Abstract
Deep learning-based variant callers are becoming the standard and have achieved superior single nucleotide polymorphisms calling performance using long reads. Here we present Clair3, which leverages two major method categories: pileup calling handles most variant candidates with speed, and full-alignment tackles complicated candidates to maximize precision and recall. Clair3 runs faster than any of the other state-of-the-art variant callers and demonstrates improved performance, especially at lower coverage.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The links to the reference genomes, truth variants, benchmarking materials and ONT data are provided in Supplementary Section 5. The commands and parameters used in this study are available in Supplementary Section 6 and Zenodo16. All analysis output, including the VCFs and running logs, is available at http://www.bio8.cs.hku.hk/clair3/analysis_result. Source data are provided with this paper.
Code availability
Clair3 is open-source software (BSD 3-Clause license), hosted by GitHub at https://github.com/HKU-BAL/Clair ref. 3, and available through Docker, Bioconda and Singularity. Clair3 is also available in Zenodo16.
References
Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987 (2018).
Luo, R., Sedlazeck, F. J., Lam, T.-W. & Schatz, M. C. A multi-task convolutional deep neural network for variant calling in single molecule sequencing. Nat. Commun. 10, 998 (2019).
Luo, R. et al. Exploring the limit of using a deep neural network on pileup data for germline variant calling. Nat. Mach. Intell. 2, 220–227 (2020).
Ahsan, M. U., Liu, Q., Fang, L. & Wang, K. NanoCaller for accurate detection of SNPs and indels in difficult-to-map regions from long-read sequencing by haplotype-aware deep neural networks. Genome Biol. 22, 261 (2021).
Shafin, K. et al. Haplotype-aware variant calling with PEPPER-Margin-DeepVariant enables high accuracy in nanopore long-reads. Nat. Methods 18, 1322–1332 (2021).
Medaka, https://github.com/nanoporetech/medaka (2018).
Patterson, M. et al. WhatsHap: weighted haplotype assembly for future-generation sequencing reads. J. Comput. Biol. 22, 498–509 (2015).
Edge, P. & Bansal, V. Longshot enables accurate variant calling in diploid genomes from single-molecule long read sequencing. Nat. Commun. 10, 4660 (2019).
Olson, N. D. et al. PrecisionFDA Truth Challenge V2: calling variants from short and long reads in difficult-to-map regions. Cell Genomics 2, 100129 (2022).
Wagner, J. et al. Benchmarking challenging small variants with linked and long reads. Cell Genomics 2, 100128 (2022).
Nanopore EPI2ME Labs, https://labs.epi2me.io/gm24385_2021.05/ (2021).
Shafin, K. et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol. 38, 1044–1053 (2020).
Krusche, P. et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat. Biotechnol. 37, 555–560 (2019).
Medaka v1.5.0, https://github.com/nanoporetech/medaka/releases/tag/v1.5.0 (2021).
PEPPER r0.7, https://github.com/kishwarshafin/pepper/releases/tag/r0.7 (2021).
Zheng, Z. et al. Symphonizing pileup and full-alignment for deep learning-based long-read variant calling. Zenodo https://doi.org/10.5281/zenodo.6637001 (2022).
He, K., Zhang, X., Ren, S. & Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 37, 1904–1916 (2015).
Rerio, https://github.com/nanoporetech/rerio (2021).
Liu, L. et al. On the variance of the adaptive learning rate and beyond. Preprint at https://arxiv.org/abs/1908.03265 (2019).
Zhang, M. R., Lucas, J., Hinton, G. & Ba, J. Lookahead optimizer: k steps forward, 1 step back. Preprint at https://arxiv.org/abs/1907.08610 (2019).
Acknowledgements
R.L. was supported by Hong Kong Research Grants Council grants GRF (17113721) and TRS (T21-705/20-N and T12-703/19-R), the Shenzhen Municipal Government General Program (JCYJ20210324134405015), the URC fund at HKU and Oxford Nanopore Technologies.
Author information
Authors and Affiliations
Contributions
R.L. conceived the study. Z.Z. and R.L. designed the algorithms, implemented Clair3, designed the experiments and wrote the paper. J.S. and S.L. developed submodules in Clair3. A.W.-S.L. and T.-W.L. evaluated the benchmarking results. All authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.L. receives research funding from ONT. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Nathan Olson, Guohua Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
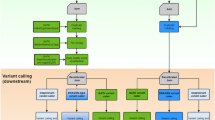
Extended Data Fig. 1 The workflow for Clair3.
The figure shows the workflow of Clair3 on how to make use of both pileup and full-alignment for variants calling, and combine the results. Pileup candidates that are above a coverage threshold and an allele frequency threshold are extracted, and then called using the pileup network. The pileup calls are grouped into variant calls and reference calls. Both groups are ranked according to variant quality (QUAL). High-quality heterozygous SNP calls are used in WhatsHap phasing to produce phased alignment for input to the full-alignment network. Low-quality pileup calls are then called again using the full-alignment network. Finally, the full-alignment calls and high-quality pileup calls are outputted. Clair3 supports both variant call format (VCF) and genomic variant call format (GVCF) output formats.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1–12 and sections 1–6 and references.
Source data
Source Data Fig. 1
Data points source data.
Source Data Fig. 2
Data points source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, Z., Li, S., Su, J. et al. Symphonizing pileup and full-alignment for deep learning-based long-read variant calling. Nat Comput Sci 2, 797–803 (2022). https://doi.org/10.1038/s43588-022-00387-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-022-00387-x
This article is cited by
-
Flexible and cost-effective genomic surveillance of P. falciparum malaria with targeted nanopore sequencing
Nature Communications (2024)
-
De novo diploid genome assembly using long noisy reads
Nature Communications (2024)
-
Combinatorial optimization of gene expression through recombinase-mediated promoter and terminator shuffling in yeast
Nature Communications (2024)
-
Combined approaches, including long-read sequencing, address the diagnostic challenge of HYDIN in primary ciliary dyskinesia
European Journal of Human Genetics (2024)
-
FixItFelix: improving genomic analysis by fixing reference errors
Genome Biology (2023)