Abstract
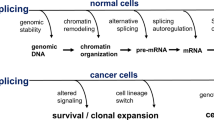
Dysregulated RNA splicing is a molecular feature that characterizes almost all tumour types. Cancer-associated splicing alterations arise from both recurrent mutations and altered expression of trans-acting factors governing splicing catalysis and regulation. Cancer-associated splicing dysregulation can promote tumorigenesis via diverse mechanisms, contributing to increased cell proliferation, decreased apoptosis, enhanced migration and metastatic potential, resistance to chemotherapy and evasion of immune surveillance. Recent studies have identified specific cancer-associated isoforms that play critical roles in cancer cell transformation and growth and demonstrated the therapeutic benefits of correcting or otherwise antagonizing such cancer-associated mRNA isoforms. Clinical-grade small molecules that modulate or inhibit RNA splicing have similarly been developed as promising anticancer therapeutics. Here, we review splicing alterations characteristic of cancer cell transcriptomes, dysregulated splicing’s contributions to tumour initiation and progression, and existing and emerging approaches for targeting splicing for cancer therapy. Finally, we discuss the outstanding questions and challenges that must be addressed to translate these findings into the clinic.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Black, D. L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336 (2003).
Reixachs-Sole, M. & Eyras, E. Uncovering the impacts of alternative splicing on the proteome with current omics techniques. Wiley Interdiscip. Rev. RNA 13, e1707 (2022).
Blencowe, B. J. Alternative splicing: new insights from global analyses. Cell 126, 37–47 (2006).
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008). This landmark study uses RNA-seq to quantify isoform expression across tissues.
Kahles, A. et al. Comprehensive analysis of alternative splicing across tumors from 8705 patients. Cancer Cell 34, 211–224.e6 (2018). This landmark study identifies splicing alterations across tumour types.
Urbanski, L. M., Leclair, N. & Anczukow, O. Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA 9, 1476 (2018).
Dvinge, H., Kim, E., Abdel-Wahab, O. & Bradley, R. K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16, 413–430 (2016).
Stanley, R. F. & Abdel-Wahab, O. Dysregulation and therapeutic targeting of RNA splicing in cancer. Nat. Cancer 3, 536–546 (2022).
Bonnal, S. C., López-Oreja, I. & Valcárcel, J. Roles and mechanisms of alternative splicing in cancer—implications for care. Nat. Rev. Clin. Oncol. 17, 457–474 (2020).
Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46 (2022).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Wilkinson, M. E., Charenton, C. & Nagai, K. RNA splicing by the spliceosome. Annu. Rev. Biochem. 89, 359–388 (2020).
Akinyi, M. V. & Frilander, M. J. At the intersection of major and minor spliceosomes: crosstalk mechanisms and their impact on gene expression. Front. Genet. 12, 700744 (2021).
Lee, Y. & Rio, D. C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 (2015).
Ellis, J. D. et al. Tissue-specific alternative splicing remodels protein–protein interaction networks. Mol. Cell 46, 884–892 (2012).
Tung, K. F., Pan, C. Y., Chen, C. H. & Lin, W. C. Top-ranked expressed gene transcripts of human protein-coding genes investigated with GTEx dataset. Sci. Rep. 10, 16245 (2020).
Howard, J. M. & Sanford, J. R. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 6, 93–110 (2015).
Geuens, T., Bouhy, D. & Timmerman, V. The hnRNP family: insights into their role in health and disease. Hum. Genet. 135, 851–867 (2016).
Papaemmanuil, E. et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011).
Yoshida, K. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011).
Harbour, J. W. et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 45, 133–135 (2013).
Martin, M. et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 45, 933–936 (2013). Together with Harbour et al. (2013), this paper identifies recurrent mutations in SF3B1 in UVM.
Imielinski, M. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012).
Lee, S. C. et al. Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell 34, 225–241.e8 (2018).
Taylor, J. et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood 136, 1477–1486 (2020).
Wang, L. et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 365, 2497–2506 (2011). This work identifies recurrent mutations in SF3B1 in CLL.
Alsafadi, S. et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 7, 10615 (2016).
Darman, R. B. et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 13, 1033–1045 (2015). This work demonstrates that recurrent SF3B1 mutations alter branch point selection.
Dalton, W. B. et al. The K666N mutation in SF3B1 is associated with increased progression of MDS and distinct RNA splicing. Blood Adv. 4, 1192–1196 (2020).
Inoue, D. et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature 574, 432–436 (2019). This work shows that SF3B1 mutations disrupt chromatin remodelling to promote tumorigenesis.
Lieu, Y. K. et al. SF3B1 mutant-induced missplicing of MAP3K7 causes anemia in myelodysplastic syndromes. Proc. Natl Acad. Sci. USA 119, 2111703119 (2022).
Clough, C. A. et al. Coordinated missplicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. Blood 139, 2038–2049 (2022).
Yoshimi, A. et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature 574, 273–277 (2019). This work demonstrates genetic and functional interactions between SRSF2 and IDH2 in leukaemia.
Robertson, A. G. et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32, 204–220.e15 (2017).
Kim, E. et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27, 617–630 (2015).
Gallardo, M. et al. hnRNP K is a haploinsufficient tumor suppressor that regulates proliferation and differentiation programs in hematologic malignancies. Cancer Cell 28, 486–499 (2015).
Graubert, T. A. et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet. 44, 53–57 (2012).
Brooks, A. N. et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS ONE 9, 87361 (2014).
Ilagan, J. O. et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 25, 14–26 (2015).
Smith, M. A. et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 21, 640–650 (2019).
Biancon, G. et al. Precision analysis of mutant U2AF1 activity reveals deployment of stress granules in myeloid malignancies. Mol. Cell 82, 1107–1122.e7 (2022).
