Abstract
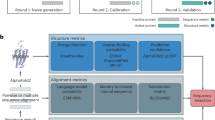
Models based on machine learning can enable accurate and fast molecular property predictions, which is of interest in drug discovery and material design. Various supervised machine learning models have demonstrated promising performance, but the vast chemical space and the limited availability of property labels make supervised learning challenging. Recently, unsupervised transformer-based language models pretrained on a large unlabelled corpus have produced state-of-the-art results in many downstream natural language processing tasks. Inspired by this development, we present molecular embeddings obtained by training an efficient transformer encoder model, MoLFormer, which uses rotary positional embeddings. This model employs a linear attention mechanism, coupled with highly distributed training, on SMILES sequences of 1.1 billion unlabelled molecules from the PubChem and ZINC datasets. We show that the learned molecular representation outperforms existing baselines, including supervised and self-supervised graph neural networks and language models, on several downstream tasks from ten benchmark datasets. They perform competitively on two others. Further analyses, specifically through the lens of attention, demonstrate that MoLFormer trained on chemical SMILES indeed learns the spatial relationships between atoms within a molecule. These results provide encouraging evidence that large-scale molecular language models can capture sufficient chemical and structural information to predict various distinct molecular properties, including quantum-chemical properties.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Data for model pretraining and fine-tuning on benchmark tasks are available at https://github.com/IBM/molformer.
Code availability
Python codes for MoLFormer training and fine-tuning, and Python notebooks for MoLFormer attention visualization, as well as instances of pretrained models, are available at https://github.com/IBM/molformer. For other enquiries contact the corresponding authors.
References
Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 50, 742–754 (2010).
Rupp, M., Tkatchenko, A., Müller, K.-R. & Von Lilienfeld, O. A. Fast and accurate modeling of molecular atomization energies with machine learning. Phys. Rev. Lett. 108, 058301 (2012).
Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988).
Goh, G. B., Hodas, N. O., Siegel, C. & Vishnu, A. SMILES2Vec: an interpretable general-purpose deep neural network for predicting chemical properties. Preprint at https://arxiv.org/abs/1712.02034 (2017).
Öztürk, H., Özgür, A. & Ozkirimli, E. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics 34, i821–i829 (2018).
Paul, A. et al. CheMixNet: mixed DNN architectures for predicting chemical properties using multiple molecular representations. Preprint at https://arxiv.org/abs/1811.08283 (2018).
Shin, B., Park, S., Kang, K. & Ho, J. C. Self-attention based molecule representation for predicting drug–target interaction. Proc. Mach. Learn. Res. 106, 230–248 (2019).
Daylight Chemical Information Systems SMARTS—a Language for Describing Molecular Patterns https://www.daylight.com/dayhtml/doc/theory/theory.smiles.html (2007).
Krenn, M., Häse, F., Nigam, A., Friederich, P. & Aspuru-Guzik, A. Self-referencing embedded strings (SELFIES): a 100% robust molecular string representation. Mach. Learn. Sci. Technol. 1, 045024 (2020).
Gao, W., Fu, T., Sun, J. & Coley, C. W. Sample efficiency matters: a benchmark for practical molecular optimization. In Thirty-Sixth Conference on Neural Information Processing Systems Datasets and Benchmarks Track (2022).
Jo, J., Kwak, B., Choi, H.-S. & Yoon, S. The message passing neural networks for chemical property prediction on SMILES. Methods 179, 65–72 (2020).
Duvenaud, D. et al. Convolutional networks on graphs for learning molecular fingerprints. In NIPS’15: Proc. 28th International Conference on Neural Information Processing Systems Vol. 2 (2015).
Defferrard, M., Bresson, X. & Vandergheynst, P. Convolutional neural networks on graphs with fast localized spectral filtering. Adv. Neural Inf. Process. Syst. 29 (2016).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. In 5th International Conference on Learning Representations (OpenReview.net, 2017).
Li, Y., Tarlow, D., Brockschmidt, M. & Zemel, R. Gated graph sequence neural networks. In 4th International Conference on Learning Representations (eds Bengio, Y. & LeCun, Y.) (OpenReview.net, 2016).
Veličković, P. et al. Graph attention networks. In International Conference on Learning Representations (2018).
Hamilton, W., Ying, Z. & Leskovec, J. Inductive representation learning on large graphs. Adv. Neural Inf. Process. Syst. 30, 1025–1035 (Curran Associates Inc., 2017).
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. Proc. Mach. Learn Res. 70, 1263–1272 (2017).
Schlichtkrull, M. et al. Modeling relational data with graph convolutional networks. In European Semantic Web Conference 593–607 (Springer, 2018).
Liao, R., Zhao, Z., Urtasun, R. & Zemel, R. S. LanczosNet: multi-scale deep graph convolutional networks. In 7th International Conference on Learning Representations (OpenReview.net, 2019).
Chen, P., Liu, W., Hsieh, C.-Y., Chen, G. & Zhang, S. Utilizing edge features in graph neural networks via variational information maximization. Preprint at https://arxiv.org/abs/1906.05488 (2019).
Kirkpatrick, P. & Ellis, C. Chemical space. Nature 432, 823–824 (2004).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30, 6000–6010 (Curran Associates Inc., 2017).
Bommasani, R. et al. On the opportunities and risks of foundation models. Preprint at https://doi.org/10.48550/ARXIV.2108.07258 (2021).
Chithrananda, S., Grand, G. & Ramsundar, B. ChemBERTa: large-scale self-supervised pretraining for molecular property prediction. Preprint at https://arxiv.org/abs/2010.09885 (2020).
Wang, Y., Wang, J., Cao, Z. & Barati Farimani, A. Molecular contrastive learning of representations via graph neural networks. Nat. Mach. Intell. 4, 279–287 (2022).
