Abstract
Randomized controlled trials (RCTs) are considered the gold standard for testing causal hypotheses in the clinical domain; however, the investigation of prognostic variables of patient outcome in a hypothesized cause–effect route is not feasible using standard statistical methods. Here we propose a new automated causal inference method (AutoCI) built on the invariant causal prediction (ICP) framework for the causal reinterpretation of clinical trial data. Compared with existing methods, we show that the proposed AutoCI allows one to clearly determine the causal variables of two real-world RCTs of patients with endometrial cancer with mature outcome and extensive clinicopathological and molecular data. This is achieved via suppressing the causal probability of non-causal variables by a wide margin. In ablation studies, we further demonstrate that the assignment of causal probabilities by AutoCI remains consistent in the presence of confounders. In conclusion, these results confirm the robustness and feasibility of AutoCI for future applications in real-world clinical analysis.
Similar content being viewed by others
Main
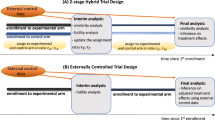
Many clinical studies are driven by the research questions that are statistical at first glance but causal by nature1. For instance2, how safe and efficient is a vaccine against viral infection? How are clinicopathological variables3 related to cancer patient survival? From the causal perspective, the common ground of these problems starts with determining the causal variables of the outcome of interest. In clinical medicine, randomized controlled trials (RCTs) are considered to be the gold standard to investigate cause–effect relationships4. In a prototypical RCT, a participant is randomly assigned to the experimental or control arm and the outcome of interest is observed. In the context of causal inference, such a randomization can be modelled with do-intervention5.
In this Article, we develop a new automated causal inference method (AutoCI) and apply this to two large-scale, practice-changing RCTs of patients with endometrial carcinoma conducted in the Netherlands from 1990–1997 (PORTEC 1; refs. 6,7) and 2002–2006 (PORTEC 2; refs. 8,9), with full clinicopathological datasets and mature outcome data. Endometrial carcinoma is the most common type of gynaecological cancer for women in developed countries10. The majority of women that are diagnosed with early stage endometrial cancer (EC) have a favourable prognosis and are treated with surgery11. Approximately 15–20% of patients have an unfavourable prognosis with a high risk of distant metastasis11. For those patients, different adjuvant therapies such as vaginal brachytherapy, external beam radiotherapy and chemoradiation are recommended on the basis of their risk group12. The two trials (PORTEC 1 and 2) used in this study made a key contribution to clinical practice by investigating how these therapies impact the risk of recurrence rates and survival6,8. According to the latest ESGO/ESTRO/ESP guidelines12 for the management of patients with endometrial carcinoma, the risk classification is based on a series of clinical and pathological variables such as tumour grading (Grade), lymphovascular space invasion (LVSI), myometrial invasion and so on, as well as molecular variables including—but not limited to—polymerase epsilon mutant EC (POLEmut), mismatch repair deficient (MMRd) EC, p53 abnormal EC (p53abn) and EC with no specific molecular profile (NSMP)12. Correlative statistical methods were used in a recent study3 to investigate the hazardous relevance of these variables to EC recurrence. There is strong evidence to suggest that these variables impact EC recurrence, but a systematic investigation to support this understanding from a modern causal inference perspective has not been performed.
Causal inference addresses the determination of cause–effect relationships from data13,14,15,16. When given either observational data or the inclusion of additional interventional data, clinical studies aim to either (1) quantify the causal effect of a treatment given the outcome13 or (2) infer the underlying causal structure of relationships between patient and treatment characteristics and relevant outcomes5,17.
The former can be well formulated as the difference between the outcome expectations conditioned on different treatments (average causal effect)13. A wide range of studies have built on this methodology, including—but not limited to—target trial specification18, target trial emulation19,20 and extending inferences from randomized trials to new target populations21.
In comparison to the causal effect identification, we refer to (2) as (causal) structure identification5. The goal of structure identification is often to learn the entire causal structure, that is, a directed acyclic graph composed of nodes and edges that connect nodes; however, this is generally a non-deterministic polynomial-time hard problem16,22. To learn the cause–effect relations without inferring the entire causal structure, invariant causal prediction (ICP)23 was proposed to determine the set of causal variables given an outcome variable. Under the ICP framework, we use the random variable concept (informally, a function that assigns a set of possible samples to a measurable quantity) and define causal variable as follows (see Supplementary Table 1 for summarized terminologies used in the paper).
Definition 1
Assume there exists a set of environments u ∈ U, given a collection of random variables X = (X1, X2, …, Xn) and an outcome variable Y, if \({{{{\bf{X}}}}}_{{S}^{* }}=({X}_{{S}_{1}^{* }},\ldots ,{X}_{{S}_{j}^{* }})\) exists with indices S* ⊆ {1, …, n} such that
Then \({{{{\bf{X}}}}}_{{S}^{* }}\) are the plausible causal variables (under U). Here, \({{{{\bf{X}}}}}_{{S}^{* }}^{u}=({X}_{{S}_{1}^{* }}^{u},\ldots ,{X}_{{S}_{j}^{* }}^{u})\) are the corresponding random variables to \({{{{\bf{X}}}}}_{{S}^{* }}=({X}_{{S}_{1}^{* }},\ldots ,{X}_{{S}_{j}^{* }})\) created under the environment u. For example, the (experimental) environment u can arise via do-intervention17 (do(X0 ≔ c)) on X0, then \({X}_{0}^{u}\) only samples the fixed value c. It is worth noting that the cause–effect relation \(f^{*}\) in equation (1) stays invariant and independent of U. Next we introduce the definition of identifiable causal variables.