Damm, F. et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood 119, 3211–3218 (2012).
Haferlach, T. et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28, 241–247 (2014).
Madan, V. et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 6, 6042 (2015).
Inoue, D. et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat. Genet. 53, 707–718 (2021). This work shows that disruption of minor intron splicing by ZRSR2 mutations promotes clonal advantage.
Wang, X., Song, X. & Yan, X. Effect of RNA splicing machinery gene mutations on prognosis of patients with MDS: a meta-analysis. Medicine 98, e15743 (2019).
Obeng, E. A. et al. Physiologic expression of Sf3b1K700E causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 30, 404–417 (2016).
Shirai, C. L. et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell 27, 631–643 (2015).
Fei, D. L. et al. Impaired hematopoiesis and leukemia development in mice with a conditional knock-in allele of a mutant splicing factor gene U2af1. Proc. Natl Acad. Sci. USA 115, 10437–E10446 (2018).
Mian, S. A. et al. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat. Commun. 6, 10004 (2015). This work shows that SF3B1 mutations are initiating events in MDS.
Fabre, M. A. et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature 606, 335–342 (2022).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Ibrahimpasic, T. et al. Genomic alterations in fatal forms of non-anaplastic thyroid cancer: identification of MED12 and RBM10 as novel thyroid cancer genes associated with tumor virulence. Clin. Cancer Res. 23, 5970–5980 (2017).
Anczuków, O. & Krainer, A. R. Splicing-factor alterations in cancers. RNA 22, 1285–1301 (2016).
Seiler, M. et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23, 282–296.e4 (2018).
Escobar-Hoyos, L. F. et al. Altered RNA splicing by mutant p53 activates oncogenic RAS signaling in pancreatic cancer. Cancer Cell 38, 198–211.e8 (2020).
Anczukow, O. et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol. 19, 220–228 (2012).
Anczukow, O. et al. SRSF1-regulated alternative splicing in breast cancer. Mol. Cell 60, 105–117 (2015).
Park, S. et al. Differential functions of splicing factors in mammary transformation and breast cancer metastasis. Cell Rep. 29, 2672–2688.e7 (2019). This work identifies the functional roles of individual SR proteins in breast cancer.
Karni, R. et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 (2007). This landmark study shows that SRSF1 is a proto-oncoprotein.
Sebestyén, E. et al. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res. 26, 732–744 (2016).
Ghigna, C. et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20, 881–890 (2005).
Das, S., Anczukow, O., Akerman, M. & Krainer, A. R. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 1, 110–117 (2012).
De Miguel, F. J. et al. Identification of alternative splicing events regulated by the oncogenic factor SRSF1 in lung cancer. Cancer Res. 74, 1105–1115 (2014).
Michlewski, G., Sanford, J. R. & Caceres, J. F. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell 30, 179–189 (2008).
Karni, R., Hippo, Y., Lowe, S. W. & Krainer, A. R. The splicing-factor oncoprotein SF2/ASF activates mTORC1. Proc. Natl Acad. Sci. USA 105, 15323–15327 (2008).
Sen, S., Langiewicz, M., Jumaa, H. & Webster, N. J. Deletion of serine/arginine-rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology 61, 171–183 (2015).
Ajiro, M., Jia, R., Yang, Y., Zhu, J. & Zheng, Z. M. A genome landscape of SRSF3-regulated splicing events and gene expression in human osteosarcoma U2OS cells. Nucleic Acids Res. 44, 1854–1870 (2016).
Wang, Z. et al. Exon-centric regulation of pyruvate kinase M alternative splicing via mutually exclusive exons. J. Mol. Cell Biol. 4, 79–87 (2012).
Kurokawa, K. et al. Downregulation of serine/arginine-rich splicing factor 3 induces G1 cell cycle arrest and apoptosis in colon cancer cells. Oncogene 33, 1407–1417 (2014).
Jia, R., Ajiro, M., Yu, L., McCoy, P. Jr & Zheng, Z. M. Oncogenic splicing factor SRSF3 regulates ILF3 alternative splicing to promote cancer cell proliferation and transformation. RNA 25, 630–644 (2019).
Freytag, M. et al. Epithelial splicing regulatory protein 1 and 2 (ESRP1 and ESRP2) upregulation predicts poor prognosis in prostate cancer. BMC Cancer 20, 1220 (2020).
Gokmen-Polar, Y. et al. Splicing factor ESRP1 controls ER-positive breast cancer by altering metabolic pathways. EMBO Rep. 20, 46078 (2019).
Shapiro, I. M. et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 7, 1002218 (2011).
Munkley, J. et al. Androgen-regulated transcription of ESRP2 drives alternative splicing patterns in prostate cancer. eLife 8, 47678 (2019).
Ishii, H. et al. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J. Biol. Chem. 289, 27386–27399 (2014).
Bechara, E. G., Sebestyen, E., Bernardis, I., Eyras, E. & Valcarcel, J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell 52, 720–733 (2013). This work demonstrates the role of RBMs in tumorigenesis.
Rintala-Maki, N. D. et al. Expression of RBM5-related factors in primary breast tissue. J. Cell Biochem. 100, 1440–1458 (2007).
Inoue, A. RBM10: structure, functions, and associated diseases. Gene 783, 145463 (2021).
Lu, W. et al. QKI impairs self-renewal and tumorigenicity of oral cancer cells via repression of SOX2. Cancer Biol. Ther. 15, 1174–1184 (2014).
Zong, F. Y. et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 10, 1004289 (2014).
Bandopadhayay, P. et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat. Genet. 48, 273–282 (2016).
Shirakihara, T. et al. TGF-β regulates isoform switching of FGF receptors and epithelial–mesenchymal transition. EMBO J. 30, 783–795 (2011).
