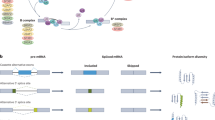
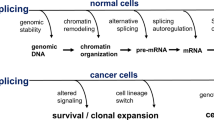
Abstract
High-throughput sequencing and functional characterization of the cancer transcriptome have uncovered cancer-specific dysregulation of RNA splicing across a variety of cancers. Alterations in the cancer genome and dysregulation of RNA splicing factors lead to missplicing, splicing alteration-dependent gene expression and, in some cases, generation of novel splicing-derived proteins. Here, we review recent advances in our understanding of aberrant splicing in cancer pathogenesis and present strategies to harness cancer-specific aberrant splicing for therapeutic intent.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nilsen, T. W. & Graveley, B. R. Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 (2010).
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Brody, E. & Abelson, J. The ‘spliceosome’: yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 228, 963–967 (1985).
Will, C. L. & Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707 (2011).
El Marabti, E., Malek, J. & Younis, I. Minor intron splicing from basic science to disease. Int. J. Mol. Sci. 22, 6062 (2021).
Agafonov, D. E. et al. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351, 1416–1420 (2016).
Fica, S. M. Cryo-EM snapshots of the human spliceosome reveal structural adaptions for splicing regulation. Curr. Opin. Struct. Biol. 65, 139–148 (2020).
Nguyen, T. H. et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 523, 47–52 (2015).
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008).
Gerstein, M. B. et al. Comparative analysis of the transcriptome across distant species. Nature 512, 445–448 (2014).
Braberg, H. et al. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154, 775–788 (2013).
Howard, J. M. & Sanford, J. R. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 6, 93–110 (2015).
Thibault, P. A. et al. hnRNP A/B proteins: an encyclopedic assessment of their roles in homeostasis and disease. Biology 10, 712 (2021).
Van Nostrand, E. L. et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020).
Schwerk, C. & Schulze-Osthoff, K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell 19, 1–13 (2005).
Jiang, Z. H., Zhang, W. J., Rao, Y. & Wu, J. Y. Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl Acad. Sci. USA 95, 9155–9160 (1998).
Wu, J. Y., Tang, H. & Havlioglu, N. Alternative pre-mRNA splicing and regulation of programmed cell death. Prog. Mol. Subcell. Biol. 31, 153–185 (2003).
Kedzierska, H. & Piekielko-Witkowska, A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 396, 53–65 (2017).
Boise, L. H. et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74, 597–608 (1993).
Dou, Z. et al. Aberrant Bcl-x splicing in cancer: from molecular mechanism to therapeutic modulation. J. Exp. Clin. Cancer Res. 40, 194 (2021).
Kim, J. H. et al. MCL-1ES, a novel variant of MCL-1, associates with MCL-1L and induces mitochondrial cell death. FEBS Lett. 583, 2758–2764 (2009).
Supek, F., Minana, B., Valcarcel, J., Gabaldon, T. & Lehner, B. Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335 (2014).
Jung, H. et al. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 47, 1242–1248 (2015).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Papaemmanuil, E. et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011).
Wang, L. et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 (2011).
Graubert, T. A. et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 44, 53–57 (2011).
Harbour, J. W. et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 45, 133–135 (2013).
Quesada, V. et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 44, 47–52 (2011).
Martin, M. et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 45, 933–936 (2013).
Karni, R. et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 (2007).
Anczukow, O. et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19, 220–228 (2012).
Danan-Gotthold, M. et al. Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic Acids Res. 43, 5130–5144 (2015).
Chen, L., Tovar-Corona, J. M. & Urrutia, A. O. Increased levels of noisy splicing in cancers, but not for oncogene-derived transcripts. Hum. Mol. Genet. 20, 4422–4429 (2011).
Kahles, A. et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 34, 211–224 (2018).
Seiler, M. et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296 (2018).
Ellis, M. J. et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012).
The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Biankin, A. V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012).
Imielinski, M. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012).
Chen, S., Benbarche, S. & Abdel-Wahab, O. Splicing factor mutations in hematologic malignancies. Blood 138, 599–612 (2021).
Cretu, C. et al. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell 64, 307–319 (2016).
Teng, T. et al. Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A–SF3b complex. Nat. Commun. 8, 15522 (2017).
Malcovati, L. et al. SF3B1-mutant MDS as a distinct disease subtype: a proposal from the International Working Group for the Prognosis of MDS. Blood 136, 157–170 (2020).