Definition 2
Following the specification of U, X = (X1, X2, …, Xn) and Y in Definition 1, if a \({{{{\bf{X}}}}}_{\bar{S}}=({X}_{{\bar{S}}_{1}},\ldots ,{X}_{{\bar{S}}_{j}})\) exists with indices \(\bar{S}\subseteq \{1,\ldots ,n\}\) such that
then \({{{{\bf{X}}}}}_{\bar{S}}\) are the identifiable causal variables (under U), as they are referred to henceforth.
Vanilla ICP23 was initially presented and verified on linear cause–effect relations. Heinze-Deml and co-workers24 next defined u as a random variable that is neither the descendant nor the parent of Y, and conducted multiple conditional independence tests on nonlinear cause–effect settings (NICP). Gamella and Heinze-Deml25 recently suggested investigating the stable set of variables instead, which is a relaxation of the set of identifiable causal variables (AICP). This progress in ICP has opened unprecedented paths to interpret complex datasets, especially those collected from RCTs. Finding which variables determine whether a treatment works or whether a patient will have a recurrence using ICP methods has great scientific potential. In application to the clinical domain, data interpretation by causal inference methods could improve our understanding of disease and aid in the design of new experiments and clinical trials; however, non-negligible efforts are required to adopt the existing ICPs to the clinical domain. This is due to: (1) the complexity and multitude of variables that are considered relevant for treatment outcomes including patient level characteristics (patient demographics, text data from clinical records), information derived from images (radiology, pathology) and molecular data (genomic sequencing); and (2) the incompatibility between the error-tolerant implementation for the simulated dataset and safe-critical application relevant for medical decisions. Candidate ICP methods therefore need to be robust against noise and need to provide meaningful outputs that can be related to clinical risk in order to inform patient stratification.
Results
Clinical variables overview
The PORTEC 1 and 2 trials6,8 recruited 714 (since 1990–1997) and 427 (since 2000–2006) patients with early stage endometrial carcinoma respectively; 305 cases from PORTEC 1 (42.7%) and 335 cases from PORTEC 2 (78.5%) with complete clinicopathological datasets were aligned and used in the experiments. Clinicopathologic characteristics of these subgroups were similar to the original trial populations (less than or equal to 17.3% absolute difference in frequency of any variable). Importantly, there was no substantial difference in the variable of interest (mean and five-year recurrence free survival (RFS)) for causal variable identification between the excluded and included patient datasets, supporting that our analysis is representative of the overall study population (Supplementary Fig. 1 and Supplementary Tables 2 and 3). Considering PORTEC 1 and 2 as the two experimental environments, we aimed to determine the causal pathological, molecular and immune-related variables of EC recurrence status.
Pathological variables
Pathological criteria tumour grading (Grade)26,27, LVSI28 and myometrial invasion26,27 are examined in the study, all of which are important indicators for an elevated risk of EC recurrence (see also ref. 12). All variables were reevaluated on formalin-fixed paraffin-embedded tumour material by specialized gynecopathologists to guarantee variable consistency for the two environments (trials).
Molecular variables
The molecular classification of EC distinguishes four subtypes with validated prognostic impact: (1) ultra-mutated EC with DNA-polymerase epsilon exonuclease domain mutations (POLEmut), with an excellent prognosis; (2) hypermutated EC with MMRd, with an intermediate prognosis; (3) copy-number-high EC with frequent TP53 mutations (p53abn), with an unfavourable prognosis; and (4) copy-number-low EC without an NSMP, with an intermediate prognosis27,29. Pathogenic POLE mutations were detected by next-generation sequencing of POLE hotspot exons30. Mismatch repair deficient (MMRd) EC and p53 status were determined by immunohistochemistry31. Cases with more than one classifying feature were classified according to the dominant molecular feature on the basis of pathogenicity32. Over-expression of L1CAM by tumour cells was assessed by immunohistochemistry using a cut-off of ≥10% for positivity, and is associated with an increased risk of metastasis and death33,34.
Immune variable
(Intraepithelial) CD8+ T-cell infiltration is an independent favourable prognostic indicator in early stage EC3. To quantify CD8+ T-cell infiltration in tissue samples of the PORTEC 1 and 2 trials, we compute CD8+ cell density derived from tissue microarrays by immunohistochemistry and image analysis3,35. Specifically, tissue microarrays capture cancer tissue samples from each patient in a highly standardized manner, allowing for the highly accurate evaluation of tumour and microenvironment-related factors in cancer samples for investigation with clinical outcomes36.
Based on existing domain knowledge and biological understanding3,29,31,37,38,39, we consider the pathological (P), molecular (M) and immune (I) variables to be the proxies of causal variables (Sprox) (see Supplementary Table 3 for more characteristics details).
Sanity-check variables
To investigate the robustness of the causal inference models, we intentionally include a randomized number as the Patient ID and the vital tissue area of each tissue microarray core (where the tissue area is the sum of randomly sampled tumour and stroma areas from each case) in our subsequent analysis as non-causal variables. Based on prior domain knowledge, the two variables should not be causally related to cancer outcomes. The inclusion of these two non-causal variables thus serves as an important benchmark for comparison of the methods presented in this study. Importantly, as the two variables are expected not to impact clinical outcome and we attempt to verify that they do not impact the outcome, this strategy can also be considered as a typical use case of negative control40 in RCT studies.