Warzecha, C. C. et al. An ESRP-regulated splicing programme is abrogated during the epithelial–mesenchymal transition. EMBO J. 29, 3286–3300 (2010). This work demonstrates the important role played by regulated splicing during EMT.
Warzecha, C. C., Shen, S., Xing, Y. & Carstens, R. P. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 6, 546–562 (2009).
Yae, T. et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat. Commun. 3, 883 (2012). This work demonstrates the role of a CD44 isoform in lung metastasis in vivo.
Sutherland, L. C., Wang, K. & Robinson, A. G. RBM5 as a putative tumor suppressor gene for lung cancer. J. Thorac. Oncol. 5, 294–298 (2010).
Jamsai, D. et al. In vivo evidence that RBM5 is a tumour suppressor in the lung. Sci. Rep. 7, 16323 (2017).
Zhao, Y. et al. The tumor suppressing effects of QKI-5 in prostate cancer: a novel diagnostic and prognostic protein. Cancer Biol. Ther. 15, 108–118 (2014).
Frampton, G. M. et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 5, 850–859 (2015).
Jung, H. et al. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 47, 1242–1248 (2015).
Mogilevsky, M. et al. Modulation of MKNK2 alternative splicing by splice-switching oligonucleotides as a novel approach for glioblastoma treatment. Nucleic Acids Res. 46, 11396–11404 (2018).
Koh, C. M. et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523, 96–100 (2015). This work identifies a MYC-driven splicing vulnerability.
Ben-Hur, V. et al. S6K1 alternative splicing modulates its oncogenic activity and regulates mTORC1. Cell Rep. 3, 103–115 (2013).
Amaral, C. L. et al. S6Ks isoforms contribute to viability, migration, docetaxel resistance and tumor formation of prostate cancer cells. BMC Cancer 16, 602 (2016).
Mei, H., Wang, Y., Fan, J. & Lin, Z. Alternative splicing of S6K1 promotes non-small cell lung cancer survival. Tumour Biol. 37, 13369–13376 (2016).
David, C. J., Chen, M., Assanah, M., Canoll, P. & Manley, J. L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463, 364–368 (2010).
Clower, C. V. et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl Acad. Sci. USA 107, 1894–1899 (2010).
Dayton, T. L., Jacks, T. & Vander Heiden, M. G. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 17, 1721–1730 (2016).
Wang, Z., Jeon, H. Y., Rigo, F., Bennett, C. F. & Krainer, A. R. Manipulation of PK-M mutually exclusive alternative splicing by antisense oligonucleotides. Open. Biol. 2, 120133 (2012).
Boise, L. H. et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74, 597–608 (1993).
Wu, L., Mao, C. & Ming, X. Modulation of Bcl-x alternative splicing induces apoptosis of human hepatic stellate cells. Biomed. Res. Int. 2016, 7478650 (2016).
Dole, M. G. et al. Bcl-xS enhances adenoviral vector-induced apoptosis in neuroblastoma cells. Cancer Res. 56, 5734–5740 (1996).
Minn, A. J., Boise, L. H. & Thompson, C. B. Bcl-x(S) anatagonizes the protective effects of Bcl-x(L). J. Biol. Chem. 271, 6306–6312 (1996).
Paronetto, M. P., Achsel, T., Massiello, A., Chalfant, C. E. & Sette, C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 176, 929–939 (2007).
Zhou, A., Ou, A. C., Cho, A., Benz, E. J. Jr & Huang, S. C. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol. Cell Biol. 28, 5924–5936 (2008).
Bielli, P., Bordi, M., Di Biasio, V. & Sette, C. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5′ splice site selection. Nucleic Acids Res. 42, 12070–12081 (2014).
Wang, Y. et al. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell 26, 374–389 (2014). This work demonstrates the role of RBMs in tumorigenesis in vivo.
Moore, M. J., Wang, Q., Kennedy, C. J. & Silver, P. A. An alternative splicing network links cell-cycle control to apoptosis. Cell 142, 625–636 (2010).
Garneau, D., Revil, T., Fisette, J. F. & Chabot, B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 280, 22641–22650 (2005).
Revil, T., Pelletier, J., Toutant, J., Cloutier, A. & Chabot, B. Heterogeneous nuclear ribonucleoprotein K represses the production of pro-apoptotic Bcl-xS splice isoform. J. Biol. Chem. 284, 21458–21467 (2009).
Cloutier, P. et al. Antagonistic effects of the SRp30c protein and cryptic 5′ splice sites on the alternative splicing of the apoptotic regulator Bcl-x. J. Biol. Chem. 283, 21315–21324 (2008).
Pabla, N., Bhatt, K. & Dong, Z. Checkpoint kinase 1 (Chk1)-short is a splice variant and endogenous inhibitor of Chk1 that regulates cell cycle and DNA damage checkpoints. Proc. Natl Acad. Sci. USA 109, 197–202 (2012).
Hu, G. et al. Clinical and functional significance of CHK1-S, an alternatively spliced isoform of the CHK1 gene, in hepatocellular carcinoma. J. Cancer 11, 1792–1799 (2020).
Wong, M. S., Shay, J. W. & Wright, W. E. Regulation of human telomerase splicing by RNA:RNA pairing. Nat. Commun. 5, 3306 (2014).
Sayed, M. E. et al. NOVA1 directs PTBP1 to hTERT pre-mRNA and promotes telomerase activity in cancer cells. Oncogene 38, 2937–2952 (2019).
Ludlow, A. T. et al. NOVA1 regulates hTERT splicing and cell growth in non-small cell lung cancer. Nat. Commun. 9, 3112 (2018).
Listerman, I., Sun, J., Gazzaniga, F. S., Lukas, J. L. & Blackburn, E. H. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res. 73, 2817–2828 (2013).
