Abstract
Despite advances in precision medicine, the clinical prospects for patients with ovarian and uterine cancers have not substantially improved. Here, we analyzed genome-scale CRISPR–Cas9 loss-of-function screens across 851 human cancer cell lines and found that frequent overexpression of SLC34A2—encoding a phosphate importer—is correlated with sensitivity to loss of the phosphate exporter XPR1, both in vitro and in vivo. In patient-derived tumor samples, we observed frequent PAX8-dependent overexpression of SLC34A2, XPR1 copy number amplifications and XPR1 messenger RNA overexpression. Mechanistically, in SLC34A2-high cancer cell lines, genetic or pharmacologic inhibition of XPR1-dependent phosphate efflux leads to the toxic accumulation of intracellular phosphate. Finally, we show that XPR1 requires the novel partner protein KIDINS220 for proper cellular localization and activity, and that disruption of this protein complex results in acidic “vacuolar” structures preceding cell death. These data point to the XPR1–KIDINS220 complex and phosphate dysregulation as a therapeutic vulnerability in ovarian cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Publicly available data used in this study include CRISPR–Cas9 loss of viability screens for 851 cancer cell lines10,11,12, cancer cell line RNA-seq expression data15, harmonized gene expression data for GTEx and TCGA datasets24 and copy number alterations for ovarian adenocarcinoma and uterine corpus endometrial carcinoma22,23. RNA-seq data following PAX8 suppression and multiplexed transcriptional profiling are available from the corresponding author upon request. RNA-seq data following PAX8 knockdown are available at https://doi.org/10.6084/m9.figshare.19125671.v1. MixSeq data following XPR1 knockdown are available at https://doi.org/10.6084/m9.figshare.19125677.v1. Source data are provided with this paper.
Code availability
Computer code to reproduce these results is available from the corresponding authors upon request.
References
Lord, C. J. & Ashworth, A. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017).
Lheureux, S., Braunstein, M. & Oza, A. M. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J. Clin. 69, 280–304 (2019).
Vaughan, S. et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat. Rev. Cancer 11, 719–725 (2011).
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Boehm, J. S. et al. Cancer research needs a better map. Nature 589, 514–516 (2021).
Costanzo, M. et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420 (2016).
Pan, J. et al. Sparse dictionary learning recovers pleiotropy from human cell fitness screens. Cell Syst. https://www.sciencedirect.com/science/article/pii/S2405471221004889 (2022).
Cheung, H. W. et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl Acad. Sci. USA 108, 12372–12377 (2011).
Price, C. et al. Genome-wide interrogation of human cancers identifies EGLN1 dependency in clear cell ovarian cancers. Cancer Res. 79, 2564–2579 (2019).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
DepMap. DepMap 21Q2 Public. figshare https://doi.org/10.6084/m9.figshare.14541774.v2 (2021).
Dempster, J. M., Rossen, J., Kazachkova, M. & Pan, J. Extracting biological insights from the Project Achilles genome-scale CRISPR screens in cancer cell lines. Preprint at bioRxiv https://doi.org/10.1101/720243 (2019).
Battini, J. L., Rasko, J. E. & Miller, A. D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl Acad. Sci. USA 96, 1385–1390 (1999).
Giovannini, D., Touhami, J., Charnet, P., Sitbon, M. & Battini, J.-L. Inorganic phosphate export by the retrovirus receptor XPR1 in metazoans. Cell Rep. 3, 1866–1873 (2013).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576 (2017).
Dempster, J. M. et al. Gene expression has more power for predicting in vitro cancer cell vulnerabilities than genomics. Preprint at bioRxiv https://doi.org/10.1101/2020.02.21.959627 (2020).
Levan, K. et al. Immunohistochemical evaluation of epithelial ovarian carcinomas identifies three different expression patterns of the MX35 antigen, NaPi2b. BMC Cancer 17, 303 (2017).
Yin, B. W. T. et al. Monoclonal antibody MX35 detects the membrane transporter NaPi2b (SLC34A2) in human carcinomas. Cancer Immun. 8, 3 (2008).
