Abstract
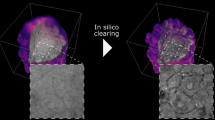
Intensity diffraction tomography (IDT) refers to a class of optical microscopy techniques for imaging the three-dimensional refractive index (RI) distribution of a sample from a set of two-dimensional intensity-only measurements. The reconstruction of artefact-free RI maps is a fundamental challenge in IDT due to the loss of phase information and the missing-cone problem. Neural fields has recently emerged as a new deep learning approach for learning continuous representations of physical fields. The technique uses a coordinate-based neural network to represent the field by mapping the spatial coordinates to the corresponding physical quantities, in our case the complex-valued refractive index values. We present Deep Continuous Artefact-free RI Field (DeCAF) as a neural-fields-based IDT method that can learn a high-quality continuous representation of a RI volume from its intensity-only and limited-angle measurements. The representation in DeCAF is learned directly from the measurements of the test sample by using the IDT forward model without any ground-truth RI maps. We qualitatively and quantitatively evaluate DeCAF on the simulated and experimental biological samples. Our results show that DeCAF can generate high-contrast and artefact-free RI maps and lead to an up to 2.1-fold reduction in the mean squared error over existing methods.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data used for reproducing the results in the manuscript are available at https://github.com/wustl-cig/DeCAF65. We visualized the pre-processed raw intensity images of the relevant samples in Figs. 1 and 4.
Code availability
The code used for reproducing the results in the manuscript is available at https://github.com/wustl-cig/DeCAF65.
References
Kim, K. et al. Three-dimensional label-free imaging and quantification of lipid droplets in live hepatocytes. Sci. Rep. 6, 36815 (2016).
Yamada, K. M. & Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 130, 601–610 (2007).
Kim, G. et al. Measurements of three-dimensional refractive index tomography and membrane deformability of live erythrocytes from pelophylax nigromaculatus. Sci. Rep. 8, 9192 (2018).
Cooper, K. L. et al. Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions. Nature 495, 375–378 (2013).
Park, Y. K., Depeursinge, C. & Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photonics 12, 578–589 (2018).
Jin, D., Zhou, R., Yaqoob, Z. & So, P. Tomographic phase microscopy: principles and applications in bioimaging. J. Opt. Soc. Am. B 34, B64–B77 (2017).
Park, Y. K. et al. Refractive index maps and membrane dynamics of human red blood cells parasitized by plasmodium falciparum. Proc. Natl Acad. Sci. USA 105, 13730–13735 (2008).
Sung, Y. et al. Optical diffraction tomography for high resolution live cell imaging. Opt. Express 17, 266–277 (2009).
Kamilov, U. S. et al. A learning approach to optical tomography. In OSA Frontiers in Optics (Optica Publishing Group, 2015); https://doi.org/10.1364/LS.2015.LW3I.1
Gbur, G. & Wolf, E. Diffraction tomography without phase information. Opt. Lett. 27, 1890–1892 (2002).
Jenkins, M. H. & Gaylord, T. K. Three-dimensional quantitative phase imaging via tomographic deconvolution phase microscopy. Appl. Opt. 54, 9213–9227 (2015).
Tian, L. & Waller, L. 3D Intensity and phase imaging from light field measurements in an LED array microscope. Optica 2, 104–111 (2015).
Chen, M., Tian, L. & Waller, L. 3D Differential phase contrast microscopy. Biomed. Opt. Express 7, 3940–3950 (2016).
Ling, R., Tahir, W., Lin, H.-Y., Lee, H. & Tian, L. High-throughput intensity diffraction tomography with a computational microscope. Biomed. Opt. Express 9, 2130–2141 (2018).
Li, J. et al. Three-dimensional tomographic microscopy technique with multi-frequency combination with partially coherent illuminations. Biomed. Opt. Express 9, 2526–2542 (2018).
Wang, Z. et al. Spatial light interference microscopy (SLIM). Opt. Express 19, 1016–1026 (2011).
Nguyen, T. H., Kandel, M. E., Rubessa, M., Wheeler, M. B. & Popescu, G. Gradient light interference microscopy for 3D imaging of unlabeled specimens. Nat. Commun. 8, 210 (2017).
Chowdhury, S. et al. High-resolution 3D refractive index microscopy of multiple-scattering samples from intensity images. Optica 6, 1211–1219 (2019).
Chen, M., Ren, D., Liu, H. Y., Chowdhury, S. & Waller, L. Multi-layer born multiple-scattering model for 3D phase microscopy. Optica 7, 394–403 (2020).
