Abstract
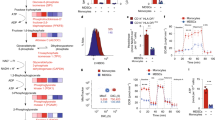
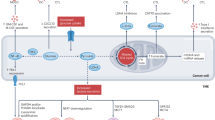
The tumour microenvironment possesses mechanisms that suppress anti-tumour immunity. Itaconate is a metabolite produced from the Krebs cycle intermediate cis-aconitate by the activity of immune-responsive gene 1 (IRG1). While it is known to be immune modulatory, the role of itaconate in anti-tumour immunity is unclear. Here, we demonstrate that myeloid-derived suppressor cells (MDSCs) secrete itaconate that can be taken up by CD8+ T cells and suppress their proliferation, cytokine production and cytolytic activity. Metabolite profiling, stable-isotope tracing and metabolite supplementation studies indicated that itaconate suppressed the biosynthesis of aspartate and serine/glycine in CD8+ T cells to attenuate their proliferation and function. Host deletion of Irg1 in female mice bearing allografted tumours resulted in decreased tumour growth, inhibited the immune-suppressive activities of MDSCs, promoted anti-tumour immunity of CD8+ T cells and enhanced the anti-tumour activity of anti-PD-1 antibody treatment. Furthermore, we found a significant negative correlation between IRG1 expression and response to PD-1 immune checkpoint blockade in patients with melanoma. Our findings not only reveal a previously unknown role of itaconate as an immune checkpoint metabolite secreted from MDSCs to suppress CD8+ T cells, but also establish IRG1 as a myeloid-selective target in immunometabolism whose inhibition promotes anti-tumour immunity and enhances the efficacy of immune checkpoint protein blockade.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
There are no restrictions in regard to the availability of materials reported in the manuscript. The data that support the findings of this study are available from the corresponding author upon reasonable request. The RNA-seq data generated in this study have been deposited in the NCBI Gene Expression Omnibus and are accessible through accession nos. GSE190028 and GSE211255. Source data are provided with this paper.
References
Chapman, N. M., Boothby, M. R. & Chi, H. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020).
Leone, R. D. & Powell, J. D. Metabolism of immune cells in cancer. Nat. Rev. Cancer 20, 516–531 (2020).
Kaymak, I., Williams, K. S., Cantor, J. R. & Jones, R. G. Immunometabolic interplay in the tumor microenvironment. Cancer Cell 39, 28–37 (2021).
Tyrakis, P. A. et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016).
Bunse, L. et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 24, 1192–1203 (2018).
Veglia, F., Sanseviero, E. & Gabrilovich, D. I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 21, 485–498 (2021).
Tarhini, A. A. et al. Immune monitoring of the circulation and the tumor microenvironment in patients with regionally advanced melanoma receiving neoadjuvant ipilimumab. PLoS ONE 9, e87705 (2014).
Sade-Feldman, M. et al. Clinical significance of circulating CD33+CD11b+HLA-DR– myeloid cells in patients with stage IV melanoma treated with ipilimumab. Clin. Cancer Res. 22, 5661–5672 (2016).
Hou, A., Hou, K., Huang, Q., Lei, Y. & Chen, W. Targeting myeloid-derived suppressor cell, a promising strategy to overcome resistance to immune checkpoint inhibitors. Front. Immunol. 11, 783 (2020).
O’Neill, L. A. J. & Artyomov, M. N. Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 19, 273–281 (2019).
Hooftman, A. & O’Neill, L. A. J. The immunomodulatory potential of the metabolite itaconate. Trends Immunol. 40, 687–698 (2019).
Kim, S. H. et al. Phenformin inhibits myeloid-derived suppressor cells and enhances the anti-tumor activity of PD-1 blockade in melanoma. J. Investig. Dermatol. 137, 1740–1748 (2017).
Michelucci, A. et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl Acad. Sci. USA 110, 7820–7825 (2013).
Condamine, T. et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 1, aaf8943 (2016).
Strelko, C. L. et al. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 133, 16386–16389 (2011).
Alshetaiwi, H. et al. Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci. Immunol. 5, eaay6017 (2020).