Zou, Y. et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun. 10, 1617 (2019).
Magtanong, L. et al. Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem. Biol. 26, 420–432 (2019).
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Cancer Genome Atlas Research Network et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013).
Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020).
Vivian, J. et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35, 314–316 (2017).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Köbel, M. et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 5, e232 (2008).
Piek, J. M. et al. Dysplastic changes in prophylactically removed Fallopian tubes of women predisposed to developing ovarian cancer. J. Pathol. 195, 451–456 (2001).
Jarboe, E. et al. Serous carcinogenesis in the fallopian tube: a descriptive classification. Int. J. Gynecol. Pathol. 27, 1–9 (2008).
Hu, Z. et al. The repertoire of serous ovarian cancer non-genetic heterogeneity revealed by single-cell sequencing of normal fallopian tube epithelial cells. Cancer Cell 37, 226–242 (2020).
Mittag, J., Winterhager, E., Bauer, K. & Grümmer, R. Congenital hypothyroid female pax8-deficient mice are infertile despite thyroid hormone replacement therapy. Endocrinology 148, 719–725 (2007).
Elias, K. M. et al. Epigenetic remodeling regulates transcriptional changes between ovarian cancer and benign precursors. JCI Insight 1, e87988 (2016).
Adler, E. K. et al. The PAX8 cistrome in epithelial ovarian cancer. Oncotarget 8, 108316–108332 (2017).
Shi, K. et al. PAX8 regulon in human ovarian cancer links lineage dependency with epigenetic vulnerability to HDAC inhibitors. eLife 8, e44306 (2019).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
DeWeirdt, P. C. et al. Genetic screens in isogenic mammalian cell lines without single cell cloning. Nat. Commun. 11, 752 (2020).
Minisola, S. et al. Tumour-induced osteomalacia. Nat. Rev. Dis. Primers 3, 17044 (2017).
Whitsett, J. A., Wert, S. E. & Weaver, T. E. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu. Rev. Med. 61, 105–119 (2010).
Lituiev, D. S. & Kiyamova, R. G. Mutations in the gene of human type IIb sodium-phosphate cotransporter SLC34A2. Zurich Open Repository and Archive https://www.zora.uzh.ch/id/eprint/44008/1/5.pdf (2010).
Wilson, M. S., Jessen, H. J. & Saiardi, A. The inositol hexakisphosphate kinases IP6K1 and -2 regulate human cellular phosphate homeostasis, including XPR1-mediated phosphate export. J. Biol. Chem. 294, 11597–11608 (2019).
Li, X. et al. Control of XPR1-dependent cellular phosphate efflux by InsP8 is an exemplar for functionally-exclusive inositol pyrophosphate signaling. Proc. Natl Acad. Sci. USA https://www.pnas.org/doi/10.1073/pnas.1908830117 (2020).
López-Sánchez, U. et al. Interplay between PFBC-associated SLC20A2 and XPR1 phosphate transporters requires inositol polyphosphates for control of cellular phosphate homeostasis. J. Biol. Chem. 295, 9366–9378 (2020).
Legati, A. et al. Mutations in XPR1 cause primary familial brain calcification associated with altered phosphate export. Nat. Genet. 47, 579–581 (2015).
Wainberg, M. et al. A genome-wide almanac of co-essential modules assigns function to uncharacterized genes. Nat. Genet. 53, 638–649 (2021).
Kim, E. et al. A network of human functional gene interactions from knockout fitness screens in cancer cells. Life Sci. Alliance 2, e201800278 (2019).
Cabrera-Poch, N., Sánchez-Ruiloba, L., Rodríguez-Martínez, M. & Iglesias, T. Lipid raft disruption triggers protein kinase C and Src-dependent protein kinase D activation and Kidins220 phosphorylation in neuronal cells. J. Biol. Chem. 279, 28592–28602 (2004).
Iglesias, T. et al. Identification and cloning of Kidins220, a novel neuronal substrate of protein kinase D. J. Biol. Chem. 275, 40048–40056 (2000).