Li, J. et al. High-speed in vitro intensity diffraction tomography. Adv. Photon. 1, 1–13 (2019).
Kak, A. C. & Slaney, M. Principles of Computerized Tomographic Imaging (IEEE, 1988).
Venkatakrishnan, S. V., Bouman, C. A. & Wohlberg, B. Plug-and-play priors for model based reconstruction. In IEEE Global Conference on Signal and Information Processing (GlobalSIP) 945–948 (2013); https://doi.org/10.1109/GlobalSIP.2013.6737048
Sreehari, S. et al. Plug-and-play priors for bright field electron tomography and sparse interpolation. IEEE Trans. Comput. Imag. 2, 408–423 (2016).
Chan, S. H., Wang, X. & Elgendy, O. A. Plug-and-play ADMM for image restoration: fixed-point convergence and applications. IEEE Trans. Comput. Imag. 3, 84–98 (2017).
Ahmad, R. et al. Plug-and-play methods for magnetic resonance imaging: using denoisers for image recovery. IEEE Signal Process. Mag. 37, 105–116 (2020).
Kang, E., Min, J. & Ye, J. C. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med. Phys. 44, e360–e375 (2017).
Jin, K. H., McCann, M. T., Froustey, E. & Unser, M. Deep convolutional neural network for inverse problems in imaging. IEEE Trans. Image Process. 26, 4509–4522 (2017).
Zhu, B., Liu, J. Z., Cauley, S. F., Rosen, B. R. & Rosen, M. S. Image reconstruction by domain-transform manifold learning. Nature 555, 487–492 (2018).
Aggarwal, H. K., Mani, M. P. & Jacob, M. MoDL: model-based deep learning architecture for inverse problems. IEEE Trans. Med. Imag. 38, 394–405 (2019).
Sun, Y., Xia, Z. & Kamilov, U. S. Efficient and accurate inversion of multiple scattering with deep learning. Opt. Express 26, 14678–14688 (2018).
Li, Y., Xue, Y. & Tian, L. Deep speckle correlation: a deep learning approach toward scalable imaging through scattering media. Optica 5, 1181–1190 (2018).
Zhang, Z. & Lin, Y. Data-driven seismic waveform inversion: A study on the robustness and generalization. IEEE Trans. Geosci. Remote Sens. 58, 6900–6913 (2020).
Wang, G., Ye, J. C. & De Man, B. Deep learning for tomographic image reconstruction. Nat. Mach. Intell. 2, 737–748 (2020).
Liang, D., Cheng, J., Ke, Z. & Ying, L. Deep magnetic resonance image reconstruction: Inverse problems meet neural networks. IEEE Signal Process. Mag. 37, 141–151 (2020).
Adler, A., Araya-Polo, M. & Poggio, T. Deep learning for seismic inverse problems: toward the acceleration of geophysical analysis workflows. IEEE Signal Process. Mag. 38, 89–119 (2021).
Matlock, A. & Tian, L. Physical model simulator-trained neural network for computational 3D phase imaging of multiple-scattering samples. Preprint at https://arxiv.org/abs/2103.15795 (2021).
Sitzmann, V., Zollhöfer, M. & Wetzstein, G. Scene Representation Networks: Continuous 3D-structure-aware neural scene representations. In Advances in Neural Information Processing Systems (NeurIPS) 1121–1132 (2019); https://doi.org/10.5555/3454287.3454388
Sitzmann, V., Martel, J. N. P., Bergman, A. W., Lindell D. B. & Wetzstein, G. Implicit neural representations with periodic activation functions. In Advances in Neural Information Processing Systems (NeurIPS) 7462–7473 (2020); https://doi.org/10.5555/3495724.3496350
Hinton, G. How to represent part-whole hierarchies in a neural network. Preprint at https://arxiv.org/abs/2102.12627 (2021).
Piala, M. & Clarck, R. TermiNeRF: ray termination prediction for efficient neural rendering. Preprint at https://arxiv.org/abs/2111.03643 (2021).
Sun, Y., Wu, Z., Xu, X., Wohlberg, B. & Kamilov, U. S. Scalable plug-and-play ADMM with convergence guarantees. IEEE Trans. Comp. Imag. 7, 849–863 (2021).
Sun, Y., Liu, J., Wohlberg, B. & Kamilov, U. Async-RED: a provably convergent asynchronous block parallel stochastic method using deep denoising priors. In International Conference on Learning Representations (ICLR) (2021).