Pont, A. R., Sadri, N., Hsiao, S. J., Smith, S. & Schneider, R. J. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol. Cell 47, 5–15 (2012).
Kang, X., Chen, W., Kim, R. H., Kang, M. K. & Park, N. H. Regulation of the hTERT promoter activity by MSH2, the hnRNPs K and D, and GRHL2 in human oral squamous cell carcinoma cells. Oncogene 28, 565–574 (2009).
DiFeo, A., Martignetti, J. A. & Narla, G. The role of KLF6 and its splice variants in cancer therapy. Drug. Resist. Updat. 12, 1–7 (2009).
Hatami, R. et al. KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Sci. Transl. Med. 5, 12 (2013).
Yea, S. et al. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology 134, 1521–1531 (2008).
Botella, L. M. et al. TGF-β regulates the expression of transcription factor KLF6 and its splice variants and promotes co-operative transactivation of common target genes through a Smad3–Sp1–KLF6 interaction. Biochem. J. 419, 485–495 (2009).
DiFeo, A. et al. A functional role for KLF6–SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 68, 965–970 (2008).
Tanaka, N., Yoshida, H., Suzuki, Y. & Harigaya, K. Relative expression of hMena11a and hMenaINV splice isoforms is a useful biomarker in development and progression of human breast carcinoma. Int. J. Oncol. 45, 1921–1928 (2014).
Oudin, M. J. et al. Characterization of the expression of the pro-metastatic MenaINV isoform during breast tumor progression. Clin. Exp. Metastasis 33, 249–261 (2016). This work characterizes the expression of MENA isoforms in primary tumours.
Goswami, S. et al. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin. Exp. Metastasis 26, 153–159 (2009).
Di Modugno, F. et al. Splicing program of human MENA produces a previously undescribed isoform associated with invasive, mesenchymal-like breast tumors. Proc. Natl Acad. Sci. USA 109, 19280–19285 (2012).
Bria, E. et al. Prognostic impact of alternative splicing-derived hMENA isoforms in resected, node-negative, non-small-cell lung cancer. Oncotarget 5, 11054–11063 (2014).
Balsamo, M. et al. The alternatively-included 11a sequence modifies the effects of Mena on actin cytoskeletal organization and cell behavior. Sci. Rep. 6, 35298 (2016).
Di Modugno, F. et al. Molecular cloning of hMena (ENAH) and its splice variant hMena+11a: epidermal growth factor increases their expression and stimulates hMena+11a phosphorylation in breast cancer cell lines. Cancer Res. 67, 2657–2665 (2007).
Philippar, U. et al. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev. Cell 15, 813–828 (2008). This work identifies MENA isoforms.
Houck, K. A. et al. The vascular endothelial growth factor family: identification of a fourth molecular species and characterization of alternative splicing of RNA. Mol. Endocrinol. 5, 1806–1814 (1991).
Bates, D. O. et al. VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res. 62, 4123–4131 (2002). This work identifies VEGF isoforms.
Woolard, J. et al. VEGF165b, an inhibitory vascular endothelial growth factor splice variant: mechanism of action, in vivo effect on angiogenesis and endogenous protein expression. Cancer Res. 64, 7822–7835 (2004).
Nowak, D. G. et al. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: a novel therapeutic strategy for angiogenesis. J. Biol. Chem. 285, 5532–5540 (2010).
Harper, S. J. & Bates, D. O. VEGF-A splicing: the key to anti-angiogenic therapeutics? Nat. Rev. Cancer 8, 880–887 (2008).
Pritchard-Jones, R. O. et al. Expression of VEGFxxxb, the inhibitory isoforms of VEGF, in malignant melanoma. Br. J. Cancer 97, 223–230 (2007).
Varey, A. H. et al. VEGF 165 b, an antiangiogenic VEGF-A isoform, binds and inhibits bevacizumab treatment in experimental colorectal carcinoma: balance of pro- and antiangiogenic VEGF-A isoforms has implications for therapy. Br. J. Cancer 98, 1366–1379 (2008).
Biselli-Chicote, P. M. et al. Overexpression of antiangiogenic vascular endothelial growth factor isoform and splicing pegulatory factors in oral, laryngeal and pharyngeal squamous cell carcinomas. Asian Pac. J. Cancer Prev. 18, 2171–2177 (2017).
Rennel, E. et al. The endogenous anti-angiogenic VEGF isoform, VEGF165b inhibits human tumour growth in mice. Br. J. Cancer 98, 1250–1257 (2008).
Robertson, C. The extracellular matrix in breast cancer predicts prognosis through composition, splicing, and crosslinking. Exp. Cell Res. 343, 73–81 (2016).
Nam, J. M., Onodera, Y., Bissell, M. J. & Park, C. C. Breast cancer cells in three-dimensional culture display an enhanced radioresponse after coordinate targeting of integrin α5β1 and fibronectin. Cancer Res. 70, 5238–5248 (2010).
Schiefner, A., Gebauer, M. & Skerra, A. Extra-domain B in oncofetal fibronectin structurally promotes fibrillar head-to-tail dimerization of extracellular matrix protein. J. Biol. Chem. 287, 17578–17588 (2012).
Fukuda, T. et al. Mice lacking the EDB segment of fibronectin develop normally but exhibit reduced cell growth and fibronectin matrix assembly in vitro. Cancer Res. 62, 5603–5610 (2002).
Borsi, L. et al. Expression of different tenascin isoforms in normal, hyperplastic and neoplastic human breast tissues. Int. J. Cancer 52, 688–692 (1992).
Briones-Orta, M. A. et al. Osteopontin splice variants and polymorphisms in cancer progression and prognosis. Biochim. Biophys. Acta 1868, 93–108A (2017).