Dalton, W. B. et al. The K666N mutation in SF3B1 is associated with increased progression of MDS and distinct RNA splicing. Blood Adv. 4, 1192–1196 (2020).
Darman, R. B. et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 13, 1033–1045 (2015).
DeBoever, C. et al. Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLoS Comput. Biol. 11, e1004105 (2015).
Boultwood, J. et al. The role of the iron transporter ABCB7 in refractory anemia with ring sideroblasts. PLoS ONE 3, e1970 (2008).
Nikpour, M. et al. The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia 27, 889–896 (2013).
Inoue, D. et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature 574, 432–436 (2019).
Clough, C. A. et al. Coordinated mis-splicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. Blood 139, 2038–2048 (2021).
Lieu, Y. K. et al. SF3B1 mutant-induced missplicing of MAP3K7 causes anemia in myelodysplastic syndromes. Proc. Natl Acad. Sci. USA 119, e2111703119 (2022).
Zhang, J. et al. Disease-causing mutations in SF3B1 alter splicing by disrupting interaction with SUGP1. Mol. Cell 76, 82–95 (2019).
Liu, Z. et al. Pan-cancer analysis identifies mutations in SUGP1 that recapitulate mutant SF3B1 splicing dysregulation. Proc. Natl Acad. Sci. USA 117, 10305–10312 (2020).
Gozani, O., Potashkin, J. & Reed, R. A potential role for U2AF–SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18, 4752–4760 (1998).
Merendino, L., Guth, S., Bilbao, D., Martinez, C. & Valcarcel, J. Inhibition of msl-2 splicing by sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402, 838–841 (1999).
Wu, S., Romfo, C. M., Nilsen, T. W. & Green, M. R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402, 832–835 (1999).
Reed, R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 3, 2113–2123 (1989).
Haferlach, T. et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28, 241–247 (2014).
Papaemmanuil, E. et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122, 3616–3627 (2013).
Ilagan, J. O. et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 25, 14–26 (2015).
Wheeler, E. C. et al. Integrative RNA-omics discovers GNAS alternative splicing as a phenotypic driver of splicing factor-mutant neoplasms. Cancer Discov. 12, 836–855 (2021).
Smith, M. A. et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 21, 640–650 (2019).
Thol, F. et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 119, 3578–3584 (2012).
Desai, P. et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24, 1015–1023 (2018).
Graveley, B. R. & Maniatis, T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1, 765–771 (1998).
Liu, H. X., Chew, S. L., Cartegni, L., Zhang, M. Q. & Krainer, A. R. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20, 1063–1071 (2000).
Schaal, T. D. & Maniatis, T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 19, 261–273 (1999).
Daubner, G. M., Clery, A., Jayne, S., Stevenin, J. & Allain, F. H. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 31, 162–174 (2012).
Kim, E. et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27, 617–630 (2015).
Zhang, J. et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl Acad. Sci. USA 112, E4726–4734 (2015).
Yoshimi, A. et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574, 273–277 (2019).
Madan, V. et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 6, 6042 (2015).
Inoue, D. et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat. Genet. 53, 707–718 (2021).
Burge, C. B., Padgett, R. A. & Sharp, P. A. Evolutionary fates and origins of U12-type introns. Mol. Cell 2, 773–785 (1998).
Tarn, W. Y., Yario, T. A. & Steitz, J. A. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA 1, 644–656 (1995).
Suzuki, H. et al. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature 574, 707–711 (2019).
Shuai, S. et al. The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature 574, 712–716 (2019).
Bousquets-Munoz, P. et al. PanCancer analysis of somatic mutations in repetitive regions reveals recurrent mutations in snRNA U2. NPJ Genom. Med. 7, 19 (2022).
Kurtovic-Kozaric, A. et al. PRPF8 defects cause missplicing in myeloid malignancies. Leukemia 29, 126–136 (2015).
Dvinge, H., Kim, E., Abdel-Wahab, O. & Bradley, R. K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16, 413–430 (2016).
Hernandez, J. et al. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 13, 466–472 (2016).
Bechara, E. G., Sebestyen, E., Bernardis, I., Eyras, E. & Valcarcel, J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell 52, 720–733 (2013).
Ghigna, C. et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20, 881–890 (2005).
Goncalves, V. et al. Phosphorylation of SRSF1 by SRPK1 regulates alternative splicing of tumor-related Rac1b in colorectal cells. RNA 20, 474–482 (2014).
