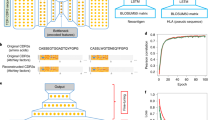
Abstract
Identifying neoepitopes that elicit an adaptive immune response is a major bottleneck to developing personalized cancer vaccines. Experimental validation of candidate neoepitopes is extremely resource intensive and the vast majority of candidates are non-immunogenic, creating a needle-in-a-haystack problem. Here we address this challenge, presenting computational methods for predicting class I major histocompatibility complex (MHC-I) epitopes and identifying immunogenic neoepitopes with improved precision. The BigMHC method comprises an ensemble of seven pan-allelic deep neural networks trained on peptide–MHC eluted ligand data from mass spectrometry assays and transfer learned on data from assays of antigen-specific immune response. Compared with four state-of-the-art classifiers, BigMHC significantly improves the prediction of epitope presentation on a test set of 45,409 MHC ligands among 900,592 random negatives (area under the receiver operating characteristic = 0.9733; area under the precision-recall curve = 0.8779). After transfer learning on immunogenicity data, BigMHC yields significantly higher precision than seven state-of-the-art models in identifying immunogenic neoepitopes, making BigMHC effective in clinical settings.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data, including model outputs and MANAFEST data, are provided in our public Mendeley repository: https://data.mendeley.com/datasets/dvmz6pkzvb. All data except MANAFEST data were collected from publicly available sources: MHCflurry-2.02, NetMHCpan-4.16, PRIME-1.013, PRIME-2.014, TESLA16, IEDB22, NEPdb23, Neopepsee24, IPD-IMGT/HLA34, IPD-MHC 2.035 and UniProt36 (accession numbers: P01899, P01900, P14427, P14426, Q31145, P01901, P01902, P04223, P14428, P01897, Q31151). Source data are provided with this paper.
Code availability
All code used in this study and the final trained models are provided in our public GitHub repository: https://github.com/KarchinLab/bigmhc ref. 41. Scikit-Learn v.1.0.2 was used to calculate performance metrics. Pandas v.1.4.2 and Numpy v.1.21.5 were used for data processing. SAM suite v.3.5 buildmodel and align2model were used to generate multiple sequence alignments. Matplotlib v.3.5.1, Seaborn v.0.12.2, py3Dmol v.2.0.1 and v.AlphaFold2 were used to generate figures.
References
Xiaoshan, S. M. et al. High-throughput prediction of MHC class I and II neoantigens with MHCnuggets. Cancer Immunol. Res. 8, 396–408 (2020).
O’Donnell, T. J., Rubinsteyn, A. & Laserson, U. MHCflurry 2.0: improved pan-allele prediction of MHC class I-presented peptides by incorporating antigen processing. Cell Syst. 11, 42–48 (2020).
Sarkizova, S. et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 38, 199–209 (2020).
Stranzl, T., Larsen, M. V., Lundegaard, C. & Nielsen, M. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics 62, 357–368 (2010).
Nielsen, M. & Andreatta, M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 8, 33 (2016).
Reynisson, B., Alvarez, B., Paul, S., Peters, B. & Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. W1, 48 (2020).
Hoof, I. et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61, 1–13 (2009).
Nielsen, M. et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B Locus protein of known sequence. PLoS One 2, e796 (2007).
Bassani-Sternberg, M. et al. Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput. Biol. 13, e1005725 (2017).
Gfeller, D. et al. The length distribution and multiple specificity of naturally presented HLA-I ligands. J. Immunol. 201, 3705–3716 (2018).
Chu, Y. et al. A transformer-based model to predict peptide–HLA class I binding and optimize mutated peptides for vaccine design. Nat. Mach. Intell. 4, 300–311 (2022).
O'Donnell, T. J. et al. MHCflurry: open-source class I MHC binding affinity prediction. Cell Syst. 7, 129–132.e124 (2018).
Schmidt, J. et al. Prediction of neo-epitope immunogenicity reveals TCR recognition determinants and provides insight into immunoediting. Cell Rep. Med. 2, 100194 (2021).
Gfeller, D. et al. Improved predictions of antigen presentation and TCR recognition with MixMHCpred2.2 and PRIME2.0 reveal potent SARS-CoV-2 CD8+ T-cell epitopes. Cell Syst. 14, 72–83.e5 (2023).
Lu, T. et al. Deep learning-based prediction of the T cell receptor–antigen binding specificity. Nat. Mach. Intell. 3, 864–875 (2021).
Wells, D. K. et al. Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell 183, 818–834 (2020).
Vaswani, A. et al. Attention is all you need. In Advances in Neural Information Processing Systems (2017).
Huang, G., Liu, Z., van der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. Proc. IEEE Conference on Computer Vision and Pattern Recognition 4700–4708 (2017).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Rego, N. & Koes, D. 3Dmol.js: molecular visualization with WebGL. Bioinformatics 31, 1322–1324 (2015).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Vita, R. et al. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343 (2018).
Xia, J. et al. NEPdb: a database of T-cell experimentally-validated neoantigens and pan-cancer predicted neoepitopes for cancer immunotherapy. Front. Immunol. 12, 644637 (2021).
Kim, S. et al. Neopepsee: accurate genome-level prediction of neoantigens by harnessing sequence and amino acid immunogenicity information. Ann. Oncol. 29, 1030–1036 (2018).
Danilova, L. et al. The Mutation-Associated Neoantigen Functional Expansion of Specific T Cells (MANAFEST) assay: a sensitive platform for monitoring antitumor immunity. Cancer Immunol. Res. 6, 888–899 (2018).
Anagnostou, V. et al. Evolution of neoantigen landscape during immune checkpoint blockade in non–small cell lung cancer. Cancer Discov. 7, 264–276 (2017).