Srebrow, A., Blaustein, M. & Kornblihtt, A. R. Regulation of fibronectin alternative splicing by a basement membrane-like extracellular matrix. FEBS Lett. 514, 285–289 (2002).
Bordeleau, F. et al. Tissue stiffness regulates serine/arginine-rich protein-mediated splicing of the extra domain B-fibronectin isoform in tumors. Proc. Natl Acad. Sci. USA 112, 8314–8319 (2015).
Martinez, N. M. & Lynch, K. W. Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol. Rev. 253, 216–236 (2013).
O’Connor, B. P. et al. Regulation of Toll-like receptor signaling by the SF3a mRNA splicing complex. PLoS Genet. 11, 1004932 (2015).
Adib-Conquy, M. et al. Up-regulation of MyD88s and SIGIRR, molecules inhibiting Toll-like receptor signaling, in monocytes from septic patients. Crit. Care Med. 34, 2377–2385 (2006).
De Arras, L. et al. Comparative genomics RNAi screen identifies Eftud2 as a novel regulator of innate immunity. Genetics 197, 485–496 (2014).
Poulikakos, P. I. et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480, 387–390 (2011).
Wang, Y. et al. The BRCA1- Δ11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 76, 2778–2790 (2016).
Ozden, O. et al. Expression of an oncogenic BARD1 splice variant impairs homologous recombination and predicts response to PARP-1 inhibitor therapy in colon cancer. Sci. Rep. 6, 26273 (2016).
Ng, K. P. et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat. Med. 18, 521–528 (2012). This work links resistance to tyrosine kinase inhibitors with BIM splicing.
Castiglioni, F. et al. Role of exon-16-deleted HER2 in breast carcinomas. Endocr. Relat. Cancer 13, 221–232 (2006).
Alajati, A. et al. Mammary tumor formation and metastasis evoked by a HER2 splice variant. Cancer Res. 73, 5320–5327 (2013).
Dehm, S. M., Schmidt, L. J., Heemers, H. V., Vessella, R. L. & Tindall, D. J. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 68, 5469–5477 (2008). This work identifies a constitutively active AR isoform that contributes to therapy resistance.
Antonarakis, E. S. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014).
Sun, S. et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J. Clin. Invest. 120, 2715–2730 (2010). This work identifies a constitutively active AR isoform that contributes to castration resistance.
Thiebaut, C. et al. The role of ERα36 in development and tumor malignancy. Int. J. Mol. Sci. 21, 4116 (2020).
Zheng, S., Asnani, M. & Thomas-Tikhonenko, A. Escape from ALL-CARTaz: leukemia immunoediting in the age of chimeric antigen receptors. Cancer J. 25, 217–222 (2019).
Sotillo, E. et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 5, 1282–1295 (2015). This work demonstrates that alternative splicing can enable escape from CAR T cell therapy.
Zheng, S. et al. Modulation of CD22 protein expression in childhood leukemia by pervasive splicing aberrations: implications for CD22-directed immunotherapies. Blood Cancer Discov. 3, 103–115 (2022).
Nakajima, H. et al. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J. Antibiot. 49, 1204–1211 (1996).
Thompson, C. F., Jamison, T. F. & Jacobsen, E. N. FR901464: total synthesis, proof of structure, and evaluation of synthetic analogues. J. Am. Chem. Soc. 123, 9974–9983 (2001).
Kaida, D. et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3, 576–583 (2007).
Osman, S. et al. Structural requirements for the antiproliferative activity of pre-mRNA splicing inhibitor FR901464. Chemistry 17, 895–904 (2011).
Albert, B. J., Sivaramakrishnan, A., Naka, T., Czaicki, N. L. & Koide, K. Total syntheses, fragmentation studies, and antitumor/antiproliferative activities of FR901464 and its low picomolar analogue. J. Am. Chem. Soc. 129, 2648–2659 (2007).
Liu, X. et al. Genomics-guided discovery of thailanstatins A, B, and C As pre-mRNA splicing inhibitors and antiproliferative agents from Burkholderia thailandensis MSMB43. J. Nat. Prod. 76, 685–693 (2013).
Corrionero, A., Minana, B. & Valcarcel, J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes. Dev. 25, 445–459 (2011). This work characterizes the mechanism of action of spliceostatin A.
Seiler, M. et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 24, 497–504 (2018).
Hong, D. S. et al. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest. N. Drugs 32, 436–444 (2014).
Eskens, F. A. et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin. Cancer Res. 19, 6296–6304 (2013).
Steensma, D. P. et al. Phase I first-in-human dose escalation study of the oral SF3B1 modulator H3B-8800 in myeloid neoplasms. Leukemia 35, 3542–3550 (2021).
O’Brien, K., Matlin, A. J., Lowell, A. M. & Moore, M. J. The biflavonoid isoginkgetin is a general inhibitor of pre-mRNA splicing. J. Biol. Chem. 283, 33147–33154 (2008).
Vanzyl, E. J. et al. The spliceosome inhibitors isoginkgetin and pladienolide B induce ATF3-dependent cell death. PLoS ONE 15, e0224953 (2020).
Yoon, S. O., Shin, S., Lee, H. J., Chun, H. K. & Chung, A. S. Isoginkgetin inhibits tumor cell invasion by regulating phosphatidylinositol 3-kinase/Akt-dependent matrix metalloproteinase-9 expression. Mol. Cancer Ther. 5, 2666–2675 (2006).
Darrigrand, R. et al. Isoginkgetin derivative IP2 enhances the adaptive immune response against tumor antigens. Commun. Biol. 4, 269 (2021).
Han, T. et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, 3755 (2017). This work identifies RBM39 as the molecular target of indisulam.
Ting, T. C. et al. Aryl sulfonamides degrade RBM39 and RBM23 by recruitment to CRL4–DCAF15. Cell Rep. 29, 1499–1510.e6 (2019).
