Abstract
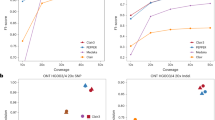
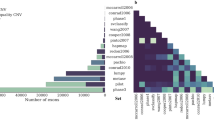
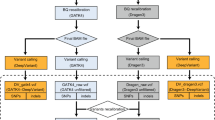
Single-molecule sequencing technologies have emerged in recent years and revolutionized structural variant calling, complex genome assembly and epigenetic mark detection. However, the lack of a highly accurate small variant caller has limited these technologies from being more widely used. Here, we present Clair, the successor to Clairvoyante, a program for fast and accurate germline small variant calling, using single-molecule sequencing data. For Oxford Nanopore Technology data, Clair achieves better precision, recall and speed than several competing programs, including Clairvoyante, Longshot and Medaka. Through studying the missed variants and benchmarking intentionally overfitted models, we found that Clair may be approaching the limit of possible accuracy for germline small variant calling using pileup data and deep neural networks. Clair requires only a conventional central processing unit (CPU) for variant calling and is an open-source project available at https://github.com/HKU-BAL/Clair.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The details of and links to the reference genomes, truth variants, ONT data, PacBio CCS data and Illumina data that support the findings of this study are available in the ‘Data sources’ section of the Supplementary Notes. The variant call format files generated by Clair in this study are available at http://www.bio8.cs.hku.hk/clair_models/VCFBenchmarked/.
Code availability
Clair is open source, and available at https://github.com/HKU-BAL/Clair. Clair is licensed under the BSD 3-Clause licence.
References
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016).
Ashley, E. A. Towards precision medicine. Nat. Rev. Genet. 17, 507–522 (2016).
Li, H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851 (2014).
Luo, R., Schatz, M. C. & Salzberg, S. L. 16GT: a fast and sensitive variant caller using a 16-genotype probabilistic model. GigaScience 6, gix045 (2017).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 43, 11–33 (2013).
Krusche, P. et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat. Biotechnol. 37, 555–560 (2019).
The long view on sequencing. Nat. Biotechnol. 36, 287 (2018).
Sedlazeck, F. J., Lee, H., Darby, C. A. & Schatz, M. C. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 19, 329–346 (2018).
Ameur, A., Kloosterman, W. P. & Hestand, M. S. Single-molecule sequencing: towards clinical applications. Trends Biotechnol. 37, 72–85 (2019).
Luo, R., Sedlazeck, F. J., Lam, T. W. & Schatz, M. C. A multi-task convolutional deep neural network for variant calling in single molecule sequencing. Nat. Commun. 10, 998 (2019).
Zook, J. M. et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. Data 3, 160025 (2016).
Zook, J. M. et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat. Biotechnol. 32, 246–251 (2014).
Zook, J. M. et al. An open resource for accurately benchmarking small variant and reference calls. Nat. Biotechnol. 37, 561–566 (2019).
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018).
Edge, P. & Bansal, V. Longshot enables accurate variant calling in diploid genomes from single-molecule long read sequencing. Nat. Commun. 10, 4660 (2019).
Sequence correction provided by ONT Research https://github.com/nanoporetech/medaka (accessed 17 November 2019).
Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162 (2019).
Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987 (2018).
Simpson, J. T. et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Meth. 14, 407 (2017).
Poplin, R. et al. DeepVariant training data https://github.com/google/deepvariant/blob/r0.9/docs/deepvariant-details-training-data.md (accessed 22 November 2019).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Smith, L. N. in 2017 IEEE Winter Conf. on Applications of Computer Vision (WACV) 464–472 (IEEE, 2017).
Lin, T.-Y., Goyal, P., Girshick, R., He, K. & Dollár, P. in Proc. IEEE Int. Conf. on Computer Vision 2980–2988 (2017).
Cleary, J. G. et al. Joint variant and de novo mutation identification on pedigrees from high-throughput sequencing data. J. Comput. Biol. 21, 405–419 (2014).
Acknowledgements
We thank S. Salzberg, M. Schatz and F. Sedlazeck for comments. R.L. was supported by the ECS (grant number 27204518) of the HKSAR government, and by the URC fund at HKU. T.-W.L., C.-L.W., Y.-S.W., C.-I.T., C.-M. Liu and C.-M. Leung were supported by the ITF (grant number ITF/331/17FP) from the Innovation and Technology Commission, HKSAR government.
Author information
Authors and Affiliations
Contributions
R.L. and T.-W.L. conceived the study. R.L, C.-L.W., Y.-S.W., C.-I.T., C.-M. Liu and C.-M. Leung analysed the data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes, Supplementary Tables 1-6, Supplementary Figure 1
Supplementary Table
The details of the FP and FN results in ONT experiment 1:168x|2:64x
Rights and permissions
About this article
Cite this article
Luo, R., Wong, CL., Wong, YS. et al. Exploring the limit of using a deep neural network on pileup data for germline variant calling. Nat Mach Intell 2, 220–227 (2020). https://doi.org/10.1038/s42256-020-0167-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-020-0167-4
This article is cited by
-
DeepSelectNet: deep neural network based selective sequencing for oxford nanopore sequencing
BMC Bioinformatics (2023)
-
Genomic variant benchmark: if you cannot measure it, you cannot improve it
Genome Biology (2023)
-
Performance analysis of conventional and AI-based variant callers using short and long reads
BMC Bioinformatics (2023)
-
nPoRe: n-polymer realigner for improved pileup-based variant calling
BMC Bioinformatics (2023)
-
A survey of algorithms for the detection of genomic structural variants from long-read sequencing data
Nature Methods (2023)