding RNA transcripts mapped to the MAGs in the soil zone metatranscriptome. RNA transcript maxima for each band and numbers in brackets are as described for Fig. 5.
Key MAG groups and selected metabolisms are represented (black arrows denote transport processes). +/− across the cytoplasmic membrane shows the low pH adaptation of an internal positive membrane potential; MR refers to metal resistance by efflux of the metals from the cell; ISC inorganic sulfur compound oxidation; and e− for extracellular electron transfer. Assignment of metabolic characteristics was aided using metagenome analysis by DRAM115 while the figure was adapted from Wu et al.116.
The oxidized zone MAG group (Fig. 5) with the highest mRNA-based activity was OxTr_Terracidiphilus that was likely involved in organic matter decomposition35 potentially coupled to the reduction of sulfate (dsvABC) as electron acceptor. The group also showed transcripts for ferrous iron oxidation (cyc1/2 and foxABYZ) found in acidophile species that preferentially grow at low pH53; adaptation to low pH54 (e.g., kdpABCDE and shc); metals resistance55 including Cu, Co-Zn-Cd, and As (copA, czcABC, and acr3 and arsBC); and sodA, bcp, and dnaX for oxidative stress56 likely due to the ferrous ion. The Ox_Gammaproteobacteria group RNA transcripts suggested involvement in iron cycling53 via ferrous iron oxidation genes typically found in acidophiles and species that grow at neutral pH (i.e., neutrophiles) along with iron reduction (e.g., dmkB, fmnB, cymA, mtrABC, and ndh2) as the terminal electron acceptor. This MAG group also had RNA transcripts annotated for acidic metal containing environments such as low pH stress54 and metals resistance55. The Acidobacteriae Ox_UBA7541 transcripts also suggested its involvement in iron cycling with low transcript numbers for acidophilic and neutrophilic ferrous oxidation and with ferric iron reductases as a terminal electron acceptor36,53 along with transcripts typical for ASS such as pH homeostasis and metals resistance. Finally, the Archaea MAG group Ox_Nitrososphaerales from the Thermoproteota phylum showed transcript based activity in iron oxidation53 and dissimilatory iron reduction53.
The transition zone MAG group (Fig. 6) with the highest mRNA-based activity was Tr_Gallionella that was suggested to mediate ferrous iron oxidation via cyc1/cyc2, foxAB, foxYZ, mtoA, and mtrB. In addition, this group had transcripts for thiosulfate oxidation involving the SOX complex (soxAX) and anaerobic sulfate reduction for energy conservation. Once again, either this suggested separate populations within the MAG group or oxic versus anoxic niches mediating sulfur cycling. In addition, transcripts were assigned for pH homeostasis; metals resistance; and oxidative stress. The Spirochaetota phylum Tr_Treponematales MAG group RNA transcripts were assigned to iron oxidation and methanogenesis from acetate (acs, pta, and ackA) for energy generation; sulfate reduction and denitrification (i.e., conversion of nitrate to N2 gas; norB) for energy conservation; and adaptation to the low pH, high concentration of metals, and oxidative stress. The Tr_Sulfuricella RNA transcripts confirmed the geochemical suggestion (Fig. 3) that the populations were involved in sulfur oxidation (fccAB for sulfide oxidation and SoxAX) coupled to nitrate reduction/denitrification (narGIY, norB, napA, and nosZ) or ferric iron reduction (dmkAB, eetB, fmnB, mtrB, and cymA). Finally, Tr_Rhodanobacter had transcripts for thiosulfate oxidation (tsdA) that was likely coupled to nitrate reduction and denitrification as terminal electron acceptors57 and to a lesser extent ferric iron reduction.