PORTEC experiments overview
As presented in Fig. 1 (top), our proposed AutoCI is composed of two key components: (1) a program synthesis language that searches the type-safe function candidates automatically, and (2) a novel causal differentiable learning scheme that determines the causal variables. In the following we first present the experimental results with emphasis on the individual components (1) and (2). We then demonstrate the overall performance on the main and ablation studies for the complete AutoCI method, confirming that the integration of program synthesis with a causal differentiable learning scheme is a critical step towards automated causal inference for clinical applications.
Top: an illustrative scheme of the proposed AutoCI. In the syntax (top left), the type T includes atomic type (ATM), function type (FUC) and abstract data type (ADT), the program prg contains neural network (NN), function composition (COMP), concatenation (CAT), filter (FILTER), predicate (PRED) and so on. In the causal differentiable learning, causal prob. indicates causal probability. The outcome variable RFS means recurrence free survival. Bottom left: the sampled numbers of type-safe functions versus generic functions. Here the size is the maximum amount of nn and PRED functions allowed during the program synthesis. Bottom middle: the learning curve of the JS for top-four type-safe functions achieved in the case with pathological, molecular and immune variables. Bottom right: the running time of determining the causal variables for P, PM and PMI. Here the proposed AutoCI utilizes the function COMP(NN, CAT(FILTER(PRED))).
Automated type-safe function search
Type-safe functions are favourable candidates to strengthen the security and reliability of critical software applications41. However, the manual verification of type-safe properties can be cumbersome, especially when examining thousands of function candidates. Figure 1 (bottom left) shows that the AutoCI can efficiently filter out a subset of promising type-safe differentiable functions (see also similar results in ref. 42), and this can be accomplished in a short period of time: 42.26 s, 118.56 s, 119.44 s for size = {3, 4, 5}. By automatically excluding a large amount of generic functions that are not type-safe, it can greatly improve the development efficiency and algorithm safety compared with manual function design. After obtaining the type-safe candidates, we execute the causal differentiable learning scheme (Fig. 1, top) on the candidates and determine the set of predicted causal variables SPRED. Here we utilize the Jaccard similarity (JS)25 as a key metric to measure the prediction accuracy,
Figure 1 (bottom middle) demonstrates the JS accuracy with the growing epochs for the top-four type-safe candidates (see also Supplementary Table 4). We conclude that the function f = COMP(NN, CAT(FILTER(PRED)))))) achieves the optimal JS score (for example, 91.9 ± 0.06%) and thus is used as the default function for further analysis. As pointed out in Valkov et al.42, HOUDINI allows us to transfer high-level modules across learning tasks. More specifically, the type-safe candidates discovered via the search algorithm are agnostic of disease-specific features for hazard analysis. Independent of the hazard analysis conducted for cancer studies, these type-safe candidates can therefore be re-used and fine-tuned to perform causal variable identification given data on the survival outcome for each patient. Depending on the JS score achieved by the candidates we determine the optimal learned type-safe model. As a result, the proposed AutoCI approach can pave the way towards an efficient causal analysis for many of the real-world cancer studies.
Determining causal variables with clear differentiation
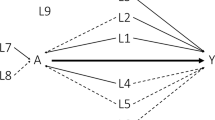
We compare the AutoCI with the state-of-the-art ICP methods, ICP and NICP. Although competitive results are achieved by AICP on the toy experiments, AICP requires the regeneration of additional interventional data in each learning step. Such a learning scheme is incompatible to the real-world RCTs setting, hence AICP is not applicable to the PORTEC experiments. As ICP and NICP explicitly accept or reject the variable of interest, we report yes or no in Table 1. For the proposed AutoCI, we report the mean of the causal probabilities for each variable. Overall, the proposed AutoCI outperforms the ICP and NICP in terms of differentiating causal and non-causal variables (≥50% versus <25% for PMI), demonstrating its advantages over the SOTA methods by a clear margin (Table 1). When examining the individual variables of interest, we can see that ICP fails to determine the proxy variables to be the causal ones, whereas for NICP all of the variables—including the sanity check ones—are considered to be causal. These results clearly contrast to the methodological comparisons of the ICP methods on the toy data (Fig. 2), which respect the normal distributions. If we decompose the proposed causal learning scheme, we witness the suppression of the causal probability on the sanity-check variables over the warm-up stage (steps 1 and 2 of the pseudo code in Fig. 1), whereas the causal variables do not show deterioration of performance. This clear differentiation of causal and non-causal variables can aid the definition of meaningful cut-offs by AutoCI on a given cohort guided by clinical expertise.
Left: the learning curve of the JS for top-four type-safe functions (finite sample setting). Middle: the learning curve of the JS for top-four type-safe functions (ABCD setting). Right: the running time of the compared methods for the finite sample and ABCD settings. Here the proposed AutoCI utilizes the function COMP(NN, CAT(FILTER(PRED))).
Hazard analysis of the individual variables
In parallel to the causal variable determination, the corresponding HR analysis on EC recurrence is also performed. In the scenario in which unknown spurious (non-causal) variables are included in the hazard analysis, the causal cut-off can help reducing the noise introduced by non-causal variables. For instance, Table 2 reports the decreased hazard of tissue area (0.90; 0.88–0.92), Patient ID (0.92; 0.91–0.94) achieved in the warm-up stage for PMI case. Without causal analysis, one may falsely conclude that larger tissue area leads to a slightly lower risk of cancer recurrence. Besides, the HR achieved within the warm-up and complete learning stage remains stable and consistent to the standard clinical interpretation, that is, the values assigned to the variable indeed correctly correspond to either poor or favourable outcomes. This learning scheme can therefore help delivering reliable outputs that are understandable for clinical experts.