Arévalo, J. C., Yano, H., Teng, K. K. & Chao, M. V. A unique pathway for sustained neurotrophin signaling through an ankyrin-rich membrane-spanning protein. EMBO J. 23, 2358–2368 (2004).
Kong, H., Boulter, J., Weber, J. L., Lai, C. & Chao, M. V. An evolutionarily conserved transmembrane protein that is a novel downstream target of neurotrophin and ephrin receptors. J. Neurosci. 21, 176–185 (2001).
Wege, S. et al. The EXS domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant Physiol. 170, 385–400 (2016).
Arpat, A. B. et al. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate: PHO1 localization to the Golgi/trans-Golgi network. Plant J. 71, 479–491 (2012).
Snyder, N. A. et al. H+ and Pi byproducts of glycosylation affect Ca2+ homeostasis and are retrieved from the Golgi complex by homologs of TMEM165 and XPR1. G3 (Bethesda) 7, 3913–3924 (2017).
Gerber, D. E. et al. Phase Ia study of anti-NaPi2b antibody-drug conjugate lifastuzumab vedotin DNIB0600A in patients with non-small cell lung cancer and platinum-resistant ovarian cancer. Clin. Cancer Res. 26, 364–372 (2020).
Marks, J. The role of SLC34A2 in intestinal phosphate absorption and phosphate homeostasis. Pflugers Arch. 471, 165–173 (2019).
He, P. et al. High phosphate actively induces cytotoxicity by rewiring pro-survival and pro-apoptotic signaling networks in HEK293 and HeLa cells. FASEB J. 35, e20997 (2021).
Di Marco, G. S. et al. Increased inorganic phosphate induces human endothelial cell apoptosis in vitro. Am. J. Physiol. Renal Physiol. 294, F1381–F1387 (2008).
Marks, M. S., Heijnen, H. F. G. & Raposo, G. Lysosome-related organelles: unusual compartments become mainstream. Curr. Opin. Cell Biol. 25, 495–505 (2013).
Ruiz, F. A., Lea, C. R., Oldfield, E. & Docampo, R. Human platelet dense granules contain polyphosphate and are similar to acidocalcisomes of bacteria and unicellular eukaryotes. J. Biol. Chem. 279, 44250–44257 (2004).
Josifova, D. J. et al. Heterozygous KIDINS220/ARMS nonsense variants cause spastic paraplegia, intellectual disability, nystagmus, and obesity. Hum. Mol. Genet. 25, 2158–2167 (2016).
McDonald, E. R. III et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577–592 (2017).
Yang, X. et al. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 (2011).
McFarland, J. M. et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. 9, 4610 (2018).
Buehler, E., Chen, Y.-C. & Martin, S. C911: a bench-level control for sequence specific siRNA off-target effects. PLoS ONE 7, e51942 (2012).
Aran, D., Sirota, M. & Butte, A. J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971 (2015).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Li, W. et al. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol. 16, 281 (2015).
Williams, S. N. et al. Phosphorus concentrations in blood, milk, feces, bone and selected fluids and tissues of growing heifers as affected by dietary phosphorus. Livest. Res. Rural. Dev. http://www.lrrd.org/lrrd3/2/florida4.htm (1991).
McFarland, J. M. et al. Multiplexed single-cell profiling of post-perturbation transcriptional responses to define cancer vulnerabilities and therapeutic mechanism of action. Nat. Commun. 11, 4296 (2020).
Stoeckius, M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018).
Li, B. et al. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat. Methods. 17, 793–798 (2020).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint at https://arxiv.org/abs/1207.3907 (2012).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Ritchie, M. E. et al. limma Powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Huttlin, E. L. et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 184, 3022–3040 (2021).
Oughtred, R. et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–D541 (2019).
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976–989 (1994).