Caushi, J. X. et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature 596, 126–132 (2021).
Anagnostou, V. et al. Multimodal genomic features predict outcome of immune checkpoint blockade in non-small-cell lung cancer. Nat. Cancer 1, 99–111 (2020).
Jones, S. et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci. Transl. Med. 7, 283ra253 (2015).
Stranzl, T., Larsen, M. V., Lundegaard, C. & Nielsen, M. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics 62, 357–368 (2010).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems (2019).
Kingma, D. P. & Ba, J. L. Adam: a method for stochastic optimization. In Third International Conference for Learning Representations (2015).
Loshchilov, I. & Hutter, F. Decoupled weight decay regularization. In Seventh International Conference for Learning Representations (2017).
Robinson, J. et al. IPD-IMGT/HLA database. Nucleic Acids Res. 48, D948–D955 (2019).
Maccari, G. et al. IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res. 45, D860–D864 (2016).
Consortium, T. U. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2022).
Hughey, R. & Krogh, A. Hidden Markov models for sequence analysis: extension and analysis of the basic method. Bioinformatics 12, 95–107 (1996).
Karplus, K. et al. What is the value added by human intervention in protein structure prediction? Proteins Struct. Funct. Bioinf. 45, 86–91 (2001).
Krogh, A., Brown, M., Mian, I. S., Sjölander, K. & Haussler, D. Hidden Markov models in computational biology: applications to protein modeling. J. Mol. Biol. 235, 1501–1531 (1994).
Kim, Y., Sidney, J., Pinilla, C., Sette, A. & Peters, B. Derivation of an amino acid similarity matrix for peptide:MHC binding and its application as a Bayesian prior. BMC Bioinf. 10, 394 (2009).
KarchinLab/bigmhc: v1.0. Zenodo https://doi.org/10.5281/zenodo.8023523 (2023).
Acknowledgements
This work was supported in part by the US National Institutes of Health grant CA121113 to V.A. and R.K., the Department of Defense Congressionally Directed Medical Research Programs grant CA190755 to V.A. and the ECOG-ACRIN Thoracic Malignancies Integrated Translational Science Center grant UG1CA233259 to V.A.
Author information
Authors and Affiliations
Contributions
B.A.A. and R.K. conceived the study and performed the experiments; Y.Y. contributed to 3D visualizations and model ideas; X.M.S. curated the MANAFEST data; D.S. and K.N.S. collected the MANAFEST dataset; B.A.A. and R.K. wrote the draft manuscript; B.A.A., V.A. and R.K. revised the manuscript; R.K. supervised the research.
Corresponding author
Ethics declarations
Competing interests
Under a licence agreement between Genentech and the Johns Hopkins University, X.M.S., R.K. and the university are entitled to royalty distributions related to the MHCnuggets technology discussed in this publication. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict-of-interest policies. V.A. has received research funding to her institution from Bristol Myers Squibb, AstraZeneca, Personal Genome Diagnostics and Delfi Diagnostics in the past 5 years. V.A. is an inventor on patent applications (63/276,525, 17/779,936, 16/312,152, 16/341,862, 17/047,006 and 17/598,690) submitted by Johns Hopkins University related to cancer genomic analyses, ctDNA therapeutic response monitoring and immunogenomic features of response to immunotherapy that have been licensed to one or more entities. Under the terms of these licence agreements, the university and inventors are entitled to fees and royalty distributions. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Reid F. Thompson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Visualization of BigMHC average attention to MHC encodings on the EL test data.
a Heatmap visualization of the average attention value for each position in the MHC pseudosequence on the EL testing dataset. The heatmap is stratified by MHC allele as rows, and separated by positive and negative testing instances. The position of each amino acid in the sequences from IPD-IMGT/HLA is provided at the bottom of each column. Darker values indicate MHC positions that are more influential on the final model output. The column of Differences depicts the Negatives values subtracted from the Positives values; thus, darker blue colours are most correctly discriminative whereas darker red attention values in this column highlight erroneous inferences. b Overlays of the Differences column from the training dataset on the MHC molecule using py3Dmol. MHC protein structure models are generated using AlphaFold.
Extended Data Fig. 2 Visualization of the average MHC attention on the EL training data.
Heatmap visualization method of Extended Data Fig. 1a applied to the EL training data.
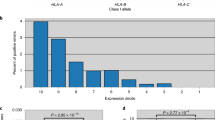
Extended Data Fig. 5 Composition of all training and evaluation datasets.
Positive and negative instances were stratified by HLA loci in the first two columns and by epitope length in the latter two columns. Positives in the EL datasets are detected by mass spectrometry, whereas negatives in the EL datasets are decoys. Both positives and negatives in the immunogenicity datasets are experimentally validated by immunogenicity assays.
Supplementary information
Supplementary Information
Supplementary discussion and Tables 1–4.
Supplementary Table 5
Results of all user-facing tools on all EL data, including training, validation and testing data.
Source data
Source Data Fig. 1
AUROC and AUPRC stratified by MHC and by MHC and epitope length for all evaluated methods on the EL test data.
Source Data Fig. 2
Mean PPVn, AUROC and AUPRC for all methods on the two immunogenicity test sets.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Albert, B.A., Yang, Y., Shao, X.M. et al. Deep neural networks predict class I major histocompatibility complex epitope presentation and transfer learn neoepitope immunogenicity. Nat Mach Intell 5, 861–872 (2023). https://doi.org/10.1038/s42256-023-00694-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-023-00694-6
This article is cited by
-
Breaking the performance ceiling for neoantigen immunogenicity prediction
Nature Cancer (2023)