B., Mildenhall et al. NeRF: Representing scenes as neural radiance fields for view synthesis. In The European Conference on Computer Vision (ECCV) 405–421 (2020); https://doi.org/10.1007/978-3-030-58452-8_24
Martin-Brualla, R. et al. NeRF in the wild: Neural radiance fields for unconstrained photo collections. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 7206–7215 (2021); https://doi.org/10.1109/CVPR46437.2021.00713
Yu, A., Ye, V., Tancik, M., & Kanazawa, A. pixelNeRF: neural radiance fields from one or few images. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2021); https://doi.org/10.1109/CVPR46437.2021.00455
K., Park et al. Nerfies: deformable neural radiance fields. In Proc. IEEE/CVF International Conference on Computer Vision (ICCV) 5845–5854 (2021); https://doi.org/10.1109/ICCV48922.2021.00581
S., Peng et al. Neural body: Implicit neural representations with structured latent codes for novel view synthesis of dynamic humans. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2021); https://doi.org/10.1109/CVPR46437.2021.00894
Li, Z., Niklaus, S., Snavely, N. & Wang, O. Neural scene flow fields for space-time view synthesis of dynamic scenes. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 6494–6504 (2021); https://doi.org/10.1109/CVPR46437.2021.00643
P. P., Srinivasan et al. NeRV: neural reflectance and visibility fields for relighting and view synthesis. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2021); https://doi.org/10.1109/CVPR46437.2021.00741
Wizadwongsa, S., Phongthawee, P., Yenphraphai, J. & Suwajanakorn, S. NeX: real-time view synthesis with neural basis expansion. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2021); https://doi.org/10.1109/CVPR46437.2021.00843
Reed, A. W. et al. Dynamic CT reconstruction from limited views with implicit neural representations and parametric motion fields. In Proc. IEEE/CVF International Conference on Computer Vision (ICCV) 2238–2248 (2021); https://doi.org/10.1109/ICCV48922.2021.00226
Matlock, A. & Tian, L. High-throughput, volumetric quantitative phase imaging with multiplexed intensity diffraction tomography. Biomed. Opt. Express 10, 6432–6448 (2019).
Wu, Z. et al. SIMBA: Scalable inversion in optical tomography using deep denoising priors. IEEE J. Sel. Topics Signal Process. 14, 1163–1175 (2020).
Wiesner, D., Svoboda, D., Maška, M. & Kozubek, M. CytoPacq: a web-interface for simulating multi-dimensional cell imaging. Bioinformatics 35, 4531–4533 (2019).
Lim, J., Ayoub, A. B., Antoine, E. E. & Psaltis, D. High-fidelity optical diffraction tomography of multiple scattering samples. Light Sci. Appl. 8, 82 (2019).
Zhu, J., Wang, H. & Tian, L. High-fidelity intensity diffraction tomography with a non-paraxial multiple-scattering model. Opt. Express 30, 32808–32821 (2022).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Kamilov, U. S. et al. Learning approach to optical tomography. Optica 2, 517–522 (2015).
Müller, T., Evans, A., Schied, C. & Keller, A. Instant neural graphics primitives with a multiresolution hash encoding. ACM Trans. Graph. 41, 102:1–102:15 (2022).
Sun, Y., Liu, J., Xie, M., Wohlberg, B. & Kamilov, U. S. CoIL: coordinate-based internal learning for tomographic imaging. IEEE Trans. Comp. Imag. 7, 1400–1412 (2021).
Tancik, M. et al. Fourier features let networks learn high frequency functions in low dimensional domains. Advances in Neural Information Processing Systems (NeurIPS) 7537–7547 (2020); https://doi.org/10.5555/3495724.3496356
Park, J. J., Florence, P., Straub, J., Newcombe, R. & Lovegrove, S. DeepSDF: learning continuous signed distance functions for shape representation. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 165–174 (2019); https://doi.org/10.1109/CVPR.2019.00025
Kingma, D. & Ba, J. Adam: a method for stochastic optimization. In International Conference on Learning Representations (ICLR) (ICLR, 2015).
Zhang, K., Zuo, W., Chen, Y., Meng, D. & Zhang, L. Beyond a Gaussian denoiser: residual learning of deep CNN for image denoising. IEEE Trans. Image Process. 26, 3142–3155 (2017).
Sun, Y. & Liu, R. wustl-cig/DeCAF (Zenodo, 2022); https://doi.org/10.5281/zenodo.6941764
Acknowledgements
This work was supported by the NSF award nos. CCF-1813910 and CCF-2043134 (to U.K.), and CCF-1813848 and EPMD-1846784 (to L.T.).