Ablation study with hidden confounders
To elaborate the robustness of AutoCI, we conducted ablation studies with the influence of confounding. This is achieved by step-wise inclusion of P and PM. As shown in Tables 1 and 2, the proposed AutoCI presents consistent advantages over the existing ICP methods in terms of learning meaningful causal probabilities for both non-causal and causal variables. For instance, the tissue area and Patient ID are determined to be 35.96% and 20.61% for P, and 24.94% and 17.48% for PM, respectively, whereas the causal probabilities of all of the proxy variables remain close to or above 50%. In the hazard analysis, the results of the confounding studies P and PM are also consistent with the results reported for the main study (PMI). For instance, L1CAM and p53abn are assigned with increased hazards for PM and PMI, indicating a poor prognostic outcome. These results are in agreement with clinical understanding27,29,33. The numerical improvements from P to PMI in the accuracy of probabilistic predictions (Fig. 3) provide complementary evidence confirming the robustness of AutoCI. When compared with the warm-up stage of AutoCI, we further observe a reduction in variance in the evaluation of the complete causal-aware AutoCI. Finally, the running time of the P, PM and PMI studies grows only mildly for the proposed method, in contrast to the dramatic increase in time complexity for ICP and NICP, where the bottleneck of the ICPs lies in the exponential increase o(2∣S∣) in search space with the growing number of variables S (equation (3)).
Discussion
In this study, we proposed a novel automated causal inference algorithm (AutoCI). Taking two large RCTs as the experimental environments, we reinterpret the clinical variables of interest from a causal perspective. The proposed AutoCI demonstrates consistent advantages compared with existing ICP methods in determining causal variables with the presence of hidden confounders. Complementary to the standard hazard analysis, AutoCI provides an automatic tool for medical data analysis to investigate causal association of patient variables from a new perspective, offering informative and critical evidence to support clinical interpretation. Specifically, the accurate determination and exclusion of the spurious (non-causal) variables is a key step to enable more precise patient stratification in the future.
Design choices of AutoCI
Dissimilar to generic algorithms, clinical algorithms must deal with unique challenges in terms of ensuring the safety43 and robustness. Error-prone algorithms can potentially lead to critical errors in medical care. Driven by the need to develop safe-critical applications, AutoCI is carried out with a type-safe program synthesis method42. By further incorporating a newly proposed causal-aware module into this framework, we are able to synthesize a subset of differentiable type-safe candidates well suited for causal-aware learning. Compared to the laborious and error-prone manual function design, this implementation improves the efficiency and safety of AutoCI. Moreover, to achieve the robustness on the real clinical tasks, we introduce a novel causal differentiable learning scheme that utilizes the Fréchet inception distance (FID)44. As a whole, the proposed AutoCI is the seamless integration of both components.
Comparison with existing ICPs
Application of the prior ICP methods has confirmed the feasibility of causal variable learning on toy experiments (Table 3 and Fig. 2). This is substantiated by the outstanding results in the absence of confounders; however, the error-tolerant implementations of prior ICPs on the synthesized experiments are not well-tailored for real clinical applications, especially in the presence of hidden confounders. Compared with ICP, AICP and NICP, AutoCI presents robust results on both toy and PORTEC experiments. With the inclusion of confounders, AutoCI demonstrated a robust differentiation between causal and non-causal variables for PORTEC, and achieves superior quantitative scores on both the finite sample and ABCD settings.
Clinical interpretations
Importantly, the hazard analysis and ranking of clinicopathological and molecular variables using AutoCI (Table 1) is generally consistent with the common biological and clinical interpretation. Taking pathological variables as an example, studies26,27,28 indeed show that grade, deep myometrial invasion and LVSI are important independent predictors of early EC recurrence. The causal probabilities provided by AutoCI thereby give additional information on the relevance of each variable for the determination of outcome, and the likelihood of each variable is consistent with domain expertise. Lymphovascular space invasion is considered to be a critical predictor independent of molecular subgroup and is ranked with the highest causal probability, while grade and myometrial invasion are indeed weaker but independent prognostic indicators.
Furthermore, AutoCI correctly identifies the prognostic associations of the molecular variables of EC, assigns the appropriate hazards for outcome and ranks the molecular subgroups in the order of causal probability that would be expected by an expert’s domain knowledge. Specifically, the molecular factors with the highest adverse risk, p53 abnormality (3.01 (PMI), 3.22 (PM)) and L1CAM over-expression (2.16 (PMI), 2.33 (PM))33 are recognized as such, whereas the POLEmut is consistently associated with a reduced risk of disease relapse as confirmed in previous studies30. Adding an immune variable further refines the model, as expected by domain expertise3, and highlights a causal relationship between cytotoxic T-cell responses and EC recurrence in early stage EC. In summary, AutoCI correctly quantifies and ranks causal pathological, molecular and immune variables for patient outcomes in the clinical trial setting.
Going beyond academic toy models, the proposed AutoCI extends the researchers current statistical toolbox with a new causal-driven method that can assign the causal likelihood to prognostic and predictive variables. As such, this method will enable identification of clinically relevant variables among the ever-increasing number of biomarkers in cancer research that show statistical correlation with clinical outcome. Hence, although the direct real-world application of this method is primarily scientific, subsequent clinical validation and development may enable better selection of (bio)markers to stratify patients for cancer treatments and prediction of prognosis.