The unoxidized zone MAG group (Fig. 7) with the highest mRNA-based activity was Un_TCS64 of the Thermoproteota class Bathyarchaeia with transcripts encoding genes involved in organic carbon oxidation potentially coupled to dissimilatory iron reduction, mttB for methanogenesis51, and metals resistance. Group Phycisphaerae Un_SG8-4 was suggested to grow via organic carbon oxidation coupled to ferric reduction along with transcripts coding for metal resistance. Archaeal phylum Halobacterota MAG group Un_ANME-1ex457252 showed RNA based activity of genes involved in anaerobic oxidation of methane via reversed methanogenesis suggested to be initiated by the mcrA encoded methyl-coenzyme M reductase58. Finally, MAG group UnTr_Desulfobacteria showed transcripts for genes involved in nitrate reduction (narHI), iron reduction, and sulfur reduction (psrA, dsvABC, ttrB, and phsA) as terminal electron acceptors
Discussion
The low pH in the oxidized and transition zones coupled with the highest concentrations of ferrous iron (although not significant) and leachable sulfur (most likely as sulfate) in the transition zone supported that metal sulfide oxidation occurred20. The results from the sulfur speciation also supported that pyrite can be preserved in the transition zone while the environment in the oxidized zone favors pyrite oxidation and thus metal mobilization20. The data also demonstrated that metals and metalloids were freed from the metal sulfides in the parent sediments and were leachable from the soil during the spring thaw such that they could cause environmental damage such as fish kills59.
In general, the populations identified as dominating the three soil zones are present in PASS and actual ASS boreal environments suggesting the data can be extrapolated to Finnish and Swedish soils. For instance, the oxidized zone Terracidiphilus from the Acetobacteraceae, Ox_UBA7541 from the Acidobacteriae, and Ox_Nitrososphaerales have been identified in Swedish60 and Finnish61 ASS. The key transition zone genus Gallionella are detected in several other boreal ASS environments in Sweden60 and Finland13,24,28 along with Australian coastal ASS drains17 while Sulfuricella are present in Finnish ASS26. In addition, Rhodanobacter are present in dredged river sediment containing sulfidic materials deposited on land nearby the Risöfladan field28 along with bioremediation experiments treating the Risöfladan ASS24,61. While fewer studies have been carried out on Scandinavian unoxidized PASS, the Desulfobacteria class has been identified in Swedish ASS environments60. Finally, despite the distance and contrasting conditions, the unoxidized zone selected for a similar microbial community as an Australian coastal ASS where 16S rRNA gene sequences were identified aligning with sulfate reducing Desulfobacterales, methanogenic Halobacteria, metal reducing Phycisphaerae, organohalide-respiring Dehalococcoidetes, and strictly anaerobic Anaerolineae16.
The dominant ferrous iron and sulfur oxidizing microbes in the transition zone were from the Gallionella and Sulfuricella genera. In contrast to the original description of Gallionella species as solely iron oxidizers62, but in agreement with results from a low pH acid mine drainage site63, the Tr_Gallionella group was also suggested to oxidize reduced sulfur compounds. In published Gallionella genomes, ferrous oxidation is attributed to MtoAB64, while the Tr_Gallionella group in this study was also suggested to code for cyc1/cyc2 that are involved in ferrous oxidation by the Acidithiobacillus genus65 and fox genes from e.g., the acidophile Sulfuracidifex metallicus66. Sulfuricella species oxidize reduced sulfur compounds at near neutral pH conditions in soils67 and freshwater lakes42 with the MAG group transcripts also supporting anaerobic sulfate and nitrate reduction for energy conservation63. The high relative abundance and activity of Gallionella and Sulfuricella species in the gradual transition of PASS to ASS in natural environments was in contrast to rapid PASS oxidation in an aerated bioreactor wherein extreme acidophiles including Acidithiobacillus and Acidocella were also identified13. In addition, rapid oxidation of the dredged river sediment mentioned above was dominated by 16S rRNA gene sequences most similar to the extremely acidophilic genus Acidithiobacillus28. The transition zone ASVs, assembled genomes, and mRNA transcripts also identified Tr_Rhodanobacter as involved in thiosulfate oxidation. Many of the transition zone MAG groups contained RNA transcripts coding for pH homeostasis54. Mechanisms to maintain a near neutral cytoplasmic pH are divided into the “first line of defense” primarily identified in acidophiles (rather than neutrophiles) that stop the influx of protons into the cell followed by the “second line of defense” that removes protons once they have entered68. For instance, the Tr_Gallionella and Tr_Treponematales groups had mRNA transcripts assigned to the “first line of defense” potassium importer genes kdpABCDE suggested to form an internal positive membrane potential to hinder proton influx in acidophiles69. Tr_Gallionella also encoded “second line of defense” genes such as arginine decarboxylase in the arginine-dependent acid resistance (ADAR) system68 plus a major histone-like protein HU that is suggested to regulate the ADAR system in Escherichia coli70. However, the lack of RNA transcripts for these genes suggested the Tr_Gallionella group was not acid stressed in the transition zone. In contrast, the oxidized zone Ox_UBA7541 group RNA transcripts not only coded for the potassium-transporting ATPase system but also shc squalene-hopene cyclase for “first line of defense” cell membrane associated pH homeostasis system71. Therefore, the MAG groups in the oxidized and transition zones were predominantly acid tolerant or moderate acidophiles with some populations potentially being adapted to lower pH values or exhibiting transcripts relate to pH stress. Finally, several oxidized and transition zone MAG groups were suggested to be adapted to metal containing environments with transcripts coding for arsenic, cadmium, cobalt–zinc–cadmium, and mercury resistance55.