Tacke, R., Chen, Y. & Manley, J. L. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc. Natl Acad. Sci. USA 94, 1148–1153 (1997).
Xiao, S. H. & Manley, J. L. Phosphorylation–dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 17, 6359–6367 (1998).
Cao, S. et al. Discovery of driver non-coding splice-site-creating mutations in cancer. Nat. Commun. 11, 5573 (2020).
Mian, S. A. et al. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat. Commun. 6, 10004 (2015).
Lee, S. C. et al. Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell 34, 225–241 (2018).
Taylor, J. et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood 136, 1477–1486 (2020).
Zhou, Q. et al. A chemical genetics approach for the functional assessment of novel cancer genes. Cancer Res. 75, 1949–1958 (2015).
Lee, S. C. et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med. 22, 672–678 (2016).
Fei, D. L. et al. Wild-type U2AF1 antagonizes the splicing program characteristic of U2AF1-mutant tumors and is required for cell survival. PLoS Genet. 12, e1006384 (2016).
Kaida, D. et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3, 576–583 (2007).
Fan, L., Lagisetti, C., Edwards, C. C., Webb, T. R. & Potter, P. M. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 6, 582–589 (2011).
Kotake, Y. et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 3, 570–575 (2007).
Hasegawa, M. et al. Identification of SAP155 as the target of GEX1A (herboxidiene), an antitumor natural product. ACS Chem. Biol. 6, 229–233 (2011).
Yokoi, A. et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 278, 4870–4880 (2011).
Cretu, C. et al. Structural basis of splicing modulation by antitumor macrolide compounds. Mol. Cell 70, 265–273 (2018).
Eskens, F. A. et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin. Cancer Res. 19, 6296–6304 (2013).
Hong, D. S. et al. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest. New Drugs 32, 436–444 (2014).
Crews, L. A. et al. RNA splicing modulation selectively impairs leukemia stem cell maintenance in secondary human AML. Cell Stem Cell 19, 599–612 (2016).
Seiler, M. et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 24, 497–504 (2018).
Steensma, D. P. et al. Phase I first-in-human dose escalation study of the oral SF3B1 modulator H3B-8800 in myeloid neoplasms. Leukemia 35, 3542–3550 (2021).
Ting, T. C. et al. Aryl sulfonamides degrade RBM39 and RBM23 by recruitment to CRL4–DCAF15. Cell Rep. 29, 1499–1510 (2019).
Han, T. et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, eaal3755 (2017).
Du, X. et al. Structural basis and kinetic pathway of RBM39 recruitment to DCAF15 by a sulfonamide molecular glue E7820. Structure 27, 1625–1633 (2019).
Faust, T. B. et al. Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15. Nat. Chem. Biol. 16, 7–14 (2020).
Bussiere, D. E. et al. Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat. Chem. Biol. 16, 15–23 (2020).
Wang, E. et al. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell 35, 369–384 (2019).
Assi, R. et al. Final results of a phase 2, open-label study of indisulam, idarubicin, and cytarabine in patients with relapsed or refractory acute myeloid leukemia and high-risk myelodysplastic syndrome. Cancer 124, 2758–2765 (2018).
Jagtap, P. K. A. et al. Identification of phenothiazine derivatives as UHM-binding inhibitors of early spliceosome assembly. Nat. Commun. 11, 5621 (2020).
Kobayashi, A. et al. Identification of a small molecule splicing inhibitor targeting UHM domains. FEBS J. 289, 682–698 (2021).
Chatrikhi, R. et al. A synthetic small molecule stalls pre-mRNA splicing by promoting an early-stage U2AF2–RNA complex. Cell Chem. Biol. 28, 1145–1157 (2021).
Mathew, R. et al. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat. Struct. Mol. Biol. 15, 435–443 (2008).
Edmond, V. et al. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 30, 510–523 (2011).
Musiani, D. et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal. 12, eaat8388 (2019).
Fong, J. Y. et al. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell 36, 194–209 (2019).
Radzisheuskaya, A. et al. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat. Struct. Mol. Biol. 26, 999–1012 (2019).
Finkel, R. S. et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 377, 1723–1732 (2017).
McDonald, C. M. et al. Open-label evaluation of eteplirsen in patients with Duchenne muscular dystrophy amenable to exon 51 skipping: PROMOVI trial. J. Neuromuscul. Dis. 8, 989–1001 (2021).