Author information
Authors and Affiliations
Contributions
The project was conceived by Y.S., R.L. and U.S.K. The code of the model was implemented by R.L. and Y.S. The experiments were designed by R.L. and Y.S. The numerical results were collected by R.L. Data acquisition and preparation were conducted by J.Z. and L.T. The manuscript was drafted by Y.S. with assistance from R.L. and J.Z. and revised by U.S.K and L.T. All of the authors reviewed manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Jan Funke, Jaejun Yoo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Reconstruction of Diatom Algae acquired by mIDT.
(a) 2D rendering obtained by accumulating all the z slices from DeCAF. Scale bar 10 μm. (b) & (d) Lateral views corresponding to the colored lines in (a). (c) & (e) Axial views at z ∈ {11, 0, 11} μm reconstructed by using DeCAF and Tikhonov, respectively. This figure illustrates the ability of DeCAF to reconstruct high-contrast RI maps for a relatively thin sample acquired by mIDT. Note how DeCAF successfully recovers the folding structure of the sample with two clear separate layers, which are barely recognizable in the Tikhonov reconstruction. Additional examples are shown in Supplementary Videos diatom-midt-decaf.mov and diatom-midt-tikhonov.mov.
Extended Data Fig. 2 Quantitative Illustration of the scalability of DeCAF due to its off-the-grid feature using the C. elegans specimen as an example.
Note how the space required to store the reconstructed sample in DeCAF is independent of the reconstruction grid.
Extended Data Fig. 3 Reconstruction of the 3D Granulocyte Phantom using DeCAF, SIMBA, and Tikhonov.
(a) From left to right, 3D volumes correspond to Groundtruth, DeCAF, SIMBA, and Tikhonov, respectively. (b) Close-up views of the reconstructions at the location shown in (a). Note how DeCAF reconstructs sharper and better quality cell images compared to both SIMBA and Tikhonov.
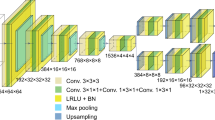
Extended Data Fig. 4 Visual illustration of the network structure and the encoding strategy used in DeCAF.
(a) The overall structure of network Mφ. (b) Illustration of positional encoding for z coordinate. (c) Illustration of radial encoding for the coordinates in the (x, z) plane.
Extended Data Fig. 5 List of algorithmic hyperparameters used by DeCAF for different biological samples.
List of algorithmic hyperparameters used by DeCAF for different biological samples.
Supplementary information
Supplementary Information
Supplementary Discussion, Figs. 1–7 and Tables 1 and 2.
Supplementary Video 1
Spirogyra algae video reconstructed by DeCAF.
Supplementary Video 2
Spirogyra algae video reconstructed by SIMBA.
Supplementary Video 3
Spirogyra algae video reconstructed by Tikhonov.
Supplementary Video 4
Diatom algae video reconstructed by DeCAF under aIDT set-up.
Supplementary Video 5
Diatom algae video reconstructed by Tikhonov under aIDT set-up.
Supplementary Video 6
Diatom algae video reconstructed by DeCAF under mIDT set-up.
Supplementary Video 7
Diatom algae video reconstructed by Tikhonov under mIDT set-up.
Supplementary Video 8
Cell video reconstructed by DeCAF (Fig. 4b).
Supplementary Video 9
Cell video reconstructed by Tikhonov (Fig. 4b).
Supplementary Video 10
Cell video reconstructed by DeCAF (Fig. 4c).
Supplementary Video 11
Cell video reconstructed by Tikhonov (Fig. 4c).
Supplementary Video 12
C. elegans video reconstructed by DeCAF (body).
Supplementary Video 13
C. elegans video reconstructed by Tikhonov (body).
Supplementary Video 14
C. elegans video reconstructed by DeCAF (head).
Supplementary Video 15
C. elegans video reconstructed by Tikhonov (head).
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, R., Sun, Y., Zhu, J. et al. Recovery of continuous 3D refractive index maps from discrete intensity-only measurements using neural fields. Nat Mach Intell 4, 781–791 (2022). https://doi.org/10.1038/s42256-022-00530-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-022-00530-3
This article is cited by
-
Single-pixel p-graded-n junction spectrometers
Nature Communications (2024)
-
Coincidence imaging for Jones matrix with a deep-learning approach
npj Nanophotonics (2024)
-
A Survey of Synthetic Data Augmentation Methods in Machine Vision
Machine Intelligence Research (2024)