Limitation
Despite the clear cut-off between non-causal and proxy variables provided by AutoCI, some of the proxy variables present borderline hazard ratios, for instance, MMRd. Due to small effect size, clinical studies45,46 usually associate MMRd with intermediate patient prognosis, not much different from the prognosis of NSMP EC; however, from the biological perspective, MMRd is highly relevant for a well-defined cascade of molecular changes in cancer cells with favourable prognostic impact as proven by a large number of well-designed experimental and translational studies31,47. The loss of DNA mismatch repair capability in cancer cells leads to a strong increase in tumour mutational burden caused by mismatch, frameshift and insertion/deletion mutations. Due to the structure of eucaryotic DNA, frameshift mutations frequently lead to the translation of truncated peptides that are highly immunogenic, and contribute to the induction of an effective anti-tumoral immune response47. Consistent with biological understanding, AutoCI identifies MMRd as causally related to outcome in the present study, although the lack of a strong association with EC recurrence requires further investigation.
Conclusion
In this study we investigate the causal variable determination among multiple RCTs and present its advantages over the compared methods on both toy and PORTEC experiments. For clinical application, further validation of this methodology in independent clinical trial datasets will be needed to ensure generalisation.
Methods
As per the AutoCI abbreviation, automated and causal are the two building blocks of the proposed method. Concretely, the automation component is implemented with a type-safe program synthesis language HOUDINI42. We also introduce a novel differentiable causal learning scheme that is built up ICP.
Type-safe program synthesis
HOUDINI42 is a typed language with a rich set of pythonic higher-order functions such as MAP, FOLD, COMP (Pythonic: map(), reduce(), lambda x: f(g(x))) and so on (Fig. 1, top left). Relying on the built-in method for program search, it allows us to efficiently search promising type-safe differentiable program candidates. Compared with other program synthesis languages48,49,50,51, HOUDINI rules out the error-prone functions that undermine the software safety and presents itself as an ideal candidate for our task (see Supplementary Table 5 for further comparison).
Despite of rich built-in functions provided by the HOUDINI, it lacks an explicit flow control mechanism. Driven by the need of integrating the causal-aware learning, we introduce the predicate module (PRED) containing the function (cau) (see also Fig. 1, top left)
where θ represents the learnable weights (normalized by sigmoid), ⊙ is the element-wise multiplication, mask is the vector containing 0 or 1 manipulated in step 5 of Fig. 1 (top), mask ⊙ sigmoid(θ) presents the causal probability for each variable. Together with the newly introduced higher-order function FILTER, we are able to synthesize type-safe causal-aware programs.
Causal differentiable learning
In Definition 1, the ICP does not make assumptions about the function \(f^{*}\) (equation (1)). In real-world applications, it is reasonable to specify the search space of \(f^{*}\). If we assume that \(f^{*}\) is differentiable, then we have a trivial extension:
where f remains differentiable with regards to all of the variables X. More importantly, the gradient norms with regards to the non (plausible) causal variables \({{{{\bf{X}}}}}_{{S}^{* c}}\) should vanish, that is, \(\parallel {\nabla }_{{S}^{* c}}f\parallel =0\). Motivated by the extension, we first assume \(f^{*}\) to be differentiable in Definition 1, then we have the claim:
Claim 1
Following the specification of U, X = (X1, X2, …, Xn) and Y in Definition 1, if \({{{{\bf{X}}}}}_{\hat{S}}=({X}_{{\hat{S}}_{1}},\ldots ,{X}_{{\hat{S}}_{j}})\) with indices \(\hat{S}\subseteq \{1,\ldots ,n\}\) are the identifiable causal variables, then there exists a differentiable function \(f({{{\bf{X}}}}):{{\mathbb{R}}}^{n}\mapsto {\mathbb{R}}\) satisfying equations (1) and (2) such that f has the maximum amount \(| {\hat{S}}^{c}|\) of variables with \(\parallel {\nabla }_{{\hat{S}}^{c}}f\parallel =0\).
From this perspective, we can reduce the ICPs to learning an invariant differentiable function f, where f has the (maximum amount of) vanishing gradient norms on the non-causal variables. Such reduction enables us to smoothly integrate the ICP into the modern differentiable learning framework.
Algorithm design. To impose the vanishing gradient norms, we seek for the mask vector in equation (5) as the solution. Initially, we assign mask = 1 for all the variables. Assume we mask a causal variable Xi by flipping maski = 0, then it should greatly disturb the learning errors across multiple environments. If this is the case, we restore maski = 1 and take Xi as the causal variable. Otherwise we reject the variable Xi and maski remains as 0, which imposes the zero gradient with respect to Xi. To quantify the disturbance when variables of interest are missing, existing ICPs use several statistical tests52,53,54. These tests suffer from capturing the nuance of distributions related to higher-dimensional clinical data. Motivated by the recent success in complex vision data55, we utilize the FID44, which is derived from the Wasserstein distance 56. More specifically, the square root of FID between Gaussian distributions is exactly W2 Wasserstein distance56, that is, satisfying three axioms (identity, symmetry and triangular inequality), whereas the statistical tests used in refs. 23,24,25 are generally not mathematical metrics. As a result, the maximum FID (mFID) of U is proposed to measure the distribution difference,
where \({{{{\mu }}}}_{u},{{{{{\mu }}}}}_{{u}^{c}}\) are the distributions with regards to the data sampled from the environment(s) {u} and {u}c. Supplementary Table 6 presents side by side comparisons between mFID, F-test + t-test23,25 and Levene-test + Wilcoxon-test24, all of which are applied for training the same type-safe function COMP(NN, CAT(FILTER(PRED))) under the proposed causal differentiable learning scheme. As displayed in Supplementary Table 6, we conclude that the proposed mFID outperforms the compared statistical tests with a clear margin. For the pseudo code of proposed algorithm please check the top plot of Fig. 1 (in the ‘causal differential learning’ box).