Acknowledgements
This work was funded in part by the Slim Initiative in Genomic Medicine for the Americas, a joint United States–Mexico project funded by the Carlos Slim Foundation (to T.R.G.), and grants from the National Cancer Institute (nos. CA242457 to T.R.G. and CA212229 to D.P.B.). We thank the Dana-Farber/Harvard Cancer Center in Boston, MA, for the use of the Specialized Histopathology Core, which provided histology and immunohistochemistry services and is supported in part by a NCI Cancer Center Support Grant (no. CA06516). We thank J. Barnett, B. Buckley, M. Veneskey, M. Cecilia-Saberi and D. Tennen for technical support.
Author information
Authors and Affiliations
Contributions
D.P.B., B.R.P., F.V. and T.R.G. initiated the project and oversaw the research plan. J.K.B, D.P.B. and B.R.P. analyzed genetic dependency data under the supervision of W.C.H., D.E.R., J.B., F.V. and T.R.G. and with support from I.F. and E.S.C. M.V.R., A.A., T.A.S., B.R.P. and D.P.B. conducted viability experiments and immunoblotting. A.A., B.P. and D.P.B. conducted the genome-scale modifier screen with analysis support from M.K., J.M.D. and J.M.M. and supervision from J.M.D. In vivo experiments were conducted by A.G. and N.D. under the supervision of F.P. Intracellular phosphate assays were performed by M.V.R. and D.P.B. PAX8 RNA-seq experiments were conducted by K.I. with analytical support from W.N.C. D.P.B. analyzed GTEx, TCGA and CCLE expression datasets with supervision from J.M.M. Multiplexed transcriptional profiling was conducted by B.R.P. and D.P.B. and was analyzed by A.W. and W.N.C. Phosphate uptake and efflux assays were conducted by D.P.B. and L.E.S. with supervision from M.M. A.A. and D.P.B. conducted coimmunoprecipitation experiments. B.R.P. conducted cellular imaging studies. D.P.B. and M.E. conducted ultrastructural analysis. D.P.B., B.R.P., F.V. and T.R.G. wrote the manuscript, and all authors edited and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
F.V. receives research funding from Novo Holdings. D.E.R. receives research funding from the Functional Genomics Consortium (Abbvie, Jannsen, Merck and Vir) and is a director of Addgene. T.R.G. previously received cash and/or equity compensation for consulting to GlaxoSmithKline, Sherlock Biosciences and FORMA Therapeutics, is currently a paid consultant for Anji Pharmaceuticals and receives research funding from Bayer HealthCare, Calico Life Sciences and Novo Holdings. W.C.H. is a consultant for ThermoFisher, Solasta, MPM Capital, iTeos, Frontier Medicines and Paraxel and is a Scientific Founder and serves on the Scientific Advisory Board for KSQ Therapeutics. All other authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks Ronny Drapkin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 XPR1 dependency is observed selectively in SLC34A2-high cancer cell lines.
a) For every cell line profiled in the Cancer Dependency Map dataset (N = 851 cancer cell lines), the degree of XPR1 essentiality is plotted on the Y-axis. The Chronos score is a scaled value of gene essentiality, where 0 is the effect of CRISPR/Cas9 genome editing and -1 is the effect of inactivation of pan-essential genes. Note that the ovarian lineage is separated into cancer subtypes. b) For every tissue type, the 10 highest SLC34A2 expressing cell lines were analyzed for their median expression of SLC34A2 (X-axis) and dependency on XPR1 (Y-axis). Note that some lineages may have less than 10 cell lines. Color encodes the correlation of SLC34A2 expression and XPR1 dependency across all cell lines within that lineage. c) Comparison of analytical methods for CRISPR/Cas9 genome-scale loss of function screens.
