Abstract
The retina is highly metabolically active, relying on glucose uptake and aerobic glycolysis. Situated in close contact to photoreceptors, a key function of cells in the retinal pigment epithelium (RPE) is phagocytosis of damaged photoreceptor outer segments (POS). Here we identify RPE as a local source of insulin in the eye that is stimulated by POS phagocytosis. We show that Ins2 messenger RNA and insulin protein are produced by RPE cells and that this production correlates with RPE phagocytosis of POS. Genetic deletion of phagocytic receptors (‘loss of function’) reduces Ins2, whereas increasing the levels of the phagocytic receptor MerTK (‘gain of function’) increases Ins2 production in male mice. Contrary to pancreas-derived systemic insulin, RPE-derived local insulin is stimulated during starvation, which also increases RPE phagocytosis. Global or RPE-specific Ins2 gene deletion decreases retinal glucose uptake in starved male mice, dysregulates retinal physiology, causes defects in phototransduction and exacerbates photoreceptor loss in a mouse model of retinitis pigmentosa. Collectively, these data identify RPE cells as a phagocytosis-induced local source of insulin in the retina, with the potential to influence retinal physiology and disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Untargeted metabolomics data can be found at www.metabolomicsworkbench.org, with project ID PR001193 or visit at https://doi.org/10.21228/M8H419. Untargeted metabolomics data were analyzed using the MetabolAnalyst v.5.0 platform (https://www.metaboanalyst.ca/). Source data are provided with this paper.
References
Swarup, A. et al. Modulating GLUT1 expression in retinal pigment epithelium decreases glucose levels in the retina: impact on photoreceptors and Müller glial cells. Am. J. Physiol. Cell Physiol. 316, C121–C133 (2019).
Kanow, M. A. et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. eLife 6, e28899 (2017).
Winkler, B. S. Glycolytic and oxidative metabolism in relation to retinal function. J. Gen. Physiol. 77, 667–692 (1981).
Punzo, C., Kornacker, K. & Cepko, C. L. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat. Neurosci. 12, 44–52 (2009).
Aït-Ali, N. et al. Rod-derived cone viability factor promotes cone survival by stimulating aerobic glycolysis. Cell 161, 817–832 (2015).
Wang, W. et al. Two-step reactivation of dormant cones in retinitis pigmentosa. Cell Rep. 15, 372–385 (2016).
Young, R. W. & Bok, D. Participation of the retinal pigment epithelium in the rod outer segment renewal process. J. Cell Biol. 42, 392–403 (1969).
Adijanto, J. et al. The retinal pigment epithelium utilizes fatty acids for ketogenesis: implications for metabolic coupling with the outer retina. J. Biol. Chem. 289, 20570–20582 (2014).
Wang, W. et al. Metabolic deregulation of the blood-outer retinal barrier in retinitis pigmentosa. Cell Rep. 28, 1323–1334 (2019).
Lynch, S. K. & Abràmoff, M. D. Diabetic retinopathy is a neurodegenerative disorder. Vis. Res. 139, 101–107 (2017).
Reiter, C. E. N. et al. Characterization of insulin signaling in rat retina in vivo and ex vivo. Am. J. Physiol. Endocrinol. Metab. 285, E763–E774 (2003).
James, C. R. & Cotlier, E. Fate of insulin in the retina: an autoradiographic study. Br. J. Ophthalmol. 67, 80–88 (1983).
Zolov, S. N. et al. Insulin-like growth factor-2 regulates basal retinal insulin receptor activity. J. Biol. Chem. https://doi.org/10.1016/j.jbc.2021.100712 (2021).
Tarchick, M. J. et al. Endogenous insulin signaling in the RPE contributes to the maintenance of rod photoreceptor function in diabetes. Exp. Eye Res. 180, 63–74 (2019).
Rajala, A., Tanito, M., Le, Y. Z., Kahn, C. R. & Rajala, R. V. S. Loss of neuroprotective survival signal in mice lacking insulin receptor gene in rod photoreceptor cells. J. Biol. Chem. 283, 19781–19792 (2008).
Edwards, R. B. & Szamier, R. B. Defective phagocytosis of isolated rod outer segments by RCS rat retinal pigment epithelium in culture. Science 197, 1001–1003 (1977).
Burstyn-Cohen, T. et al. Genetic dissection of TAM receptor-ligand interaction in retinal pigment epithelial cell phagocytosis. Neuron 76, 1123–1132 (2012).
Penberthy, K. K. et al. Context-dependent compensation among phosphatidylserine-recognition receptors. Sci. Rep. 7, 14623 (2017).