Cordes, T. et al. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J. Biol. Chem. 291, 14274–14284 (2016).
Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019).
Rodriguez, P. C. et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 64, 5839–5849 (2004).
Sullivan, L. B., Gui, D. Y. & Vander Heiden, M. G. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat. Rev. Cancer 16, 680–693 (2016).
Ma, E. H. et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. 25, 345–357 (2017).
Ron-Harel, N. et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 24, 104–117 (2016).
Kelly, B. & Pearce, E. L. Amino assets: how amino acids support immunity. Cell Metab. 32, 154–175 (2020).
Daniels, B.P. et al. The nucleotide sensor ZBP1 and kinase RIPK3 induce the enzyme IRG1 to promote an antiviral metabolic state in neurons. Immunity 50, 64–76 (2019).
Lo, J. A. et al. Epitope spreading toward wild-type melanocyte-lineage antigens rescues suboptimal immune checkpoint blockade responses. Sci. Transl. Med. 13, 581 (2021).
Smith, C. et al. IDO is a nodal pathogenic driver of lung cancer and metastasis development. Cancer Discov. 2, 722–735 (2012).
Yu, J. et al. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J. Immunol. 190, 3783–3797 (2013).
Weiss, J. M. et al. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J. Clin. Invest. 128, 3794–3805 (2018).
Luan, H. H. & Medzhitov, R. Food fight: role of itaconate and other metabolites in antimicrobial defense. Cell Metab. 24, 379–387 (2016).
Zasłona, Z. & O’Neill, L. A. J. Cytokine-like roles for metabolites in immunity. Mol. Cell 78, 814–823 (2020).
Shen, H. et al. The human knockout gene CLYBL connects itaconate to vitamin B12. Cell 171, 771–782 (2017).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018).
Bambouskova, M. et al. Electrophilic properties of itaconate and derivatives regulate the IκBζ–ATF3 inflammatory axis. Nature 556, 501–504 (2018).
Bambouskova, M. et al. Itaconate confers tolerance to late NLRP3 inflammasome activation. Cell Rep. 34, 108756 (2021).
Hooftman, A. et al. The immunomodulatory metabolite itaconate modifies NLRP3 and inhibits inflammasome activation. Cell Metab. 32, 468–478 (2020).
Runtsch, M. C. et al. Itaconate and itaconate derivatives target JAK1 to suppress alternative activation of macrophages. Cell Metab. 34, 487–501 (2022).
DeNicola, G. M. et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 47, 1475–1481 (2015).
Yuan, P. et al. Phenformin enhances the therapeutic benefit of BRAF(V600E) inhibition in melanoma. Proc. Natl Acad. Sci. USA 110, 18226–18231 (2013).
Jenkins, M. H. et al. Multiple murine BRaf V600Emelanoma cell lines with sensitivity to PLX4032. Pigment Cell Melanoma Res. 27, 495–501 (2014).
Huffaker, T. B. et al. A Stat1 bound enhancer promotes Nampt expression and function within tumor associated macrophages. Nat. Commun. 12, 2620 (2021).
Alghamri, M. S. et al. G-CSF secreted by mutant IDH1 glioma stem cells abolishes myeloid cell immunosuppression and enhances the efficacy of immunotherapy. Sci. Adv. 7, eabh3243 (2021).
Olsen, R. R. et al. ASCL1 represses a SOX9+ neural crest stem-like state in small cell lung cancer. Genes Dev. 35, 847–869 (2021).
Acknowledgements
We thank Emily Robitschek and members of the Zheng Laboratory for helpful discussion on the manuscript. This work is supported by US National Institutes of Health (NIH, nos. R21CA227588 and R01CA219814), the Melanoma Research Alliance, the Elsa U. Pardee Foundation and funds from MGH (to B.Z.); and by NIH (nos. R01CA163591 and DP1DK113643) and Stand Up to Cancer (no. SU2CAACR-DT-20-16) (to J.D.R.). Y.Z. is funded by NCI-R50CA232985. M.P.M. is supported by NIH/NCI grant nos. R01CA151588, R01CA198074 and U01CA-224145 and by the University of Michigan Cancer Center Support Grant (no. NCI P30CA046592).