Several poorly characterized populations including the archaeal MAG groups Un_ANME-1ex4572 and Un_TCS64 dominated the unoxidized zone but were not identified in the 16S rRNA gene amplicons (potentially due to the PCR bias mentioned above). The unoxidized zone MAG groups were suggested to grow via anaerobic energy conservation including organic carbon oxidation by Un_TCS64 plus Un_SG8-4 populations that may have been retained in the sediments from their marine origins46,72 and UnTr_Anaerolineae. Unoxidized zone populations potentially also carried out sulfate reduction by e.g., UnTr_Desulfobacteria that contained transcripts coding for genes involved in nitrate, iron, and sulfur reduction. Once the inorganic electron acceptors for energy conservation had been consumed in the unoxidized zone, then a transition may have occurred whereby the deeper community altered to favor methanogenic populations such as Un_TCS6458. The community within the unoxidized zone may also have mediated anaerobic oxidation of methane (AOM)73 by the methanotrophic (i.e., methane consuming) Archaeal Un_ANME-1ex4572 group from the Halobacterota. However, AOM only occurs during symbiosis between a methanotrophic population in association with, e.g., a sulfate reducing population such as UnTr_Desulfobacteria. In support of AOM occurring in the unoxidized zone, MAG groups UnTr_GIF9 and UnTr_34-128 that aligned with the Dehalococcoidia and Caldatribacteriota, respectively strongly correlate with AOM rates in Baltic Sea sediments74.
In conclusion, the three soil zones had highly distinct populations and activities supporting hypothesis one that the microbial communities altered during PASS conversion to ASS. The transition zone Gallionella population contained RNA transcripts for a typical acidophile first line of defense potassium uptake system suggesting at least some of the transition zone populations supported the hypothesis two of moderate acidophiles mediating PASS conversion to ASS. In contrast, anaerobic metabolic processes dominated the unoxidized zone with methanogenic methane being consumed during AOM.
Methods
Study site and soil sampling
Soil was sampled in mid-August 2017 from typical farmland acid sulfate soil in the area at the Risöfladan experimental field in Vaasa, Finland (63° 02’ 50.22“N, 21° 42’ 41.85“E; Fig. 1). The oxidized (OX), transition (TR), and unoxidized (UN) soil zones were exposed using a mechanical digger, surfaces that had come into contact with the scoop aseptically scraped away, and soil from the three zones collected (see Supplementary Methods for details). Samples were analyzed for pH, oxidation–reduction potential, conductivity, ferrous iron, multi-elements, and sulfur speciation as described in the Supplementary Methods. Three replicates from the OX, TR, and UN zones were used in all analyses.