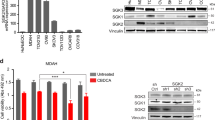
Extended Data Fig. 2 Validation of SLC34A2 and XPR1 protein levels and viability defects upon shRNA induction.
a) Validation of SLC34A2 in cell lines using immunohistochemistry. N = 1 experiment. b) Five days after viral transduction of the indicated sgRNA in the indicated cell lines stably expressing Cas9, cells were harvested and XPR1 levels were analyzed by immunoblotting. Note that irrelevant lanes were cropped out for clarity, but that this image represents a single blot at a single exposure. N = 1 technical replicate of at least N = 5 representative experiments. c) Three days after induction of shRNA, protein levels were measured in cellular lysates by protein simple. Protein levels normalized to vinculin and the untreated (-Dox) conditions are expressed below each band. Note that shXPR1 reagents effectively suppress XPR1 protein levels but shSeed reagents do not. N = 1 technical replicate of at least N = 5 representative experiments. d) Colony formation assay to measure the long-term (14 day) penetrance and viability effect of suppression of XPR1 using shRNA in IGROV1 and OVISE cells. N = 3 technical replicates of at least N = 2 representative experiments. e) Growth curves of SLC34A2-expressing cell lines after suppression of XPR1. In 96-well plates, confluency of the indicated cell lines was assessed every 4 hours. N = 3 technical replicates of at least N = 2 representative experiments. f) Six days after induction of shXPR1 in the indicated cell lines, cells were stained with DAPI to distinguish live and dead cells and Annexin V to distinguish non- and pre-apoptotic cells. N = 2 flow cytometric analyses of at least 10,000 cells, representative of N = 2 experiments.
Extended Data Fig. 3 In vivo CRISPR/Cas9 competition assays for target validation in mouse xenografts.
a) sgRNA abundance in SNGM tumor xenografts was evaluated by PCR and next-generation sequencing analysis, and the fold change compared to the early time point is shown as a heatmap for all of the negative control genes as well as any gene with a > 4 fold change in abundance in any of the screens. Each tumor/replicate is shown as an individual column, N = 1 transduction. b) Same as in d, but with the OVISE cancer cell line.
Extended Data Fig. 4 SLC34A2 in ovarian cancer is likely driven by PAX8.
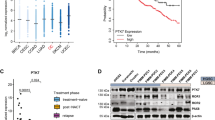
a) Using the combined GTEx, TCGA, and CCLE dataset, the differential expression of SLC34A2 in each tissue relative to the average of all tissues is compared. The relevant gynecological tissues (fallopian tube, ovary, and uterus) are highlighted in teal. The false discovery rate (FDR) was calculated using a two-sided Wilcoxon ranked sum test comparing each group to the average expression across all groups and correcting for multiple comparisons using Bonferoni’s method. The Cancer Genome Atlas abbreviations used include: LUAD = Lung adenocarcinoma; THCA = Thyroid carcinoma; KRP = Kidney renal cell papillary carcinoma; LUSC = Lung squamous cell carcinoma; OV = Ovarian serous cystadenocarcinoma; UCEC = Uterine corpus endometrial carcinoma. b) The expression of PAX8 and SLC34A2 mRNA in the indicated tissues is plotted. The pearson correlation within these samples is indicated. c) Expression of PAX8 across the indicated tissues was compared as in Fig. 3a. See methods for exact N values. Boxplots are drawn indicating the first and third quartiles, and whiskers span to the largest value within 1.5x the interquartile range. d) Immunoblot validation of CRISPR-interference mediated suppression of PAX8. N = 1 technical replicate, representative of N = 2 independent experiments. e) Gene expression - relative to un-perturbed, parental cell lines profiled in parallel - of reported PAX8 target genes (see main text) after stable overexpression of WT-PAX8 (‘PAX8 O/E’) and/or induction of PAX8-target (sg4) or control (sg9) sgRNA and dCas9-KRAB. Data represents a single experiment. N = 1 replicate. f) XPR1 expression across all tissues in TCGA and GTEx, with ovarian and uterine tissues highlighted in teal. Boxplots are drawn as in b. g) XPR1 copy number heatmap for a ~2.5 Mb region of chromosome 1 indicating XPR1 amplification in TCGA Uterine Corpus Endometrial Carcinoma20. Each patient sample is represented by a horizontal line. Red indicates copy gain and blue indicates copy loss. Data are a subset of the samples rank-ordered by highest copy gain to indicate both focal and chromosome arm-level gains.