Walker, A. R. et al. Phase 3 randomized trial of chemotherapy with or without oblimersen in older AML patients: CALGB 10201 (Alliance). Blood Adv. 5, 2775–2787 (2021).
Sha, D. et al. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov. 10, 1808–1825 (2020).
Marabelle, A. et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 21, 1353–1365 (2020).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Turajlic, S. et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 18, 1009–1021 (2017).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Jayasinghe, R. G. et al. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 23, 270–281 (2018).
Laumont, C. M. et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci. Transl. Med. 10, eaau5516 (2018).
Smart, A. C. et al. Intron retention is a source of neoepitopes in cancer. Nat. Biotechnol. 36, 1056–1058 (2018).
Wang, R. F. et al. A breast and melanoma-shared tumor antigen: T cell responses to antigenic peptides translated from different open reading frames. J. Immunol. 161, 3598–3606 (1998).
Robbins, P. F. et al. The intronic region of an incompletely spliced gp100 gene transcript encodes an epitope recognized by melanoma-reactive tumor-infiltrating lymphocytes. J. Immunol. 159, 303–308 (1997).
Ouspenskaia, T. et al. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat. Biotechnol. 40, 209–217 (2021).
Lu, S. X. et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell 184, 4032–4047 (2021).
North, K. et al. Synthetic introns enable splicing factor mutation-dependent targeting of cancer cells. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01224-2 (2022).
Monteys, A. M. et al. Regulated control of gene therapies by drug-induced splicing. Nature 596, 291–295 (2021).
Steijger, T. et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10, 1177–1184 (2013).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015).
Zhang, C., Zhang, B., Lin, L. L. & Zhao, S. Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genomics 18, 583 (2017).
De Paoli-Iseppi, R., Gleeson, J. & Clark, M. B. Isoform age—splice isoform profiling using long-read technologies. Front. Mol. Biosci. 8, 711733 (2021).
Simpson, J. T. et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017).
Lorenz, D. A., Sathe, S., Einstein, J. M. & Yeo, G. W. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26, 19–28 (2020).
Chen, S. Y., Deng, F., Jia, X., Li, C. & Lai, S. J. A transcriptome atlas of rabbit revealed by PacBio single-molecule long-read sequencing. Sci Rep. 7, 7648 (2017).
Kim, J. A. et al. Genome-wide transcriptome profiling of the medicinal plant Zanthoxylum planispinum using a single-molecule direct RNA sequencing approach. Genomics 111, 973–979 (2019).
Roach, N. P. et al. The full-length transcriptome of C. elegans using direct RNA sequencing. Genome Res. 30, 299–312 (2020).
Lagarde, J. et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 49, 1731–1740 (2017).
Gupta, I. et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 36, 1197–1202 (2018).
Singh, M. et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun. 10, 3120 (2019).
Acknowledgements
R.F.S. was supported, in part, by MSK-ICTTP T32-CA009207 and an American Society of Hematology Research Training Award for Fellows. O.A.-W. is supported by the Leukemia & Lymphoma Society, NIH R01s CA251138, HL128239 and CA242020, NIH/NCI 1P50 254838-01 and the Edward P. Evans MDS Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
O.A.-W. has served as a consultant for H3B Biomedicine, Foundation Medicine, Inc., Merck, Prelude Therapeutics and Janssen and is on the Scientific Advisory Board of Envisagenics, Inc., AIChemy, Harmonic Discovery, Inc., and Pfizer Boulder. O.A.-W. has received prior research funding from H3B Biomedicine, Nurix Therapeutics and LOXO Oncology unrelated to the current manuscript. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks James Manley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Stanley, R.F., Abdel-Wahab, O. Dysregulation and therapeutic targeting of RNA splicing in cancer. Nat Cancer 3, 536–546 (2022). https://doi.org/10.1038/s43018-022-00384-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-022-00384-z
This article is cited by
-
Variations of intronic branchpoint motif: identification and functional implications in splicing and disease
Communications Biology (2023)
-
Structure prediction of novel isoforms from uveal melanoma by AlphaFold
Scientific Data (2023)
-
RNA splicing dysregulation and the hallmarks of cancer
Nature Reviews Cancer (2023)
-
Aberrant Cyclin D1 splicing in cancer: from molecular mechanism to therapeutic modulation
Cell Death & Disease (2023)
-
Proteomics Analysis of Polyphyllin D-Treated Triple-Negative Breast Cancer Cells Reveal the Anticancer Mechanisms of Polyphyllin D
Applied Biochemistry and Biotechnology (2023)