Proof of concept
For the sake of concept validation, we first conduct experiments on toy datasets. We compare the proposed AutoCI to the SOTA methods ICP, NICP and AICP. Specifically, we follow the two experimental protocols presented in AICP25: finite sample setting and ABCD setting57. The former presents the ideal scenario where the same amount of data (1,000) are sampled from both observational and experimental (interventional) environments, whereas the latter simulates a more realistic case where limited experimental data (10) are collected in conjunction with a large amount of observational data (1,000). The data of both settings are generated from randomly chosen linear structural causal models. In our experiments, 400 structural causal models are tested to guarantee the reliability of our results. For the compared ICP methods, we applied the optimal strategies discussed in the paper and parameters are fine-tuned to the experiments. Specifically, careful parallelization and code optimization is also performed for ICP methods. We use 16 cores of CPU Intel(R) Core(TM) i7-7820X CPU @ 3.60 GHz to train the ICP methods in parallel and the GPU NVIDIA TITAN V (12 GB) to train the AutoCI. For the proposed AutoCI, we use the standard Adam optimization58 at a learning rate of 0.02 throughout the experiments. For the warm-up stage (steps 1 and 2 of Fig. 1) of causal differentiable learning we adopt eight epochs. The batch size is set to be 64 for all the experiments. We calibrate the λ = 5, 1 for toy and PORTEC on a small subset of unused data. For the toy experiment, we apply the MSE loss to supervise the learning process and report the result obtained by training the AutoCI one time, where the non-causal variable is determined to be the one with 0 causal probability (equation (5)). For the PORTEC experiment, we utilize the partial likelihood to learn the hazard coefficient59. To fully utilize the PORTEC patient data and incorporate into the differentiable cox model59, the molecular subtype variables POLEmut, MMRd and p53abn are assigned with 1 if present else (including NSMP) 0. To guarantee the representativeness of the PORTEC results, we independently train the AutoCI 64 times and average the causal probability for each variable. Complementary to JS score, we also report FWER = P(SPRED ⊈ Sprox) (type-I error).
As shown in Table 3, our AutoCI achieved competitive JS and FWER scores compared to the ICP methods. Clearly, the proposed method is more resistant to the influence of hidden confounders and all the results reach >90% JS accuracy. This is achieved by the optimal type-safe function COMP(NN, CAT(FILTER(PRED))) (Fig. 2, left and middle; Supplementary Tables 7 and 8). Such advantages also confirm the effectiveness of the proposed causal learning scheme with the utilization of mFID metric. Similar to the PORTEC experiments, due to the exhaustive subset research required in equation (3), the time complexity of ICP and NICP raises dramatically from ABCD to finite sample settings (Fig. 2, right).
Ethics statement
The PORTEC study protocols were approved by the Dutch Cancer Society and by the medical ethics committees at participating centers. Both studies were conducted in accordance with the principles of the Declaration of Helsinki. All patients provided informed consent for study participation. The PORTEC 1 trial was registered at the Daniel Den Hoed Cancer Center (DDHCC) Trial Office. The PORTEC 2 trial was registered at ClinicalTrials.gov under the identifier NCT00376844.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The PORTEC dataset analysed in this study is not publicly available due to restrictions by privacy laws. The dataset and tumour material are currently available to the members of the international TransPORTEC consortium, which is open to requests for sharing of the data and materials after receipt and evaluation of a scientific proposal. Requests should be addressed to the corresponding authors. Please contact N.H. at n.horeweg@lumc.nl for more details. Depending on the specific research proposal, the TransPORTEC consortium will determine when, for how long, for which specific purposes, and under which conditions the requested data can be made available, subject to ethical consent.
Code availability
The code used to generate the data of the toy experiments is available at https://github.com/juangamella/aicp. Our code is implemented with PyTorch and publicly accessible at https://github.com/CTPLab/AutoCI, which is released under the MIT licence.
References
Pearl, J. Causal inference in the health sciences: a conceptual introduction. Health Serv. Outcomes Res. Methodol. 2, 189–220 (2001).
Voysey, M. et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397, 99–111 (2021).
Horeweg, N. et al. Prognostic integrated image-based immune and molecular profiling in early-stage endometrial cancer. Cancer Immunol. Res. 8, 1508–1519 (2020).
Hariton, E. & Locascio, J. J. Randomised controlled trials—the gold standard for effectiveness research. BJOG 125, 1716 (2018).
Peters, J., Janzing, D. & Schölkopf, B. Elements of Causal Inference (MIT Press, 2017).
Creutzberg, C. L. et al. Surgery and postoperative radiotherapy versus surgery alone for patients with stage-1 endometrial carcinoma: multicentre randomised trial. Lancet 355, 1404–1411 (2000).
Creutzberg, C. L. et al. Fifteen-year radiotherapy outcomes of the randomized PORTEC-1 trial for endometrial carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 81, e631–e638 (2011).
Nout, R. A. et al. Vaginal brachytherapy versus pelvic external beam radiotherapy for patients with endometrial cancer of high-intermediate risk (PORTEC-2): an open-label, non-inferiority, randomised trial. Lancet 375, 816–823 (2010).
Wortman, B. et al. Ten-year results of the PORTEC-2 trial for high-intermediate risk endometrial carcinoma: improving patient selection for adjuvant therapy. Br. J. Cancer 119, 1067–1074 (2018).