Extended Data Fig. 5 A genome-scale CRISPR/Cas9 screen validates the relationship between XPR1 dependency in the context of high expression of SLC34A2.
a) Outline of the experimental method and analysis for a genome-scale, dual-knock-out modifier screen. OVISE (without Cas9 expression) is engineered to stably express sgRNA targeting XPR1 (the ‘anchor’ sgRNA). Upon introduction of ‘all-in-one’ lentivirus, containing both Cas9 ORF and a second sgRNA, both genes are simultaneously inactivated by Cas9. We used three achor sgRNA: one targeting a gene desert on chromosome 2 (sgChr2-2) and two targeting XPR1 (sgXPR1_1 and sgXPR1_2) and infecting the cells with the Brunello genome-scale sgRNA library. 15 days after infection, cells were harvested, genomic DNA was isolated, and sgRNA barcodes were quantified with next generation sequencing. See methods for full experimental and analytical details. b) Western confirmation of dual-knock-out of XPR1 and SLC34A2. The three cell lines used in the genome-scale screen were infected with ‘all-in-one’ lentivirus expressing control-, XPR1-, or SLC34A2-targeting sgRNA. Note that in the sgXPR1 ‘anchor’ cell lines, XPR1 is suppressed with the control virus, indicating that the first infection provides XPR1-targeting sgRNA and the second infection provides Cas9 protein. NIC stands for ‘no-infection control’. N = 1 technical replicate representative of N = 3 independent transductions. c) Arm-level results of the genome-scale modifier screen. See methods for full analysis details. Beta-scores represent the extent to which a gene was enriched or depleted relative to the initial plasmid representation. An XPR1-positive and control-neutral score represents a likely rescue gene (that is SLC34A2 and ARNT). XPR1-positive and control-positive scores represent genes with profound viability defects without specificity for XPR1 (for example RANBP17). N = 1 transduction per anchor condition, expanded and cultured as N = 2 independent replicates.
Extended Data Fig. 6 The XPR1 dependency is not affected by phosphate levels in the tissue culture medium.
a) The concentration of phosphate in the growth medium of DepMap cell lines does not determine the extent of XPR1 dependency. Concentrations of phosphate were estimated from manufacturer formulations (see methods) and the pearson correlation between growth medium phosphate and XPR1 dependency is indicated. b) Experimental procedure for manipulating tissue culture medium and assessing its effect on XPR1 dependency. The same parental cancer cell line was engineered to express firefly luciferase and Cas9, or Renilla luciferase alone. After a one-week adaptation to lowered phosphate, the two variants were mixed together and infected with sgRNA-encoding lentivirus. After selection for lentivirus-infected cell lines, the initial representation of Cas9:parental cells was determined by measuring the ratio of Firefly:Renilla luciferase using a DualGlo assay (Promega). One week after infection (Day 16 of the protocol), the extent to which genetic perturbation was detrimental to cell viability was determined using the DualGlo assay.c) The XPR1 dependency is maintained in a low phosphate medium condition. SNGM and ES2 were profiled in the assay outlined in panel b. Note that the CERES score - displayed below the plot - represents the viability defect of the cell line 21 days after knock-out of XPR1 and growth in the indicated growth medium. N = 5 technical replicates representative of N = 2 experiments. d) The viability of cells (as measured by total protein content) was measured in parallel with total phosphate as in Fig. 4d. N = 3 technical replicates representative of N = 4 independent experiments per cell line.
Extended Data Fig. 7 Transcriptional profiling reveals a phosphate-related homeostatic response after XPR1 inactivation.