Bussiere, D. E. et al. Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat. Chem. Biol. 16, 15–23 (2020).
Tari, M. et al. U2AF65 assemblies drive sequence-specific splice site recognition. EMBO Rep. 20, 47604 (2019).
Stepanyuk, G. A. et al. UHM–ULM interactions in the RBM39–U2AF65 splicing-factor complex. Acta Crystallogr. D. Struct. Biol. 72, 497–511 (2016).
Loerch, S., Maucuer, A., Manceau, V., Green, M. R. & Kielkopf, C. L. Cancer-relevant splicing factor CAPERα engages the essential splicing factor SF3b155 in a specific ternary complex. J. Biol. Chem. 289, 17325–17337 (2014).
Kralovicova, J. et al. PUF60-activated exons uncover altered 3′ splice-site selection by germline missense mutations in a single RRM. Nucleic Acids Res. 46, 6166–6187 (2018).
Mai, S. et al. Global regulation of alternative RNA splicing by the SR-rich protein RBM39. Biochim. Biophys. Acta 1859, 1014–1024 (2016).
Xu, Y., Nijhuis, A. & Keun, H. C. RNA-binding motif protein 39 (RBM39): an emerging cancer target. Br. J. Pharmacol. 179, 2795–2812 (2020).
Hamid, O. et al. A randomized, open-label clinical trial of tasisulam sodium versus paclitaxel as second-line treatment in patients with metastatic melanoma. Cancer 120, 2016–2024 (2014).
Bezzi, M. et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 27, 1903–1916 (2013).
Radzisheuskaya, A. et al. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat. Struct. Mol. Biol. 26, 999–1012 (2019).
Fong, J. Y. et al. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell 36, 194–209.e9 (2019).
Gammons, M. V. et al. Topical antiangiogenic SRPK1 inhibitors reduce choroidal neovascularization in rodent models of exudative AMD. Invest. Ophthalmol. Vis. Sci. 54, 6052–6062 (2013).
Hatcher, J. M. et al. SRPKIN-1: a covalent SRPK1/2 inhibitor that potently converts VEGF from pro-angiogenic to anti-angiogenic isoform. Cell Chem. Biol. 25, 460–470.e6 (2018).
Batson, J. et al. Development of potent, selective SRPK1 inhibitors as potential topical therapeutics for neovascular eye disease. ACS Chem. Biol. 12, 825–832 (2017).
Sakuma, M., Iida, K. & Hagiwara, M. Deciphering targeting rules of splicing modulator compounds: case of TG003. BMC Mol. Biol. 16, 16 (2015).
Babu, N. et al. Phosphoproteomic analysis identifies CLK1 as a novel therapeutic target in gastric cancer. Gastric Cancer 23, 796–810 (2020).
Uzor, S. et al. CDC2-like (CLK) protein kinase inhibition as a novel targeted therapeutic strategy in prostate cancer. Sci. Rep. 11, 7963 (2021).
Sohail, M. et al. A novel class of inhibitors that target SRSF10 and promote p53-mediated cytotoxicity on human colorectal cancer cells. Nar. Cancer 3, 19 (2021).
Hluchy, M. et al. CDK11 regulates pre-mRNA splicing by phosphorylation of SF3B1. Nature 609, 829–834 (2022).
Sheridan, C. First small-molecule drug targeting RNA gains momentum. Nat. Biotechnol. 39, 6–8 (2021).
Baranello, G. et al. Risdiplam in type 1 spinal muscular atrophy. N. Engl. J. Med. 384, 915–923 (2021). This work demonstrates that risdiplam increases SMN protein levels in patients with spinal muscular atrophy.
Sivaramakrishnan, M. et al. Binding to SMN2 pre-mRNA–protein complex elicits specificity for small molecule splicing modifiers. Nat. Commun. 8, 1476 (2017).
Costales, M. G., Childs-Disney, J. L., Haniff, H. S. & Disney, M. D. How we think about targeting RNA with small molecules. J. Med. Chem. 63, 8880–8900 (2020).
Umuhire Juru, A. & Hargrove, A. E. Frameworks for targeting RNA with small molecules. J. Biol. Chem. 296, 100191 (2021).
Havens, M. A. & Hastings, M. L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 44, 6549–6563 (2016).
Bennett, C. F. & Swayze, E. E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293 (2010).
Kim, Y. et al. Enhanced potency of GalNAc-conjugated antisense oligonucleotides in hepatocellular cancer models. Mol. Ther. 27, 1547–1557 (2019).
Scharner, J., Qi, S., Rigo, F., Bennett, C. F. & Krainer, A. R. Delivery of GalNAc-conjugated splice-switching ASOs to non-hepatic cells through ectopic expression of asialoglycoprotein receptor. Mol. Ther. Nucleic Acids 16, 313–325 (2019).
Juliano, R. L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 44, 6518–6548 (2016).
Li, Z. et al. Pro-apoptotic effects of splice-switching oligonucleotides targeting Bcl-x pre-mRNA in human glioma cell lines. Oncol. Rep. 35, 1013–1019 (2016).
Sun, Y., Yan, L., Guo, J., Shao, J. & Jia, R. Downregulation of SRSF3 by antisense oligonucleotides sensitizes oral squamous cell carcinoma and breast cancer cells to paclitaxel treatment. Cancer Chemother. Pharmacol. 84, 1133–1143 (2019).
Leclair, N. K. et al. Poison exon splicing regulates a coordinated network of SR protein expression during differentiation and tumorigenesis. Mol. Cell 80, 648–665.e9 (2020). This work identifies functional roles for poison exons in tumorigenesis.
Denichenko, P. et al. Specific inhibition of splicing factor activity by decoy RNA oligonucleotides. Nat. Commun. 10, 1590 (2019).