Author information
Authors and Affiliations
Contributions
H.Z., D.T., L.Y., Y.Z., J.D.R. and B.Z. designed experiments. H.Z., D.T., L.Y., X.X. and Y.Z. performed experiments. J.C., T.J., A.Y.F., L.G., L.C. and K.D.S. performed computational analysis. D.T.F., K.T.F. and G.M.B. provided patient samples. J.M.A., M.P.M., G.M.B., K.T.F., D.L., J.D.R. and B.Z. provided supervision. All of the authors interpreted data and discussed results. B.Z. wrote the paper with input from all of the authors.
Corresponding author
Ethics declarations
Competing interests
K.T.F. serves on the Board of Directors of Clovis Oncology, Strata Oncology, Kinnate, Checkmate Pharmaceuticals and Scorpion Therapeutics; on the Scientific Advisory Boards of PIC Therapeutics, Apricity, Tvardi, ALX Oncology, xCures, Monopteros, Vibliome and Soley Therapeutics; and as consultant to Takeda and Transcode Therapeutics. G.M.B. has sponsored research agreements with Olink Proteomics, Palleon Therapeutics, InterVenn Biosciences and Takeda Oncology; she was on scientific advisory boards for Novartis, Merck, Nektar Therapeutics, Iovance and Ankyra Therapeutics; and she consults for InterVenn Biosciences, Ankyra Therapeutics and Merck. J.D.R. is a paid adviser and/or stockholder in Colorado Research Partners, L.E.A.F. Pharmaceuticals, Rafael Pharmaceuticals and its subsidiaries, Empress and Agios Pharmaceuticals; is a paid consultant of Pfizer; and is a founder, director and stockholder of Farber Partners and Serien Therapeutics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Ilaria Elia, Luke O’Neill and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Alfredo Giménez-Cassina, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 MDSCs secrete itaconate.
(A) Secretion of various metabolites from naïve BM cells as detected by targeted LC-MS/MS analysis (n = 3 biologically independent samples). (B) Ratios of secretion rates of itaconate and various TCA cycle related metabolites from BM-MDSCs versus naïve BM cells (n = 3 biologically independent samples). MDSCs or BM cells were reseeded and cultured in fresh medium for 24 hours and polar metabolites in the supernatants were analyzed by targeted LC-MS-based metabolomics. (C) Secretion rates of itaconate by RAW264.7 cultured with or without cancer conditioned medium (n = 3 biologically independent samples). (D) Expression of IRG1 protein in RAW264.7 cultured with or without cancer conditioned medium. Blots are representative of 3 independent experiments. (E) Quantification of the percentage of LOX + IRG1− and LOX1 + IRG1 + MDSCs in human melanoma tumor specimens for Fig. 1E. (F) Immunofluorescence analysis of IRG1 and macrophage marker CD163, T cell marker CD3 and B cell marker B220 in human melanoma tumor specimens. Data shown are representatives of 12 biologically independent human participants. P values were calculated by paired two- tailed t-tests. Data are represented as mean ± SEM.
Extended Data Fig. 2 Expression of Irg1 is largely confined to granulocytic myeloid populations within the tumors.
Interrogation of publicly available single cell RNA-Seq data analysis to determine cell specific expression of Irg1 within the indicated experimental murine tumors. (A) UMAP showing cells expressing Arg1 and Cxcr2 in mouse breast cancer tumors shown in Fig. 1F. (B) UMAP showing cells expressing Irg1 in mouse B16F10 subcutaneous injected tumors. (C) Violin plots showing levels of Irg1 expression in each cluster from mouse B16F10 subcutaneous injected tumors. (D) UMAP showing cells expressing Irg1 in an oncogenic transgenic glioma mouse model. (E) Violin plots showing levels of Irg1 expression in each cluster from an oncogenic transgenic glioma mouse model. (F) UMAP showing cells expressing Irg1 in a mouse small cell lung cancer model. (G) Violin plots showing levels of Irg1 expression in each cluster from a mouse small cell lung cancer model.