a) Experimental workflow to determine the transcriptional profile of XPR1 inactivation across many different cancer cell lines. See methods for full details; N = 1 transduction event for panels b-g. b) The total number of cells per cell line de-multiplexed from the 10X scRNAseq library. c) The total number of unique transcripts measured for each cell, as measured by unique molecular identifiers (UMIs). Box plots represent the 1st – 3rd quartiles, with whiskers representing the minimum and the maximum. The exact N for each sample in c is indicated in panel b. d) UMAP projection of the 2,501 cells from the indicated cell lines (determined by SNP profiles) and perturbations (indicated by cell-surface antibody ‘hash-tag’ labeling). Arrows indicate the degree to which the average transcriptional profile changes between the control sgRNA and the sgXPR1 infection condition. e) Summary of transcriptional effects across cell lines after inactivation of XPR1. Middle, the log-fold change of the top 500 differentially expressed genes after regressing out the effect of cell cycle. Left, summary annotations for each cell line include XPR1 dependency (XPR1 CERES), the overall transcriptional change (average log2 fold-change), and the degree of cell cycle arrest observed in the single-cell data (ΔG0/G1). The pearson correlation of these values is shown above the heatmap. Right, the pearson correlation of the top 500 differentially expressed genes between each cell line. f) Differentially expressed genes - after correcting for cell cycle - in the less sensitive cell lines (COV413a, JHOS4, OVCAR4, HEC6, and JHUEM1). Significance was assessed by the limma-voom pipeline using a two-tailed statistical test (see methods). g) Same as in f, but for the highly correlated cell lines RMG1, IGROV1, and OVISE. h) Four days after induction of shXPR1_2 (IGROV1) or shXPR1_4 (OVISE) using doxycycline, the amount of secreted FGF23 was measured in the conditioned medium using ELISA. N = 2 technical replicates representative of N = 3 independent experiments. i) 72 hours after treatment with the XPR1 inhibitor XRBD, the indicated proteins were detected using immunoblot. N = 1 technical replicate representative of N = 2 independent experiments. j) Top, western blot analysis of SLC34A2 and XPR1 in the SLC34A2-high yet XPR1-insensitive lung cancer cell lines, five days after infection with lentivirus expressing the indicated sgRNA. Bottom, viability of the indicated cancer cell lines was assessed using Cell Titer Glo after a five day XRBD treatment to inhibit XPR1. Points represent the mean of N = 3 technical replicates; error bars represent standard error of the mean. Data are representative of N = 2 independent experiments.
Extended Data Fig. 8 Open-reading frames of XPR1 resistant to CRISPR/Cas9-mediated gene editing.
a) Immunofluorescent localization of XPR1 mutants using the V5 epitope tag. Left, WT XPR1 localizes to the secretory pathways as well as puncta within the cytoplasm. Middle, XPR1 (short) staining appears more diffuse. Note similar localization patterns between L218S and wildtype XPR1 alleles. Scale bar = 200 µm. N = 1 experiment representative of N = 2 independent transductions. b) Western blot validation of guide-resistant ORF. OVISE.Cas9 cells (parental, left, or overexpressing the WT XPR1 ORF, right, used in Fig. 3e) were infected with the indicated sgRNA and harvested 5 days after infection. The XPR1 ORF includes a mutation to block binding of sgXPR1_2 but not sgXPR1_1. Note the inactivation of both endogenous and overexpression ORF with sgXPR1_1 and only endogenous XPR1 with sgXPR1_2. N = 1 experiment representative of N = 2 independent transductions.
Extended Data Fig. 9 KIDINS220 is a unique partner protein of the XPR1 phosphate efflux complex.