RNA/DNA preparation and sequencing
RNA and DNA were extracted, processed, quality controlled, and sequencing libraries prepared as described in the Supplementary Methods. The RNA concentrations after DNase treatment and rRNA depletion as well as the total amount of RNA and DNA are in Supplementary Table S4 and detailed in Högfors-Rönnholm et al.31. All metagenome and metatranscriptome sequencing was performed by the Joint Genome Institute in Walnut Creek, CA, USA on the Illumina NovaSeq platform generating 2 × 151 bp sequences. The metagenomes and metatranscriptomes were assembled, binned, mapped, and processed as described in the Supplementary Methods.
16S rRNA gene amplicon sequencing and analysis
Two-step 16S rRNA gene PCR amplification (V3–V4 region) using primers from Hugerth et al.75 and sequencing at the SciLifeLab, Sweden (2 × 300 bp) were carried out as previously described24,31. ASVs were derived using the DADA2 pipeline76 and annotated against the GTDB bacterial small subunit collection (release 86)77. Additional downstream analysis is described in the Supplementary Methods.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The 16S rRNA gene amplicons are available from the NCBI BioProject ID PRJNA52414478. The raw sequencing data are available from the NCBI Sequence Read Archive under accession numbers SRP1850821679, SRP1850841780, SRP1850861881, SRP1850871982, SRP1850882083, SRP1850892184, SRP1850922285, SRP1850912386 and SRP1852232487 for metagenomes and SRP1852222588, SRP1850942689, SRP1850932790, SRP1852252891, SRP1852242992, SRP1852263093, SRP1852273194, SRP1852283295 and SRP1852293396 for metatranscriptomes as detailed in Högfors-Rönnholm et al.31 and summarized in Supplementary Table S5. The final assembled contigs are available from the Integrated Microbial Genomes and Microbiomes (IMG/M) portal under ID numbers 330003167197, 330003167298, 330003167099, 3300031669100, 3300031578101, 3300031673102, 3300031566103, 3300031565104 and 3300031539105 for metagenomes and 3300030718106, 3300030716107, 3300030712108, 3300030713109, 3300030710110, 3300030711111, 3300030714112, 3300030719113 and 3300030717114 for metatranscriptomes, and summarized in Supplementary Table S5.
Code availability
All software versions and scripts needed to reproduce the results are specified in the Materials and Methods section and in the Supplementary Methods.
References
Rickard, D. & Luther, G. W. Chemistry of iron sulfides. Chem. Rev. 107, 514–562 (2007).
Karimian, N., Johnston, S. G. & Burton, E. D. Iron and sulfur cycling in acid sulfate soil wetlands under dynamic redox conditions: a review. Chemosphere 197, 803–816 (2018).
Sullivan, L. A., Ward, N. J., Bush, R. T. & Burton, E. D. Improved identification of sulfidic soil materials by a modified incubation method. Geoderma 149, 33–38 (2009).
Schippers, A. & Sand, W. Bacterial leaching of metal sulfides proceeds by two indirect mechanisms via thiosulfate or via polysulfides and sulfur. Appl. Environ. Microbiol. 65, 319–321 (1999).
Ljung, K., Maley, F., Cook, A. & Weinstein, P. Acid sulfate soils and human health-A Millennium Ecosystem Assessment. Environ. Int. 35, 1234–1242 (2009).
Stephens, F. J. & Ingram, M. Two cases of fish mortality in low pH, aluminium rich water. J Fish Dis. 29, 765–770 (2006).
Sutela, T. & Vehanen, T. The effects of acidity and aluminium leached from acid-sulphate soils on riverine fish assemblages. Boreal Environ. Res. 22, 385–391 (2017).
Macdonald, B. C. T., Denmead, O. T., White, I. & Melville, M. D. Natural sulfur dioxide emissions from sulfuric soils. Atmos. Environ. 38, 1473–1480 (2004).
Bonnefoy, V. & Holmes, D. S. Genomic insights into microbial oxidation and iron homeostasis in extremely acidic environments. Environ. Microbiol. 14, 1597–1611 (2012).
Dopson, M. & Johnson, D. B. Biodiversity, metabolism and applications of acidophilic sulfur-metabolizing micro-organisms. Environ. Microbiol. 14, 2620–2631 (2012).
Vera, M., Schippers, A. & Sand, W. Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation–part A. Appl. Microbiol. Biotechnol. 97, 7529–7541 (2013).