Soares, M. B. et al. RNA-mediated gene duplication: the rat preproinsulin I gene is a functional retroposon. Mol. Cell. Biol. 5, 2090–2103 (1985).
Anderson, M. S. et al. Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002).
Iacovelli, J. et al. Generation of Cre transgenic mice with postnatal RPE-specific ocular expression. Invest. Ophthalmol. Vis. Sci. 52, 1378–1383 (2011).
Van Cauter, E., Mestrez, F., Sturis, J. & Polonsky, K. S. Estimation of insulin secretion rates from C-peptide levels. Comparison of individual and standard kinetic parameters for C-peptide clearance. Diabetes 41, 368–377 (1992).
Wang, S. et al. Deciphering primate retinal aging at single-cell resolution. Protein Cell https://doi.org/10.1007/s13238-020-00791-x (2020).
Giddings, S. J., Chirgwin, J. & Permutt, M. A. The effects of fasting and feeding on preproinsulin messenger RNA in rats. J. Clin. Invest. 67, 952–960 (1981).
Boland, B. B. et al. β-cell control of insulin production during starvation-refeeding in male rats. Endocrinology 159, 895–906 (2018).
Unger, R. H., Eisentraut, A. M. & Madison, L. L. The effects of total starvation upon the levels of circulating glucagon and insulin in man. J. Clin. Invest. 42, 1031–1039 (1963).
Giddings, S. J. & Carnaghi, L. R. The two nonallelic rat insulin mRNAs and pre-mRNAs are regulated coordinately in vivo. J. Biol. Chem. 263, 3845–3849 (1988).
Duvillié, B. et al. Phenotypic alterations in insulin-deficient mutant mice. Proc. Natl Acad. Sci. USA 94, 5137–5140 (1997).
Leroux, L. et al. Compensatory responses in mice carrying a null mutation for Ins1 or Ins2. Diabetes 50, S150 (2001).
Law, A.-L. et al. Cleavage of Mer tyrosine kinase (MerTK) from the cell surface contributes to the regulation of retinal phagocytosis. J. Biol. Chem. 290, 4941–4952 (2015).
Rajala, R. V. S. & Anderson, R. E. Light regulation of the insulin receptor in the retina. Mol. Neurobiol. 28, 123–138 (2003).
Sachdeva, M. M. Retinal neurodegeneration in diabetes: an emerging concept in diabetic retinopathy. Curr. Diab. Rep. 21, 65 (2021).
Bok, D. & Hall, M. O. The role of the pigment epithelium in the etiology of inherited retinal dystrophy in the rat. J. Cell Biol. 49, 664–682 (1971).
LaVail, M. M. Rod outer segment disk shedding in rat retina: relationship to cyclic lighting. Science 194, 1071–1074 (1976).
Nandrot, E. F. et al. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking αvβ5 integrin. J. Exp. Med. 200, 1539–1545 (2004).
Lu, Q. et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398, 723–728 (1999).
D’Cruz, P. M. et al. Mutation of the receptor tyrosine kinase gene MerTK in the retinal dystrophic RCS rat. Hum. Mol. Genet. 9, 645–651 (2000).
Ryeom, S. W., Sparrow, J. R. & Silverstein, R. L. CD36 participates in the phagocytosis of rod outer segments by retinal pigment epithelium. J. Cell Sci. 109, 387–395 (1996).
Cai, B. et al. MerTK cleavage limits proresolving mediator biosynthesis and exacerbates tissue inflammation. Proc. Natl Acad. Sci. USA 113, 6526–6531 (2016).
Villa, E. et al. mTORC1 stimulates cell growth through SAM synthesis and m6A mRNA-dependent control of protein synthesis. Mol. Cell https://doi.org/10.1016/j.molcel.2021.03.009 (2021).
Sinha, T., Ikelle, L., Naash, M. I. & Al-Ubaidi, M. R. The intersection of serine metabolism and cellular dysfunction in retinal degeneration. Cells https://doi.org/10.3390/cells9030674 (2020).
Ducker, G. S. & Rabinowitz, J. D. One-carbon metabolism in health and disease. Cell Metab. 25, 27–42 (2017).
Rajala, A., Wang, Y. & Rajala, R. V. S. Activation of oncogenic tyrosine kinase signaling promotes insulin receptor-mediated cone photoreceptor survival. Oncotarget 7, 46924–46942 (2016).
Venkatesh, A. et al. Activated mTORC1 promotes long-term cone survival in retinitis pigmentosa mice. J. Clin. Invest. 125, 1446–1458 (2015).