Gadgil, A. & Raczynska, K. D. U7 snRNA: a tool for gene therapy. J. Gene Med. 23, 3321 (2021).
Rogalska, M. E. et al. Therapeutic activity of modified U1 core spliceosomal particles. Nat. Commun. 7, 11168 (2016).
Wang, Y., Cheong, C. G., Hall, T. M. & Wang, Z. Engineering splicing factors with designed specificities. Nat. Methods 6, 825–830 (2009).
Cox, D. B. T. et al. RNA editing with CRISPR–Cas13. Science 358, 1019–1027 (2017).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14 (2018). This work uses RNA-targeting Cas to manipulate alternative splicing.
Du, M., Jillette, N., Zhu, J. J., Li, S. & Cheng, A. W. CRISPR artificial splicing factors. Nat. Commun. 11, 2973 (2020).
Gapinske, M. et al. CRISPR–SKIP: programmable gene splicing with single base editors. Genome Biol. 19, 107 (2018).
Banas, K. et al. Exon skipping induced by CRISPR-directed gene editing regulates the response to chemotherapy in non-small cell lung carcinoma cells. Gene Ther. 29, 357–367 (2022).
Thomas, J. D. et al. RNA isoform screens uncover the essentiality and tumor-suppressor activity of ultraconserved poison exons. Nat. Genet. 52, 84–94 (2020). This work reveals tumour-suppressive roles for poison exons.
Smith, C. C. et al. Alternative tumour-specific antigens. Nat. Rev. Cancer 19, 465–478 (2019).
Vauchy, C. et al. CD20 alternative splicing isoform generates immunogenic CD4 helper T epitopes. Int. J. Cancer 137, 116–126 (2015).
Oka, M. et al. Aberrant splicing isoforms detected by full-length transcriptome sequencing as transcripts of potential neoantigens in non-small cell lung cancer. Genome Biol. 22, 9 (2021).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Frankiw, L., Baltimore, D. & Li, G. Alternative mRNA splicing in cancer immunotherapy. Nat. Rev. Immunol. 19, 675–687 (2019).
Slansky, J. E. & Spellman, P. T. Alternative splicing in tumors—a path to immunogenicity? N. Engl. J. Med. 380, 877–880 (2019).
Hoyos, L. E. & Abdel-Wahab, O. Cancer-specific splicing changes and the potential for splicing-derived neoantigens. Cancer Cell 34, 181–183 (2018).
Bowling, E. A. et al. Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell 184, 384–403.e21 (2021). This work shows that inhibition of the SF3b complex triggers antiviral signalling.
Lu, S. X. et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell 184, 4032–4047.e31 (2021). This work demonstrates that splicing modulation triggers neoantigen production and synergizes with immune checkpoint blockade.
De Paoli-Iseppi, R., Gleeson, J. & Clark, M. B. Isoform age—splice isoform profiling using long-read technologies. Front. Mol. Biosci. 8, 711733 (2021).
Veiga, D. F. T. et al. A comprehensive long-read isoform analysis platform and sequencing resource for breast cancer. Sci. Adv. 8, eabg6711 (2022). This work identifies full-length isoforms in primary tumours using LR-seq.
Tian, L. et al. Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol. 22, 310 (2021).
Gupta, I. et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 36, 1197–1202 (2018).
Hardwick, S. A. et al. Single-nuclei isoform RNA sequencing unlocks barcoded exon connectivity in frozen brain tissue. Nat. Biotechnol. 40, 1082–1092 (2022).
Joglekar, A. et al. A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun. 12, 463 (2021).
Mou, H. et al. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 18, 108 (2017).
Urbanski, L. et al. MYC regulates a pan-cancer network of co-expressed oncogenic splicing factors. Cell Rep. 41, 11704 (2022).
Hsu, T. Y. et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525, 384–388 (2015). This work identifies a MYC-driven splicing vulnerability.
Rossbach, O. et al. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol. Cell Biol. 29, 1442–1451 (2009).
Lareau, L. F., Inada, M., Green, R. E., Wengrod, J. C. & Brenner, S. E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446, 926–929 (2007). This landmark study identifies conserved poison exons in genes encoding SR proteins.
Ciesla, M. et al. Oncogenic translation directs spliceosome dynamics revealing an integral role for SF3A3 in breast cancer. Mol. Cell 81, 1453–1468.e12 (2021).
Kralovicova, J., Moreno, P. M., Cross, N. C., Pego, A. P. & Vorechovsky, I. Antisense oligonucleotides modulating activation of a nonsense-mediated RNA decay switch exon in the ATM gene. Nucleic Acid. Ther. 26, 392–400 (2016).
Mercatante, D. R., Bortner, C. D., Cidlowski, J. A. & Kole, R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J. Biol. Chem. 276, 16411–16417 (2001).
Taylor, J. K., Zhang, Q. Q., Wyatt, J. R. & Dean, N. M. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat. Biotechnol. 17, 1097–1100 (1999).
Liu, J. et al. Overcoming imatinib resistance conferred by the BIM deletion polymorphism in chronic myeloid leukemia with splice-switching antisense oligonucleotides. Oncotarget 8, 77567–77585 (2017).
Anczuków, O. et al. BRCA2 deep intronic mutation causing activation of a cryptic exon: opening toward a new preventive therapeutic strategy. Clin. Cancer Res. 18, 4903–4909 (2012).
Nielsen, T. O., Sorensen, S., Dagnaes-Hansen, F., Kjems, J. & Sorensen, B. S. Directing HER4 mRNA expression towards the CYT2 isoform by antisense oligonucleotide decreases growth of breast cancer cells in vitro and in vivo. Br. J. Cancer 108, 2291–2298 (2013).