Huang, L. N., Kuang, J. L. & Shu, W. S. Microbial ecology and evolution in the acid mine drainage model system. Trends Microbiol. 24, 581–593 (2016).
Wu, X. et al. Microbial community potentially responsible for acid and metal release from an Ostrobothnian acid sulfate soil. FEMS Microbiol. Ecol. 84, 555–563 (2013).
Ohba, H. & Owa, N. Vertical distribution of physico-chemical properties and number of sulfur-oxidizing bacteria in the buried layer of soil profiles with marine-reduced sulfur compounds. Soil Sci. Plant Nutr. 51, 379–388 (2005).
Arkesteyn, G. J. M. W. Pyrite oxidation in acid sulphate soils: the role of microorganisms. Plant Soil 54, 119–134 (1980).
Ling, Y.-C. et al. Distribution of iron- and sulfate-reducing bacteria across a coastal acid sulfate soil (CASS) environment: implications for passive bioremediation by tidal inundation. Front. Microbiol. https://doi.org/10.3389/fmicb.2015.00624 (2015).
Stroud, J. L., Low, A., Collins, R. N. & Manefield, M. Metal(loid) bioaccessibility dictates microbial community composition in acid sulfate soil horizons and sulfidic drain sediments. Environ. Sci. Technol. 48, 8514–8521 (2014).
Su, J.-Q. et al. Metagenomic assembly unravel microbial response to redox fluctuation in acid sulfate soil. Soil Biol. Biochem. 105, 244–252 (2017).
Ling, Y. C., Gan, H. M., Bush, M., Bush, R. & Moreau, J. W. Time-resolved microbial guild responses to tidal cycling in a coastal acid-sulfate system. Environ. Chem. 15, 2–17 (2018).
Boman, A., Frojdo, S., Backlund, K. & Åström, M. E. Impact of isostatic land uplift and artificial drainage on oxidation of brackish-water sediments rich in metastable iron sulfide. Geochim. Cosmochim. Acta. 74, 1268–1281 (2010).
Åström, M., Österholm, P., Bärlund, I. & Tattari, S. Hydrochemical effects of surface liming, controlled drainage and lime-filter drainage on boreal acid sulfate soils. Water Air Soil Pollut. 179, 107–116 (2007).
Boman, A., Astrom, M. & Frojdo, S. Sulfur dynamics in boreal acid sulfate soils rich in metastable iron sulfide-the role of artificial drainage. Chem. Geol. 255, 68–77 (2008).
Dalhem, K., Engblom, S., Stén, P. & Österholm, P. Subsurface hydrochemical precision treatment of a coastal acid sulfate soil. Appl. Geochem. 100, 352–362 (2019).
Högfors-Rönnholm, E. et al. Chemical and microbiological evaluation of novel chemical treatment methods for acid sulfate soils. Sci. Total Environ. 625, 39–49 (2018).
Nordmyr, L., Boman, A., Åström, M. & Österholm, P. Estimation of leakage of chemical elements from boreal acid sulphate soils based on a geochemical and hydrochemical approach. Boreal Environ. Res. 11, 261–271 (2006).
Wu, X. et al. Impact of mitigation strategies on microbial community from an Ostrobothnian acid sulfate soil. Sci. Total Environ. 526, 215–221 (2015).
Österholm, P. et al. Groundwater management of acid sulfate soils using controlled drainage, by-pass flow prevention, and subsurface irrigation on a boreal farmland. Acta Agric. Scand. Sect. B. Soil Plant Sci. 65, 110–120 (2015).
Johnson, A. et al. Dredging and deposition of metal sulfide rich river sediments results in rapid conversion to acid sulfate soil materials. Sci. Total Environ. 813, 151864 (2022).
Dopson, M. & Lindström, E. B. Analysis of community composition during moderately thermophilic bioleaching of pyrite, arsenical pyrite, and chalcopyrite. Microb. Ecol. 48, 19–28 (2004).