Akalu, Y. T. et al. Tissue-specific modifier alleles determine MerTK loss-of-function traits. eLife 11, e80530 (2022).
Fan, Y. et al. Thymus-specific deletion of insulin induces autoimmune diabetes. EMBO J. 28, 2812–2824 (2009).
Li, S. et al. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis 41, 73–80 (2005).
Xin-Zhao Wang, C., Zhang, K., Aredo, B., Lu, H. & Ufret-Vincenty, R. L. Novel method for the rapid isolation of RPE cells specifically for RNA extraction and analysis. Exp. Eye Res. 102, 1–9 (2012).
Fernandez-Godino, R., Garland, D. L. & Pierce, E. A. Isolation, culture and characterization of primary mouse RPE cells. Nat. Protoc. 11, 1206–1218 (2016).
Venkatesh, A., Ma, S., Langellotto, F., Gao, G. & Punzo, C. Retinal gene delivery by rAAV and DNA electroporation. Curr. Protoc. Microbiol. https://doi.org/10.1002/9780471729259.mc14d04s28 (2013).
Wei, H., Xun, Z., Granado, H., Wu, A. & Handa, J. T. An easy, rapid method to isolate RPE cell protein from the mouse eye. Exp. Eye Res. 145, 450–455 (2016).
Sheybani, N. D. et al. ImmunoPET-informed sequence for focused ultrasound-targeted mCD47 blockade controls glioma. J. Control. Release 331, 19–29 (2021).
Zhong, M. & Kundu, B. K. Optimization of a model corrected blood input function from dynamic FDG-PET images of small animal heart in vivo. IEEE Trans. Nucl. Sci. 60, 3417–3422 (2013).
Huang, Q., Massey, J. C., Mińczuk, K., Li, J. & Kundu, B. K. Non-invasive determination of blood input function to compute rate of myocardial glucose uptake from dynamic FDG PET images of rat heart in vivo: comparative study between the inferior vena cava and the left ventricular blood pool with spill over and partial volume corrections. Phys. Med. Biol. 64, 165010 (2019).
Tsugawa, H. et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526 (2015).
Tsugawa, H. et al. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 16, 295–298 (2019).
Narendran, S. et al. Nucleoside reverse transcriptase inhibitors and Kamuvudines inhibit myloid-β induced retinal pigmented epithelium degeneration. Signal Transduct. Target Ther. 6, 1–9 (2021).
Acknowledgements
We thank members of the Ravichandran laboratory as well as B. Gelfand and R. Lindsay for input and critical reading of the manuscript. We thank E. Thorpe and I. Tabas for the MerTKCR mice, J. Mandell for human retinal samples and E. Nandrot and R. Rajala for advice on specific protocols. Figure schematics were created using BioRender images as templates. This work was supported by grants to K.S.R. from the National Institute of General Medical Sciences (R35GM122542), National Institute of Allergy and Infectious Diseases (R01AI159551), the National Heart, Lung and Blood Institute (P01HL120840), the Center for Cell Clearance/University of Virginia and Bone and Joint Canada Investigator funds from the Washington University School of Medicine to J.I.E. via NEI F32 (EY031211) and T32AI7496-23. J.A. is a cofounder of DiceRx, iVeena Holdings, iVeena Delivery Systems, and Inflammasome Therapeutics; has been a consultant for Abbvie, Boehringer-Ingelheim, Olix Pharmaceuticals, Retinal Solutions, and Saksin LifeSciences; and is named as an inventors on patent applications filed by his university, all unrelated to this work. J.A. has received support from the UVA Strategic Investment Fund, National Institutes of Health (NIH) grants (R01EY028027, R01EY029799, R01EY031039, R01AG082108), DuPont Guerry, III, Professorship, and a gift from Mr. and Mrs. Eli W. Tullis.