Sung, H. et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
van den Heerik, A. S. V., Horeweg, N., de Boer, S. M., Bosse, T. & Creutzberg, C. L. Adjuvant therapy for endometrial cancer in the era of molecular classification: radiotherapy, chemoradiation and novel targets for therapy. Int. J. Gynecol. Cancer 31, 594–604 (2021).
Concin, N. et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 31, 12–39 (2021).
Hernán, M. A. & Robins, J. M. Causal Inference: What If (CRC, 2020).
Zenil, H., Kiani, N. A., Zea, A. A. & Tegnér, J. Causal deconvolution by algorithmic generative models. Nat. Mach. Intell. 1, 58–66 (2019).
Prosperi, M. et al. Causal inference and counterfactual prediction in machine learning for actionable healthcare. Nat. Mach. Intell. 2, 369–375 (2020).
Luo, Y., Peng, J. & Ma, J. When causal inference meets deep learning. Nat. Mach. Intell. 2, 426–427 (2020).
Pearl, J. et al. Causal inference in statistics: an overview. Stat. Surv. 3, 96–146 (2009).
Hernán, M. A., Sauer, B. C., Hernández-Díaz, S., Platt, R. & Shrier, I. Specifying a target trial prevents immortal time bias and other self-inflicted injuries in observational analyses. J. Clin. Epidemiol. 79, 70–75 (2016).
Dickerman, B. A., García-Albéniz, X., Logan, R. W., Denaxas, S. & Hernán, M. A. Avoidable flaws in observational analyses: an application to statins and cancer. Nat. Med. 25, 1601–1606 (2019).
Caniglia, E. C. et al. Emulating a target trial of antiretroviral therapy regimens started before conception and risk of adverse birth outcomes. AIDS 32, 113 (2018).
Dahabreh, I. J., Robertson, S. E., Steingrimsson, J. A., Stuart, E. A. & Hernan, M. A. Extending inferences from a randomized trial to a new target population. Stat. Med. 39, 1999–2014 (2020).
Zhu, S., Ng, I. & Chen, Z. Causal Discovery with Reinforcement Learning (ICLR, 2019).
Peters, J., Bühlmann, P. & Meinshausen, N. Causal inference by using invariant prediction: identification and confidence intervals. J. R. Stat. Soc. B 78, 947–1012 (2016).
Heinze-Deml, C., Peters, J. & Meinshausen, N. Invariant causal prediction for nonlinear models. J. Causal Inference https://doi.org/10.1515/jci-2017-0016 (2018).
Gamella, J. L. & Heinze-Deml, C. Active invariant causal prediction: experiment selection through stability. Adv. Neural Inf. Process. Syst. 33, 15464–15475 (2020).
Scholten, A. N. et al. Postoperative radiotherapy for stage 1 endometrial carcinoma: long-term outcome of the randomized PORTEC trial with central pathology review. Int. J. Radiat. Oncol. Biol. Phys. 63, 834–838 (2005).
Stelloo, E. et al. Improved risk assessment by integrating molecular and clinicopathological factors in early-stage endometrial cancer—combined analysis of the PORTEC cohorts. Clin. Cancer Res. 22, 4215–4224 (2016).
Bosse, T. et al. Substantial lymph-vascular space invasion (LVSI) is a significant risk factor for recurrence in endometrial cancer—a pooled analysis of PORTEC 1 and 2 trials. Eur. J. Cancer 51, 1742–1750 (2015).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Church, D. N. et al. Prognostic significance of pole proofreading mutations in endometrial cancer. J. Natl Cancer Inst. 107, 402 (2015).
Stelloo, E. et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Modern Pathol. 28, 836–844 (2015).
Vermij, L., Smit, V., Nout, R. & Bosse, T. Incorporation of molecular characteristics into endometrial cancer management. Histopathology 76, 52–63 (2020).
Bosse, T. et al. L1 cell adhesion molecule is a strong predictor for distant recurrence and overall survival in early stage endometrial cancer: pooled PORTEC trial results. Eur. J. Cancer 50, 2602–2610 (2014).
Van Gool, I. C. et al. Prognostic significance of L1CAM expression and its association with mutant p53 expression in high-risk endometrial cancer. Modern Pathol. 29, 174–181 (2016).
Koelzer, V. H., Sirinukunwattana, K., Rittscher, J. & Mertz, K. D. Precision immunoprofiling by image analysis and artificial intelligence. Virchows Arch. 474, 511–522 (2019).
Zlobec, I., Koelzer, V. H., Dawson, H., Perren, A. & Lugli, A. Next-generation tissue microarray (NGTMA) increases the quality of biomarker studies: an example using CD3, CD8, and CD45RO in the tumor microenvironment of six different solid tumor types. J. Transl. Med. 11, 1–7 (2013).
Creutzberg, C. L. et al. Nomograms for prediction of outcome with or without adjuvant radiation therapy for patients with endometrial cancer: a pooled analysis of PORTEC-1 and PORTEC-2 trials. Int. J. Radiat. Oncol. Biol. Phys. 91, 530–539 (2015).
Karnezis, A. N. et al. Evaluation of endometrial carcinoma prognostic immunohistochemistry markers in the context of molecular classification. J. Pathol. Clin. Res. 3, 279–293 (2017).
Talhouk, A. et al. Molecular subtype not immune response drives outcomes in endometrial carcinoma. Clin. Cancer Res. 25, 2537–2548 (2019).
Lipsitch, M., Tchetgen, E. T. & Cohen, T. Negative controls: a tool for detecting confounding and bias in observational studies. Epidemiology 21, 383 (2010).