Li, L. et al. Targeting the ERG oncogene with splice-switching oligonucleotides as a novel therapeutic strategy in prostate cancer. Br. J. Cancer 123, 1024–1032 (2020).
Bruno, I. G., Jin, W. & Cote, G. J. Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements. Hum. Mol. Genet. 13, 2409–2420 (2004).
Lin, J. et al. Induced-decay of glycine decarboxylase transcripts as an anticancer therapeutic strategy for non-small-cell lung carcinoma. Mol. Ther. Nucleic Acids 9, 263–273 (2017).
Karras, J. G., McKay, R. A., Lu, T., Dean, N. M. & Monia, B. P. Antisense inhibition of membrane-bound human interleukin-5 receptor-α chain does not affect soluble receptor expression and induces apoptosis in TF-1 cells. Antisense Nucleic Acid. Drug Dev. 10, 347–357 (2000).
Shieh, J. J., Liu, K. T., Huang, S. W., Chen, Y. J. & Hsieh, T. Y. Modification of alternative splicing of Mcl-1 pre-mRNA using antisense morpholino oligonucleotides induces apoptosis in basal cell carcinoma cells. J. Invest. Dermatol. 129, 2497–2506 (2009).
Shiraishi, T., Eysturskarth, J. & Nielsen, P. E. Modulation of mdm2 pre-mRNA splicing by 9-aminoacridine-PNA (peptide nucleic acid) conjugates targeting intron–exon junctions. BMC Cancer 10, 342 (2010).
Dewaele, M. et al. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J. Clin. Invest. 126, 68–84 (2016).
Ghigna, C. et al. Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed: therapeutic potential of bifunctional oligonucleotides and indole derivatives. RNA Biol. 7, 495–503 (2010).
Zammarchi, F. et al. Antitumorigenic potential of STAT3 alternative splicing modulation. Proc. Natl Acad. Sci. USA 108, 17779–17784 (2011). This work demonstrates that ASOs can be used to manipulate splicing in in vivo cancer models.
Izaguirre, D. I. et al. PTBP1-dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol. Carcinog. 51, 895–906 (2012).
Shen, H., Zheng, X., Luecke, S. & Green, M. R. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 24, 2389–2394 (2010).
Dvinge, H. & Bradley, R. K. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 7, 45 (2015).
Mehmood, A. et al. Systematic evaluation of differential splicing tools for RNA-seq studies. Brief. Bioinform 21, 2052–2065 (2020).
Shen, S. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl Acad. Sci. USA 111, 5593–5601 (2014).
Katz, Y., Wang, E. T., Airoldi, E. M. & Burge, C. B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods 7, 1009–1015 (2010).
Vaquero-Garcia, J. et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. eLife 5, 11752 (2016).
Trincado, J. L. et al. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 19, 40 (2018).
Li, Y. I. et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 50, 151–158 (2018).
Leger, A. et al. RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat. Commun. 12, 7198 (2021).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01483-z (2022).
Acknowledgements
The authors thank members of the Bradley and Anczukow laboratories for helpful discussions. O.A. was supported by the National Institutes of Health (NIH)/National Cancer Institute (NCI) (R01 CA248317 and P30 CA034196) and NIH/National Institute of General Medical Sciences (NIGMS) (R01 GM138541). R.K.B. was supported, in part, by the NIH/NCI (R01 CA251138), NIH/National Heart, Lung, and Blood Institute (NHLBI) (R01 HL128239 and R01 HL151651) and the Blood Cancer Discoveries Grant programme through the Leukaemia & Lymphoma Society, Mark Foundation for Cancer Research and Paul G. Allen Frontiers Group (8023-20). R.K.B. is a Scholar of The Leukaemia & Lymphoma Society (1344-18) and holds the McIlwain Family Endowed Chair in Data Science.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
R.K.B. is an inventor on patent applications filed by Fred Hutchinson Cancer Center related to modulating splicing for cancer therapy. O.A. is an inventor on a patent application filed by The Jackson Laboratory related to modulating splicing factors.
Peer review
Peer review information
Nature Reviews Cancer thanks Maria Carmo-Fonseca, Polly Leilei Chen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Branch point
-
A nucleotide that performs a nucleophilic attack on the 5′ splice site (5′SS) in the first step of splicing.
- K homology (KH) domain
-
A protein domain that can bind RNA and is found in various RNA-binding proteins (RBPs), including splicing factors.
- Polypyrimidine tract
-
A pyrimidine (C or T)-rich sequence motif upstream of many 3′ splice sites (3′SSs) that is bound by the U2AF2 subunit of the U2AF heterodimer to facilitate 3′SS recognition.
- RNA splicing
-
A post-transcriptional mechanism that mediates the removal of introns from a pre-mRNA transcript and the ligation of exons to form a mature mRNA.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bradley, R.K., Anczuków, O. RNA splicing dysregulation and the hallmarks of cancer. Nat Rev Cancer 23, 135–155 (2023). https://doi.org/10.1038/s41568-022-00541-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41568-022-00541-7
This article is cited by
-
Full-length isoform concatenation sequencing to resolve cancer transcriptome complexity
BMC Genomics (2024)
-
Roles and mechanisms of aberrant alternative splicing in melanoma — implications for targeted therapy and immunotherapy resistance
Cancer Cell International (2024)
-
Metabolic heterogeneity in clear cell renal cell carcinoma revealed by single-cell RNA sequencing and spatial transcriptomics
Journal of Translational Medicine (2024)
-
RBM10 C761Y mutation induced oncogenic ASPM isoforms and regulated β-catenin signaling in cholangiocarcinoma
Journal of Experimental & Clinical Cancer Research (2024)
-
RNAi-based drug design: considerations and future directions
Nature Reviews Drug Discovery (2024)