Saglam, E. S., Akcay, M., Colak, D. N., Inan Bektas, K. & Belduz, A. O. Generation of acid mine drainage around the Karaerik copper mine (Espiye, Giresun, NE Turkey): implications from the bacterial population in the Acisu effluent. Extremophiles 20, 673–685 (2016).
Högfors-Rönnholm, E. et al. Metagenomes and metatranscriptomes from boreal potential and actual acid sulfate soil materials. Sci. Data 6, 207 (2019).
Lopez-Fernandez, M., Broman, E., Wu, X., Bertilsson, S. & Dopson, M. Investigation of viable taxa in the deep terrestrial biosphere suggests high rates of nutrient recycling. FEMS Microbiol. Ecol. https://doi.org/10.1093/femsec/fiy1121 (2018).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017).
Conradie, T. A. & Jacobs, K. Distribution patterns of Acidobacteriota in different fynbos soils. PLoS ONE 16, e0248913 (2021).
Garcia-Fraile, P., Benada, O., Cajthaml, T., Baldrian, P. & Llado, S. Terracidiphilus gabretensis gen. nov., sp. nov., an abundant and active forest soil Acidobacterium important in organic matter transformation. Appl. Environ. Microbiol. 82, 560–569 (2016).
Epihov, D. Z. et al. Legume–microbiome interactions unlock mineral nutrients in regrowing tropical forests. Proc. Natl Acad. Sci. USA 118, e2022241118 (2021).
Pinto, A. J., Sharp, J. O., Yoder, M. J. & Almstrand, R. Draft genome sequences of two novel Acidimicrobiaceae members from an acid mine drainage biofilm metagenome. Genome Announc. 4, e01563-01515 (2016).
Otoguro, M., Yamamura, H. & Quintana, E. T. in The Prokaryotes: Actinobacteria (eds Rosenberg, E. et al.) 1011–1045 (Springer Berlin Heidelberg, 2014).
Grettenberger, C. L. & Hamilton, T. L. Metagenome assembled genomes of novel taxa from an acid mine drainage environment. Appl. Environ. Microbiol. 87, e00772-21 (2021).
Prosser, J. I. & Nicol, G. W. in Bergey’s Manual of Systematics of Archaea and Bacteria (eds Whitman, W. B. et al.) (John Wiley & Sons, 2016), https://doi.org/10.1002/9781118960608.obm00123.
Pedersen, K. in Encyclopedia of Geobiology (eds J. Reitner & V. Thiel) 411–412 (Springer Netherlands, 2011).
Kojima, H. & Fukui, M. Sulfuricella denitrificans gen. nov., sp. nov., a sulfur-oxidizing autotroph isolated from a freshwater lake. Int. J. Syst. Evol. Bacteriol. 60, 2862–2866 (2010).
Green, S. J. et al. Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl. Environ. Microbiol. 78, 1039–1047 (2012).
Kelly, D. P. & Wood, A. P. Confirmation of Thiobacillus denitrificans as a species of the genus Thiobacillus, in the beta-subclass of the Proteobacteria, with strain NCIMB 9548 as the type strain. Int. J. Syst. Evol. Microbiol. 50, 547–550 (2000).
Gupta, R. S., Mahmood, S. & Adeolu, M. A phylogenomic and molecular signature based approach for characterization of the phylum Spirochaetes and its major clades: proposal for a taxonomic revision of the phylum. Front. Microbiol. 4, 217 (2013).
Fukunaga, Y. et al. Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 55, 267–275 (2009).
Sonthiphand, P., Hall, M. W. & Neufeld, J. D. Biogeography of anaerobic ammonia-oxidizing (anammox) bacteria. Front. Microbiol. 5, 399–399 (2014).
Löffler, F. E. et al. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 63, 625–635 (2013).
Yang, Y. et al. Roles of organohalide-respiring Dehalococcoidia in carbon cycling. mSystems 5, e00757-00719 (2020).
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017).
Xiang, X. et al. Distribution of Bathyarchaeota communities across different terrestrial settings and their potential ecological functions. Sci. Rep. 7, 45028 (2017).