Yang, J. & Hawblitzel, C. Safe to the last instruction: automated verification of a type-safe operating system. In Proc. 31st ACM SIGPLAN Conference on Programming Language Design and Implementation 99–110 (ACM, 2010).
Valkov, L., Chaudhari, D., Srivastava, A., Sutton, C. & Chaudhuri, S. HOUDINI: lifelong learning as program synthesis. In 32nd Conferece on Neural Information Processing Systems 8687–8698 (NeurIPS, 2018).
Allen, B. The role of the FDA in ensuring the safety and efficacy of artificial intelligence software and devices. J. Am. College Radiol. 16, 208–210 (2019).
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B. & Hochreiter, S. Gans trained by a two time-scale update rule converge to a local Nash equilibrium. In 31st Conference on Neural Information Processing Systems 6626–6637 (NeurIPS, 2017).
Smyth, E. C. et al. Mismatch repair deficiency, microsatellite instability, and survival: an exploratory analysis of the medical research council adjuvant gastric infusional chemotherapy (MAGIC) trial. JAMA Oncol. 3, 1197–1203 (2017).
León-Castillo, A. et al. Molecular classification of the PORTEC-3 trial for high-risk endometrial cancer: impact on prognosis and benefit from adjuvant therapy. J. Clin. Oncol. 38, 3388–3397 (2020).
Kloor, M. & von Knebel Doeberitz, M. The immune biology of microsatellite-unstable cancer. Trends Cancer 2, 121–133 (2018).
Gaunt, A. L., Brockschmidt, M., Kushman, N. & Tarlow, D. Differentiable Programs with Neural Libraries 1213–1222 (ICLR, 2017).
Mao, J., Gan, C., Kohli, P., Tenenbaum, J. B. & Wu, J. The Neuro-Symbolic Concept Learner: Interpreting Scenes, Words, and Sentences From Natural Supervision (ICLR, 2018).
Vedantam, R. et al. Probabilistic Neural Symbolic Models for Interpretable Visual Question Answering 6428–6437 (ICLR, 2019).
Ellis, K. et al. Dreamcoder: growing generalizable, interpretable knowledge with wake-sleep Bayesian program learning. Preprint at https://arxiv.org/abs/2006.08381 (2020).
Pfanzagl, J. & Sheynin, O. Studies in the history of probability and statistics XLIV a forerunner of the t-distribution. Biometrika 83, 891–898 (1996).
Levene, H. in Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling 279–292 (Stanford Univ. Press, 1961).
Wilcoxon, F. Individual Comparisons by Ranking Methods: Breakthroughs in Statistics 196–202 (Springer, 1992).
Lucic, M., Kurach, K., Michalski, M., Gelly, S. & Bousquet, O. Are GANs created equal? A large-scale study. Adv. Neural Inf. Process. Syst. 31, 1–10(2018).
Villani, C. Optimal Transport: Old and New Vol. 338 (Springer, 2008).
Agrawal, R., Squires, C., Yang, K., Shanmugam, K. & Uhler, C. ABCD-strategy: budgeted experimental design for targeted causal structure discovery. In 22nd International Conference on Artificial Intelligence and Statistics 3400–3409 (National Science Foundation, 2019).
Kingma, D. P. & Ba, J. Adam: A Method for Stochastic Optimization (ICLR, 2015).
Kvamme, H., Borgan, Ø. & Scheel, I. Time-to-event prediction with neural networks and Cox regression. J. Mach. Learn. Res. 20, 1–30 (2019).
Acknowledgements
We convey our gratitude to all clinicians and technicians that participated in the PORTEC 1 and 2 trials (registration no. ISRCTN16228756), and all scientists, pathologists and patients involved in the data processing and analysis. The PORTEC 1 and 2 trials were supported by the grants from the Dutch Cancer Society (grant nos. CKTO 90–01 and CKTO 2001–04, respectively). Molecular profiling was supported by the grants from the Dutch Cancer Society (grant nos. KWF UL2012-5447 and KWF/YIG 10418, respectively). V.H.K. reports a grant from the Promedica Foundation (grant no. F-87701-41-01) during the conduct of the study. N.H. reports grants from the Dutch Cancer Society (grant nos. KWF-2021-13400, KWF-2021-13404) during the conduct of the study.
Author information
Authors and Affiliations
Contributions
J.Q.W. and V.H.K. conceived the research idea. J.Q.W. implemented the algorithm and ran the experiments. J.Q.W., V.H.K. and N.H. analysed the data and results. J.Q.W. and V.H.K. wrote the manuscript. N.H. and V.H.K. reviewed the manuscript and supervised this study. R.A.N., I.M.J.-S., J.J.J., L.C.H.W.L., E.M.v.d.S.-B., T.B., C.L.C., M.d.B., H.W.N., T.B. and V.T.H.B.M.S. provided the PORTEC 1 and 2 trials data. C.L.C. and V.T.H.B.M.S. conceptualized and designed the PORTEC 1 and 2 study. R.A.N., T.B., C.L.C. and V.T.H.B.M.S. supervised the PORTEC 1 and 2 study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Miquel Porta and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The consort diagram presenting the process of patient selection.
Abbreviations: QC - quality control, IHC - immunohistochemistry, EEC- endometrioid endometrial carcinoma, NEEC- non-endometrioid endometrial carcinoma.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Tables 1–8.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, J.Q., Horeweg, N., de Bruyn, M. et al. Automated causal inference in application to randomized controlled clinical trials. Nat Mach Intell 4, 436–444 (2022). https://doi.org/10.1038/s42256-022-00470-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-022-00470-y
This article is cited by
-
Causal inference in the medical domain: a survey
Applied Intelligence (2024)