Abstract
Macrophages rely on tightly integrated metabolic rewiring to clear dying neighboring cells by efferocytosis during homeostasis and disease. Here we reveal that glutaminase-1-mediated glutaminolysis is critical to promote apoptotic cell clearance by macrophages during homeostasis in mice. In addition, impaired macrophage glutaminolysis exacerbates atherosclerosis, a condition during which, efficient apoptotic cell debris clearance is critical to limit disease progression. Glutaminase-1 expression strongly correlates with atherosclerotic plaque necrosis in patients with cardiovascular diseases. High-throughput transcriptional and metabolic profiling reveals that macrophage efferocytic capacity relies on a non-canonical transaminase pathway, independent from the traditional requirement of glutamate dehydrogenase to fuel ɑ-ketoglutarate-dependent immunometabolism. This pathway is necessary to meet the unique requirements of efferocytosis for cellular detoxification and high-energy cytoskeletal rearrangements. Thus, we uncover a role for non-canonical glutamine metabolism for efficient clearance of dying cells and maintenance of tissue homeostasis during health and disease in mouse and humans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. Topological analyses are available at https://artyomovlab.wustl.edu/shiny/gam/ and at biotest.hematometabolism.science upon request. Raw and processed sequencing data are deposited on the Gene Expression Omnibus under accession code GSE183176. Source data are provided with this paper.
References
Kojima, Y., Weissman, I. L. & Leeper, N. J. The role of efferocytosis in atherosclerosis. Circulation 135, 476–489 (2017).
Elliott, M. R. & Ravichandran, K. S. The dynamics of apoptotic cell clearance. Dev. Cell 38, 147–160 (2016).
Wang, Y. et al. Mitochondrial fission promotes the continued clearance of apoptotic cells by macrophages. Cell 171, 331–345 (2017).
Yurdagul, A. Jr. et al. Macrophage metabolism of apoptotic cell-derived arginine promotes continual efferocytosis and resolution of Injury. Cell Metab. 31, 518–533 (2020).
Bosurgi, L. et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science 356, 1072–1076 (2017).
Han, C. Z. & Ravichandran, K. S. Metabolic connections during apoptotic cell engulfment. Cell 147, 1442–1445 (2011).
DeBerardinis, R. J. & Cheng, T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29, 313–324 (2010).
O’Neill, L. A. & Pearce, E. J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016).
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Morioka, S. et al. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 563, 714–718 (2018).
Zhang, S. et al. Efferocytosis fuels requirements of fatty acid oxidation and the electron transport chain to polarize macrophages for tissue repair. Cell Metab. 29, 443–456 (2019).
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Nicklin, P. et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136, 521–534 (2009).
Tabas, I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat. Rev. Immunol. 10, 36–46 (2010).
Grabner, R. et al. Lymphotoxin β receptor signaling promotes tertiary lymphoid organogenesis in the aorta adventitia of aged ApoE−/− mice. J. Exp. Med. 206, 233–248 (2009).
Jais, A. et al. Heme oxygenase-1 drives metaflammation and insulin resistance in mouse and man. Cell 158, 25–40 (2014).
Vats, D. et al. Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 (2006).
Perales-Clemente, E. et al. Restoration of electron transport without proton pumping in mammalian mitochondria. Proc. Natl Acad. Sci. USA 105, 18735–18739 (2008).
Fernández-Ayala, D. J. et al. Expression of the Ciona intestinalis alternative oxidase (AOX) in Drosophila complements defects in mitochondrial oxidative phosphorylation. Cell Metab. 9, 449–460 (2009).
Chandel, N. S. Evolution of mitochondria as signaling organelles. Cell Metab. 22, 204–206 (2015).
Caron, E. & Hall, A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science 282, 1717–1721 (1998).
A-Gonzalez, N. et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity 31, 245–258 (2009).