Mand, T. D. & Metcalf, W. W. Energy conservation and hydrogenase function in methanogenic archaea, in particular the genus Methanosarcina. Microbiol. Mol. Biol. Rev. https://doi.org/10.1128/mmbr.00020-00019 (2019).
Garber, A. I. et al. FeGenie: a comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front. Microbiol. https://doi.org/10.3389/fmicb.2020.00037 (2020).
Slonczewski, J. L., Fujisawa, M., Dopson, M. & Krulwich, T. A. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv. Microb. Physiol. 55, 1–79 (2009).
Dopson, M. & Holmes, D. S. Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl. Microbiol. Biotechnol. 98, 8133 (2014).
Galaris, D., Barbouti, A. & Pantopoulos, K. Iron homeostasis and oxidative stress: an intimate relationship. Biochim. Biophys. Acta Mol. Cell Res. 1866, 118535 (2019).
Lee, C. S., Kim, K. K., Aslam, Z. & Lee, S.-T. Rhodanobacter thiooxydans sp. nov., isolated from a biofilm on sulfur particles used in an autotrophic denitrification process. Int. J. Syst. Evol. Microbiol. 57, 1775–1779 (2007).
Qi, Y. L. et al. Comparative genomics reveals thermal adaptation and a high metabolic diversity in “Candidatus Bathyarchaeia”. Msystems https://doi.org/10.1128/mSystems.00252-00221 (2021).
Roos, M. & Åström, M. Gulf of Bothnia receives high concentrations of potentially toxic metals from acid sulphate soils. Boreal Environ. Res. 11, 383–388 (2006).
Christel, S. et al. Comparison of boreal acid sulfate soil microbial communities in oxidative and reductive environments. Res. Microbiol. https://doi.org/10.1016/j.resmic.2019.06.002 (2019).
Högfors-Rönnholm, E. et al. Biodegraded peat and ultrafine calcium carbonate result in retained metals and higher microbial diversities in boreal acid sulfate soil. Soil Ecol. Lett. 2, 120–130 (2020).
Hallbeck, L. & Pedersen, K. in The Prokaryotes: Alphaproteobacteria and Betaproteobacteria (eds Rosenberg, E. et al.) 853–858 (Springer Berlin Heidelberg, 2014).
Kadnikov, V. V. et al. A novel uncultured bacterium of the family Gallionellaceae: description and genome reconstruction based on metagenomic analysis of microbial community in acid mine drainage. Microbiology 85, 449–461 (2016).
Emerson, D. et al. Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front. Microbiol. https://doi.org/10.3389/fmicb.2013.00254 (2013).
Quatrini, R. et al. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10, 394 (2009).
Bathe, S. & Norris, P. R. Ferrous iron- and sulfur-induced genes in Sulfolobus metallicus. Appl. Environ. Microbiol. 73, 2491–2497 (2007).
Hayakawa, A. et al. Sulfur-based denitrification in streambank subsoils in a headwater catchment underlain by marine sedimentary rocks in Akita, Japan. Front. Environ. Sci. https://doi.org/10.3389/fenvs.2021.664488 (2021).
González-Rosales, C., Vergara, E., Dopson, M., Valdés, J. H. & Holmes, D. S. Integrative genomics sheds light on evolutionary forces shaping the Acidithiobacillia class acidophilic lifestyle. Front. Microbiol. https://doi.org/10.3389/fmicb.2021.822229 (2022).
Buetti-Dinh, A., Dethlefsen, O., Friedman, R. & Dopson, M. Transcriptomic analysis reveals how a lack of potassium ions increases Sulfolobus acidocaldarius sensitivity to pH changes. Microbiology 162, 1422–1434 (2016).
Bi, H. K., Sun, L. L., Fukamachi, T., Saito, H. & Kobayashi, H. HU participates in expression of a specific set of genes required for growth and survival at acidic pH in Escherichia coli. Curr. Microbiol. 58, 443–448 (2009).
Mykytczuk, N. C. S., Trevors, J. T., Ferroni, G. D. & Leduc, L. G. Cytoplasmic membrane fluidity and fatty acid composition of Acidithiobacillus ferrooxidans in response to pH stress. Extremophiles 14, 427–441 (2010).