Extended Data Fig. 3 Itaconate suppresses CD8 + T cell proliferation.
(A)Uptake of Itaconate from media (5 mM) by CD3/CD28 activated CD4 + T or CD8 + T cells at 6 hours of incubation (n = 3 biologically independent samples). (B)Effects of itaconate on the cell viability of CD3/CD28 activated CD8 + T cell death (n = 3 biologically independent experiments). (C)Effects of itaconate on the proliferation of A375 melanoma cells (n = 6 biologically independent samples). (D)Effects of itaconate on the proliferation of BP01 melanoma cells (n = 6 biologically independent samples). (E)Itaconate (5 mM) inhibited the proliferation of mouse CD8 + T cells previously activated by CD3/CD28 as measured by cell number counting (n = 3 biologically independent experiments). (F)Effects of various concentration of itaconate (5 mM) on the proliferation of CD3/CD28-activated human CD8 + T cells as measured by CFSE dilution (n = 5 biologically independent experiments). (G)Effects of itaconate (5 mM) on the proliferation of CD3/CD28-activated human CD8 + T cells as measured by cell number counting (n = 3 biologically independent experiments). (H) Effects of itaconate on JAK-STAT signaling in CD3/CD28 activated CD8 + T cells. Cells were treated with NaCl or itaconate (5 mM) for 48 hours before lysates were collected for Western blotting analyses with indicated antibodies. Blots are representative of 3 independent experiments. (I) Effects of itaconate of various concentrations on the levels p-STAT3 and total STAT3 proteins in CD3/CD28 activated CD8 + T cells. Cells were treated with NaCl or itaconate for 48 hours, before lysates were collected for Western blotting analyses with indicated antibodies. Blots are representative of 3 independent experiments. (J) GSEA enrichment plots depicting the enrichment of IFNγ-response gene sets in mouse CD8 + T cells treated with itaconate. (K-L) GSEA of RNA-seq data reveals enriched pathways in mouse CD8 + T cells (K) and CD4 + T cells (L) treated with 5 mM itaconate (ITA) versus NaCl (Ctrl) (n = 3 biologically independent samples). (M) Relative MFI expression of 5-hmC in mouse CD3/CD28-activated CD8 + T cells treated with 3 mM NaCl (Ctrl) or itaconate for 24 hours (n = 5 biologically independent experiments). (N) Representative flow cytometry plots of 5-hmC expression for (M). For all panels, p values were calculated by paired two- tailed t-tests. Data are mean ± SEM.
Extended Data Fig. 4 Itaconate suppresses CD8 + T cell activation.
(A-C) Relative percentage of CD69 (A), IFNγ, TNFα, IL-2, (B), Granzyme B and Perforin (C) expressing cells in mouse CD8 + T cells treated with 5 mM NaCl (Ctrl) or itaconate for 3 days in the presence of anti-CD3/CD28, as analyzed by flow cytometry (n = 6 biologically independent experiments). (D-H) Representative flow cytometry plots for expression of CD69 (D), Granzyme B (E), Perforin (F), IFNγ(G), TNFα(H) and IL-2 (I) in mouse CD8 + T cells treated with 5 mM NaCl (Ctrl) or itaconate for 3 days in the presence of anti-CD3/CD28. For all panels, p values were calculated by paired two- tailed t-tests. Data are mean ± SEM.
Extended Data Fig. 5 Itaconate impedes aspartate and serine/glycine biosynthesis in CD8 + T cells.
(A) Levels of various glycolytic intermediates in CD8 + T cells exposed to exogenous itaconate (ITA) or NaCl (Ctrl), as revealed by metabolite profiling analyses (n = 3 biologically independent samples). (B) Levels of various nucleotides in CD8 + T cells exposed to exogenous itaconate or NaCl (Ctrl), as revealed by metabolite profiling analyses (n = 3 biologically independent samples). (C) Incorporation of 13 C to Aspartate from [U-13C]-glucose in CD8 + T cells exposed to exogenous itaconate or NaCl (Ctrl) (n = 4 biologically independent samples). (D) Effects of nucleosides supplementation on the proliferation of mouse CD8 + T cells treated with 3 mM exogenous itaconate, as measured by CFSE dilution (n = 3 biologically independent experiments). (E) Representative CFSE intensity histograms from flow cytometry analysis for (D). (F) Effects of aspartate (Asp) and serine (Ser) supplementation on the ability of itaconate (3 mM) treated mouse CD8 + T cells on killing MC38-OVA cells (n = 5 biologically independent experiments). For all panels, p values were calculated by paired two- tailed t-tests. Data are mean ± SEM.