a) Genetic dependency correlations to XPR1 dependency across 851 cancer cell lines was assessed by pearson correlation test and corrected for multiple comparisons using the Benjamini-Hochberg method. Genes with significantly correlated dependency profiles are highlighted, as are proteins with known connection to XPR1 regulation. b) The viability defect of the indicated cancer cell lines after KIDINS220 inactivation was evaluated as in Fig. 1c. N = 3 technical replicates representative of at least N = 2 independent transductions per cell line. c) SLC34A2 was inactivated in EMTOKA and OVISE, and the KIDINS220 dependency was evaluated as in b. N = 3 technical replicates representative of at least N = 2 independent transductions per cell line.d) The interacting partners of XPR1 and KIDINS220 were downloaded from the BioGrid and Bioplex databases and compared. Proteins present in the XPR1 or KIDINS220 interactomes are highlighted as text.e) Left, the mRNA expression of XPR1 and KIDINS220 is shown for the fifteen tissues with the highest correlation in expression. The line represents linear regression for these samples (N = 2,799). Right, the Pearson correlation for those tissues, highlighting the diverse tissues in which there is a high correlation between XPR1 and KIDINS220 expression. f) Mutants of XPR1 used in this manuscript. XPR1 WT refers to the 696 amino acid protein produced by NM_004736 (the only isoform detected by RT-PCR of OVISE mRNA), while XPR1 (short) refers to the 631 amino acid product of NM_001135669. All constructs have C-terminal V5 tags for immunoprecipitation, western blotting, and immunofluorescent detection. g) XPR1 or Luciferase (Luc) were overexpressed in 293 T cells and immunoprecipitated using the V5 tag and analyzed by targeted immunoblot or for total protein. Proteins were extracted from this gel and identified using mass spectrometry, the results of which are shown in Fig. 6c. N = 1 replicate of N = 2 independent transfections. h) Cas9 + sgRNA targeting XPR1 or KIDINS220 were transfected into 293 T cells, and clones were isolated that lacked expression of the target proteins. For XPR1 inactivated cells, the XPR1 ORF was re-expressed, and the relative levels of the indicated proteins were detected by immunoblot. At least N = 2 clonal populations were profiled for each inactivation condition. i) Five days after infection with the indicated sgRNA targeting XPR1 or KIDINS220, free inorganic intracellular phosphate was determined as in Fig. 4d. N = 3 technical replicates of N = 3 independent transductions.
Supplementary information
Supplementary Table 1
A file containing multiple tabs: sgRNA sequences; shRNA sequences; RT–PCR primers; a compilation of phosphate concentrations in different tissue culture media; results of the XPR1 modifier screen (Fig. 4a); and RT–PCR validation of FGF23 elevation and SLC34A2 suppression following XPR1 suppression.
Supplementary Video 1
Time-course video demonstrating vacuole development in OVISE cancer cells following XPR1 inactivation.
Source data
Source Data Fig. 1
Numerical data.
Source Data Fig. 2
Numerical data.
Source Data Fig. 3
Numerical data.
Source Data Fig. 4
Numerical data.
Source Data Fig. 5
Numerical data.
Source Data Fig. 6
Numerical data.
Source Data Fig. 6
Blot data.
Source Data Extended Data Fig. 1
Numerical data.
Source Data Extended Data Fig. 2
Numerical data.
Source Data Extended Data Fig. 2
Blot data.
Source Data Extended Data Fig. 3
Numerical data.
Source Data Extended Data Fig. 4
Numerical data.
Source Data Extended Data Fig. 4
Blot data.
Source Data Extended Data Fig. 5
Numerical data.
Source Data Extended Data Fig. 5
Blot data.
Source Data Extended Data Fig. 6
Numerical data.
Source Data Extended Data Fig. 7
Numerical data.
Source Data Extended Data Fig. 7
Blot data.
Source Data Extended Data Fig. 8
Blot data.
Source Data Extended Data Fig. 9
Numerical data.
Source Data Extended Data Fig. 9
Blot data.
Rights and permissions
About this article
Cite this article
Bondeson, D.P., Paolella, B.R., Asfaw, A. et al. Phosphate dysregulation via the XPR1–KIDINS220 protein complex is a therapeutic vulnerability in ovarian cancer. Nat Cancer 3, 681–695 (2022). https://doi.org/10.1038/s43018-022-00360-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-022-00360-7
This article is cited by
-
Is XPR1 mediating phosphate efflux?
Pflügers Archiv - European Journal of Physiology (2024)
-
XPR1: a regulator of cellular phosphate homeostasis rather than a Pi exporter
Pflügers Archiv - European Journal of Physiology (2024)
-
Scalable genetic screening for regulatory circuits using compressed Perturb-seq
Nature Biotechnology (2023)
-
Inositol pyrophosphates activate the vacuolar transport chaperone complex in yeast by disrupting a homotypic SPX domain interaction
Nature Communications (2023)
-
Functional genomics of complex cancer genomes
Nature Communications (2022)