Su, J., Lu, Y., Pan, S., Wen, B. & Liu, Y. RoFormer: enhanced transformer with rotary position embedding. Preprint at https://arxiv.org/abs/2104.09864 (2021).
Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Liu, Y. et al. RoBERTa: a robustly optimized BERT pretraining approach. Preprint at https://arxiv.org/abs/1907.11692 (2019).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the NAACL: HLT Vol 1, 4171–4186 (Association for Computational Linguistics, 2019).
Katharopoulos, A., Vyas, A., Pappas, N. & Fleuret, F. Transformers are RNNs: fast autoregressive transformers with linear attention. Proc. Mach. Learn. Res. 119, 5156–5165 (2020).
Hu, W. et al. Strategies for pre-training graph neural networks. In 8th International Conference on Learning Representations (OpenReview.net, 2020).
Liu, S., Demirel, M. F. & Liang, Y. N-gram graph: simple unsupervised representation for graphs, with applications to molecules. In NIPS’19: Proc. 33rd International Conference on Neural Information Processing Systems (eds Wallach, H. et al.) 8464–8476 (Curran Associates, Inc., 2019).
Chen, T., Kornblith, S., Norouzi, M. & Hinton, G. A simple framework for contrastive learning of visual representations. Proc. Mach. Learn. Res. 119, 1597–1607 (2020).
Oord, A. V. D., Li, Y. & Vinyals, O. Representation learning with contrastive predictive coding. Preprint at https://arxiv.org/abs/1807.03748 (2018).
Liu, S. et al. Pre-training molecular graph representation with 3D geometry. In International Conference on Learning Representations (2022).
Klicpera, J., Groß, J. & Günnemann, S. Directional message passing for molecular graphs. In 8th International Conference on Learning Representations (OpenReview.net, 2020).
Fang, X. et al. Geometry-enhanced molecular representation learning for property prediction. Nat. Mach. Intell. 4, 127–134 (2022).
Altae-Tran, H., Ramsundar, B., Pappu, A. S. & Pande, V. Low data drug discovery with one-shot learning. ACS Cent. Sci. 3, 283–293 (2017).
Xiong, Z. et al. Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J. Med. Chem. 63, 8749–8760 (2019).
Schütt, K. et al. SchNet: a continuous-filter convolutional neural network for modeling quantum interactions. Adv. Neural Inf. Process. Syst. 992–1002 (Curran Associates Inc., 2017).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. USA 118, e2016239118 (2021).
Vig, J. et al. BERTology meets biology: interpreting attention in protein language models. In 9th International Conference on Learning Representations (OpenReview.net, 2021).
Urbina, F., Lentzos, F., Invernizzi, C. & Ekins, S. Dual use of artificial-intelligence-powered drug discovery. Nat. Mach. Intell. 4, 189–191 (2022).
Choromanski, K. et al. Rethinking attention with Performers. In Proc. 9th International Conference on Learning Representations (OpenReview.net, 2021).
Shaw, P., Uszkoreit, J. & Vaswani, A. Self-attention with relative position representations. In Proc. NAACL-HLT 464–468 (Association for Computational Linguistics, 2018).
Raffel, C. et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 21, 1–67 (2020).
Ke, G., He, D. & Liu, T.-Y. Rethinking positional encoding in language pre-training. In 9th International Conference on Learning Representations (OpenReview.net, 2021).
Kim, S. et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109 (2018).
Irwin, J. J. & Shoichet, B. K. ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 (2005).
Schwaller, P. et al. Molecular transformer: a model for uncertainty-calibrated chemical reaction prediction. ACS Cent. Sci. 5, 1572–1583 (2019).
Beltagy, I., Peters, M. E. & Cohan, A. Longformer: the long-document transformer. Preprint at https://arxiv.org/abs/2004.05150 (2020).
Kitaev, N., Kaiser, L. & Levskaya, A. Reformer: the efficient transformer. In 8th International Conference on Learning Representations (OpenReview.net, 2020).
Wang, S., Li, B. Z., Khabsa, M., Fang, H. & Ma, H. Linformer: self-attention with linear complexity. Preprint at https://arxiv.org/abs/2006.04768 (2020).
You, Y. et al. Large batch optimization for deep learning: training BERT in 76 minutes. In 8th International Conference on Learning Representations (OpenReview.net, 2020).
Lu, C. et al. Molecular property prediction: a multilevel quantum interactions modeling perspective. Proc. AAAI 33, 1052–1060 (2019).
Yang, K. et al. Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 59, 3370–3388 (2019).
Acknowledgement
We thank IBM Research for supporting this work.
Author information
Authors and Affiliations
Contributions
All authors conceived the project, developed the MoLFormer framework and designed experiments. J.R., B.B., V.C. and I.P. performed model training, fine-tuning and inference experiments. I.P. and P.D. performed attention map analyses. All authors analysed the results and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ross, J., Belgodere, B., Chenthamarakshan, V. et al. Large-scale chemical language representations capture molecular structure and properties. Nat Mach Intell 4, 1256–1264 (2022). https://doi.org/10.1038/s42256-022-00580-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-022-00580-7
This article is cited by
-
Reinvent 4: Modern AI–driven generative molecule design
Journal of Cheminformatics (2024)
-
Bidirectional generation of structure and properties through a single molecular foundation model
Nature Communications (2024)
-
Extrapolative prediction of small-data molecular property using quantum mechanics-assisted machine learning
npj Computational Materials (2024)
-
Probabilistic generative transformer language models for generative design of molecules
Journal of Cheminformatics (2023)
-
Neural scaling of deep chemical models
Nature Machine Intelligence (2023)