Author information
Authors and Affiliations
Contributions
J.I.E. and K.S.R. designed all experiments and wrote and composed the manuscript. J.I.E. performed most of the experiments. S.K. performed all immunoblots. K.P. performed RNA-seq of MerTK KO mice. L.K. assisted with eye dissections. Y.N. and J.A. conducted ERG experiments. S.G. provided input and generated figures. S.P., V.S. and B.K. carried out PET experiments. M.R., C.L. and E.U. provided conceptual advice and/or helped with specific experiments. A.R.M. helped with the RPE phagocytosis assay. J. Pinney analyzed RNA-seq data and mined public datasets. M.S. assisted with ELISAs. B.B. performed glucose tolerance assays. N.W. performed metabolomics. Y.F. provided Ins2f/f mice. T.B.-C. conducted mouse insulin immunofluorescence assays. J. Perry analyzed metabolomics data and gave conceptual advice.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Christoph Schmitt in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
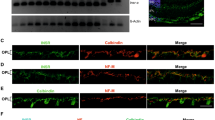
Extended Data Fig. 1 Expression of Ins2 mRNA and Protein Within the RPE Cells of the Eye.
a. RT–PCR of Ins2 from mRNA isolated from different mouse tissues. n = 3 mice used for each tissue. **p < .01, one-way ANOVA on ranks. b. Flow cytometry panels showing YFP+ gating strategy for isolated RPE from Best1-Cre/YFPflox-STOP-flox mice. c. Representative image of two independent experiments showing immunofluorescence analysis at 40x magnification on mouse retina from Rhodopsin-Cre Ins2f/f mice using antibodies against Cre (green) (left). RT–PCR of Ins2 mRNA in RPE of Rhodopsin-Cre Ins2f/f shows no change in the expression of the Ins2 transcript compared to control (right). n = 2 mice used for each genotype. d. Representative image of immunofluorescence analysis at 60x magnification on human retina using antibodies against C-peptide (magenta) with brightfield microscopy. Merge shows that C-peptide, a byproduct of insulin secretion and a metric of insulin release, is present and located in the retina. Plots are presented as in Fig. 1.
Extended Data Fig. 2 Data Mining / Reanalysis of scRNAseq Data from Macaque Retinas Reveals Ins2 mRNA Expression in RPE and RPE-like Cells.
a. UMAP plot showing different cell types with the RPE-like cell clusters labeled. b. Violin plot showing normalized expression levels of TPM2, a RPE cell gene marker, for all samples. The clusters with the highest TPM2 expression overlay with the clusters we defined as containing RPE-like cells. c. Violin plot showing normalized expression levels of INS in one young male monkey sample (YM2). The detectable expression of INS in YM2 was restricted to RPE-like cells.
Extended Data Fig. 3 Ins1 mRNA and Insulin Receptor Phosphorylation in Ins2 KO mice.
a. RT–PCR of Ins1 fold change in mRNA of isolated RPE from control and Ins2 KO mice. n = 8 mice used for each genotype. b. Insulin receptor was immunoprecipitated from various tissue lysates of control and Ins2 KO mice that were starved overnight. Phosphorylation status of the InsR was probed using phospho-specific InsR antibodies via immunoblotting and quantified as the ratio of p-InsR to total InsR. n = 2 mice for control vs Ins2 KO. Plots are presented as in Fig. 1.
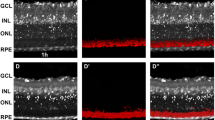
Extended Data Fig. 4 InsR Phosphorylation Decreases in the Retina of Ins2 KO, but not Control, Streptozotocin (STZ) Treated Mice.
a. Schematic of streptozotocin (STZ) treatment. b. ELISA measuring C-peptide levels in blood of control, control STZ-treated, and Ins2 KO STZ-treated mice starved mice. N = 6 mice for control and N = 4 mice for STZ-treated Control and STZ-treated Ins2 KO. *p < .05, one-way ANOVA with Dunnet’s multiple comparisons test. c. Insulin receptor (InsR) was immunoprecipitated from retina tissue lysates of control, control STZ-treated, and Ins2KO STZ-treated mice, starved overnight (left) or control and control STZ-treated mice fed ad libitum (right). Phosphorylation status of the InsR was probed using phospho-specific antibodies (Tyr1150/1151) via immunoblotting and quantified as the ratio of p-InsR to total InsR, normalized to untreated WT mice. N = 3 mice for all conditions. *p < 05, one-way ANOVA with Tukey’s multiple comparison’s test. Plots are presented as in Fig. 1.