Lloyd, K. G. et al. Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218 (2013).
Zhang, X., Yuan, Z. & Hu, S. Anaerobic oxidation of methane mediated by microbial extracellular respiration. Environ. Microbiol. Rep. 13, 790–804 (2021).
Iasakov, T. R. et al. The Baltic Sea methane pockmark microbiome: The new insights into the patterns of relative abundance and ANME niche separation. Mar. Environ. Res. 173, 105533 (2022).
Hugerth, L. W. et al. DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies. Appl. Environ. Microbiol. 80, 5116–5123 (2014).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581 (2016).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. https://doi.org/10.1038/nbt.4229 (2018).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/bioproject:PRJNA524144 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185082 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185084 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185086 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185087 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185088 (2019).
NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRP185089 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185092 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185091 (2019).
NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRP185223 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185222 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185094 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185093 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185225 (2019).
NCBI Sequence Read Archive, https://identifiers.org/ncbi/insdc.sra:SRP185224 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185226 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185227 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185228 (2019).
NCBI Sequence Read Archive. https://identifiers.org/ncbi/insdc.sra:SRP185229 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300031671 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300031672 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300031670 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300031669 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300031578 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300031673 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300031566 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300031565 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300031539 (2019).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300030718 (2018).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300030716 (2018).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300030712 (2018).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300030713 (2018).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300030710 (2018).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300030711 (2018).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300030714 (2018).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300030719 (2018).
Integrated Microbial Genomes & Microbiomes portal. https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=3300030717 (2018).
Shaffer, M. et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 48, 8883–8900 (2020).
Wu, X. et al. Microbial metagenomes from three aquifers in the Fennoscandian shield terrestrial deep biosphere reveal metabolic partitioning among populations. ISME J. 10, 1192–1203 (2016).
Acknowledgements
A portion of this research was performed under the Facilities Integrating Collaborations for User Science (FICUS) initiative and used resources at the DOE Joint Genome Institute and the Environmental Molecular Sciences Laboratory, which are DOE Office of Science User Facilities. Both facilities are sponsored by the Office of Biological and Environmental Research and operated under Contract Nos. DE-AC02-05CH11231 (JGI) and DE-AC05-76RL01830 (EMSL). E.H.-R. acknowledges the Nessling Foundation (grant number 201700273 and 201800502) and Svensk-Österbottniska Samfundet r.f. (grant number 18/0604) for financial support. E.H.-R. and S.E. further acknowledge the financial support by the European Regional Development Fund via the Interreg Botnia-Atlantica program to the project “Sustainable treatment of coastal deposited sulfide soils (STASIS).” M.D. acknowledges The Swedish Research Council Formas (grant number 2018-00760) and the Geological Survey of Sweden (grant number 36-1878/2017) for financial support. M.D further acknowledges the Science for Life Laboratory (SciLifeLab) and the National Genomics Infrastructure (NGI). The computations were enabled by resources (SNIC 2021/22-628) provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX at Uppsala University partially funded by the Swedish Research Council through grant agreement no. 2018-05973. D.L. and D.B. acknowledges the Swedish Research Council infrastructure project Biodiversity Atlas Sweden (VR, grant number 2017-00688) and the marine strategic research environment EcoChange (Formas) for financial support. Jussi Hyvönen is gratefully acknowledged for geochemical analyses.
Author information
Authors and Affiliations
Contributions
E.H.-R. and M.D. conceived the study. E.H.-R. and S.E. took the samples. E.H.-R., S.C., M.L.-F., and P.Ö. prepared nucleic acids and analyzed samples. D.L., D.B., S.C., and T.L. analyzed the data. E.H.-R. and M.D. wrote the manuscript. All authors commented upon and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth & Environment thanks Chamindra Vithana and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary handling editor: Clare Davis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Högfors-Rönnholm, E., Lundin, D., Brambilla, D. et al. Gallionella and Sulfuricella populations are dominant during the transition of boreal potential to actual acid sulfate soils. Commun Earth Environ 3, 304 (2022). https://doi.org/10.1038/s43247-022-00642-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-022-00642-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.