Extended Data Fig. 6 Loss of Irg1 enhances anti-tumor immunity of CD8 + T cells.
(A) Spider plot of D4M3A syngeneic tumors grown in WT and Irg1-/- mice (n = 7 mice). (B) Tumor mass of D4M3A syngeneic tumors grown in WT and Irg1-/- mice (n = 7 mice). (C) Pictures of D4M3A syngeneic tumors grown in WT and Irg1-/- mice. (D-E) Measurement of itaconate and lactate in tumor tissues (D) and interstitial fluids (E) showed measurable amounts of itaconate only in tumors grown in WT animals (n = 14 for tumor tissues from both groups, n = 12 for interstitial fluid samples from WT mice, n = 9 interstitial fluid samples from for Irg1-/- mice). (F) Representative flow plots of MDSCs and CD8 + T cells from D4M3A tumors grown in WT and Irg1-/- mice. (G) Secretion of itaconate by WT BM-MDSCs versus Irg1-/- BM-MDSCs (n = 3 biologically independent samples). (H) Uptake of itaconate from the culture media of WT BM-MDSCs by CD3/CD28 activated CD8 + T cells (n = 3 biologically independent samples). (I) MC38 syngeneic tumors grown in Irg1-/- mice exhibited significantly slower grow compared to those in WT mice (n = 8 mice). p values were calculated by unpaired two-tailed t-test (B,D,E,G), two-way ANOVA (I). Data are represented as mean ± SEM.
Extended Data Fig. 7 Loss of Irg1 enhances the efficacy of anti-PD-1 immune checkpoint blockade.
(A) Depletion of CD8 + T cells restored D4M3A tumor growth in Irg1-/- mice (n = 7 mice). P value were calculated by two-sided Tukey’s multiple comparisons test. (B) Spider plot of D4M3A syngeneic tumors grown in WT and Irg1-/- mice and treated with anti-PD-1 or isotype control IgG (n = 6 mice). (C-F) Expression of IL6 (C), TGFβ1(D), CXCL10 (E) and IL10 (F) in D4M3A tumor tissues from WT and Irg1-/- mice that were treated with anti-PD-1 or isotype control IgG (n = 5 biologically independent samples). For all panels, data are represented as mean ± SEM.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Table 1.
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 1
Unprocessed immunoblots.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 1
Statistical Source Data.
Source Data Extended Data Fig. 1
Unprocessed immunoblots.
Source Data Extended Data Fig. 3
Statistical Source Data.
Source Data Extended Data Fig. 3
Unprocessed immunoblots.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 6
Statistical Source Data.
Source Data Extended Data Fig. 7
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, H., Teng, D., Yang, L. et al. Myeloid-derived itaconate suppresses cytotoxic CD8+ T cells and promotes tumour growth. Nat Metab 4, 1660–1673 (2022). https://doi.org/10.1038/s42255-022-00676-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-022-00676-9
This article is cited by
-
Monocytic myeloid-derived suppressor cells as an immune indicator of early diagnosis and prognosis in patients with sepsis
BMC Infectious Diseases (2024)
-
Cancer cell metabolism and antitumour immunity
Nature Reviews Immunology (2024)
-
Metabolic engineering for optimized CAR-T cell therapy
Nature Metabolism (2024)
-
Metabolite itaconate in host immunoregulation and defense
Cellular & Molecular Biology Letters (2023)
-
Amino acid positions near the active site determine the reduced activity of human ACOD1 compared to murine ACOD1
Scientific Reports (2023)