Extended Data Fig. 5 RPE Phagocytosis Increases in Starved Mice vs Fed and is not Changed in Ins2 KO Mice.
a. Quantification of OS phagocytosis by the RPE 2 hours after light onset using immunohistochemistry. Quantification of phagocytosis was measured by the amount of Rhodopsin immunoreactivity in the RPE divided by pixels and presented as phagosomes per area on the y-axis (left). Quantification of the number of phagosomes in RPE was done by counting Rhodopsin puncta in the RPE (right). n = 3 mice used for each condition. *p ≤ .05 paired two-tailed t-test. b. Western blot against Rhodopsin on isolated RPE protein lysates from fed and starved mice at 8 am and 10 am (left). Right panel is quantification of the blot to evaluate POS degradation showing 10 am band intensity as a percent of 8 am (peak phagocytosis) band intensity (right). n = 2 mice used for each time point. c. Insulin receptor was immunoprecipitated from lysates of retina from control and MerTK KO mice that were starved overnight. The lysates were probed for InsR phosphorylation or GLUT4 levels by immunoblotting. C-peptide 2 levels were determined by ELISA. N = 6 mice for each condition. *p < .05 paired two-tailed t-test. d. Schematic of WT and cleavage-resistant ‘gain of function’ MerTKCR mice with altered cleavage sites indicated (left). e. Phagocytosis quantification of ingested photoreceptor outer segments using flow cytometry on isolated RPE stained with antibody against rhodopsin from Control and Ins2 KO mice two hours after light onset. n = 7 mice used for each genotype. Plots are presented as in Fig. 1.
Extended Data Fig. 6 Loss of Ins2 leads to decreased eye glucose uptake as measured by Dynamic in vivo PET.
a. Quantification of Dynamic in vivo PET imaging for 18[F]FDG glucose uptake in the eyes of control and Ins2 KO mice after 18[F]FDG injection.
Extended Data Fig. 7 Untargeted Metabolomics Reveals Changes in Global Metabolism of Retinas from WT vs Ins2 Knockout Mice.
a. Enrichment analysis of metabolites from untargeted metabolomics for control over Ins2 KO retinas (using KEGG database). Data are representative of four biological replicates. b. Impact analysis of specific metabolic pathways down (left) and up (right) in control vs Ins2 KO retinas using KEGG database. Pathway significance was determined using Fisher’s Exact Tests.
Extended Data Fig. 8 Untargeted Metabolomics of RPE from WT and Ins2 Knockout Mice reveals Minimal Changes in Global metabolism.
a. Volcano plot from untargeted metabolomics of RPE representing statistically enriched or reduced metabolites in the RPE of control mice relative to those of Ins2 KO using Fisher’s exact test. False Discovery rate (FDR, p < 0.1) is shown in red, absolute value of the Log2 fold change of 0.3 (1.2 fold change) in orange, and those in blue meet both criteria. Data are representative of 4 biological replicates. b. Enrichment analysis of metabolites using KEGG database from untargeted metabolomics for control over Ins2 knockout RPE cells. c. Impact analysis of specific metabolic pathways in control vs Ins2 KO RPE using KEGG database. Pathway significance was determined using Fisher’s Exact Tests.
Extended Data Fig. 9 a-wave Amplitude in the Retina of Fed. or Starved Control and Ins2 KO Mice.
a. Schematic of scotopic electroretinogram (ERG) on mouse retina to detect visual processing in response to light (top). Quantification of a-wave amplitude in ERG of WT and Ins2 KO mice under fed and starved condition (bottom). n = 8 mice for WT, n = 4 mice for Ins2 KO mice.
Supplementary information
Supplementary Information
Flow-gating method.
Supplementary Video 1
Dynamic PET imaging of control (left) and Ins2 KO (right) mice.
Source data
Source Data Fig. 1
Source data for graphs.
Source Data Fig. 2
Source data for graphs.
Source Data Fig. 2
Western blots – uncropped gel images.
Source Data Fig. 3
Source data for graphs.
Source Data Fig. 3
Western blots – uncropped gel images.
Source Data Fig. 4
Source data for graphs.
Source Data Fig. 5
Source data for graphs.
Source Data Extended Data Fig. 1
Source data for graphs.
Source Data Extended Data Fig. 3
Source data for graphs.
Source Data Extended Data Fig. 3
Western blots – uncropped gel images.
Source Data Extended Data Fig. 4
Source data for graphs.
Source Data Extended Data Fig. 4
Western blots – uncropped gel images.
Source Data Extended Data Fig. 5
Source data for graphs.
Source Data Extended Data Fig. 5
Western blots – uncropped gel images.
Source Data Extended Data Fig. 6
Source data for graphs.
Source Data Extended Data Fig. 9
Source data for graphs.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Iker Etchegaray, J., Kelley, S., Penberthy, K. et al. Phagocytosis in the retina promotes local insulin production in the eye. Nat Metab 5, 207–218 (2023). https://doi.org/10.1038/s42255-022-00728-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-022-00728-0
This article is cited by
-
The mouse retinal pigment epithelium mounts an innate immune defense response following retinal detachment
Journal of Neuroinflammation (2024)