Yvan-Charvet, L. et al. ABCA1 and ABCG1 protect against oxidative stress-induced macrophage apoptosis during efferocytosis. Circ. Res. 106, 1861–1869 (2010).
Viaud, M. et al. Lysosomal cholesterol hydrolysis couples efferocytosis to anti-inflammatory oxysterol production. Circ. Res. 122, 1369–1384 (2018).
Tavakoli, S. et al. Characterization of macrophage polarization states using combined measurement of 2-deoxyglucose and glutamine accumulation: implications for imaging of atherosclerosis. Arterioscler Thromb. Vasc. Biol. 37, 1840–1848 (2017).
Johnson, M. O. et al. Distinct regulation of TH17 and TH1 cell differentiation by glutaminase-dependent metabolism. Cell 175, 1780–1795 (2018).
Papathanassiu, A. E. et al. BCAT1 controls metabolic reprogramming in activated human macrophages and is associated with inflammatory diseases. Nat. Commun. 8, 16040 (2017).
O’Neill, L. A. & Artyomov, M. N. Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 19, 273–281 (2019).
Liu, P. S. et al. α-Ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994 (2017).
Scialò, F., Fernández-Ayala, D. J. & Sanz, A. Role of mitochondrial reverse electron transport in ROS signaling: potential roles in health and disease. Front. Physiol. 8, 428 (2017).
Murphy, M. P. How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 (2009).
Lapuente-Brun, E. et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 340, 1567–1570 (2013).
Guarás, A. et al. The CoQH2/CoQ ratio serves as a sensor of respiratory chain efficiency. Cell Rep. 15, 197–209 (2016).
Stipanuk, M.H and Caudill, M.A. Biochemical, Physiological and Molecular Aspects of Human Nutrition 3rd edn (Elsevier/Saunders, 2013).
Jin, H. et al. Integrative multiomics analysis of human atherosclerosis reveals a serum response factor-driven network associated with intraplaque hemorrhage. Clin. Transl. Med 11, e458 (2021).
Gautier, E. L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Devos, M. et al. Keratinocyte expression of A20/TNFAIP3 controls skin inflammation associated with atopic dermatitis and psoriasis. J. Invest. Dermatol. 139, 135–145 (2019).
Aidoud, N. et al. A combination of lipidomics, MS imaging, and PET scan imaging reveals differences in cerebral activity in rat pups according to the lipid quality of infant formulas. FASEB J. 32, 4776–4790 (2018).
Acknowledgements
We thank B. Caraveo for computional development of DreamBio, a new topological tool for Integrated Network Analysis. N. Grandchamp provided Lenti-ONE AOX vector through GEG Tech. We thank S. Fernandez for the noninvasive study of atheroma plaques by ultrasound echography as part of the European Center for Research in Imaging (Cerimed), F. Labret for assistance with flow cytometry, V. Corcelle for assistance in animal facilities and M. Irondelle for assistance with confocal microscopy. This work was supported by grants from the Fondation de France, ANR and the European Research Council consolidator program (ERC2016COG724838) to L.Y.-C. CCMA electron microscopy equipment was funded by the Région Sud - Provence-Alpes-Côte d’Azur, the Conseil Départemental des Alpes Maritimes and the GIS-IBiSA.
Author information
Authors and Affiliations
Contributions
L.Y.-C. conceived the project, designed the experiments and wrote the manuscript. J.M., S.I. and A.D. performed most of the molecular, histological and in vivo experiments. A.S., J.G., M.S., M.A., N.V., A.C., A.S. and A.G. helped with the experimental design and assisted with the data analysis. F.O., T.B. and J.C.M. provided access to platform facilities and assisted with the data acquisition and analysis. S.C., J.M., I.G.S., P.M. and S.R. provided transgenic mice and intellectual discussion. J.C.S. and E.A.L.B. aided in the design and analysis of human studies. R.R. and E.L.G., E.B.T., M.N.A. provided scientific advice and helped with the experimental design. L.Y.-C. also designed and supervised the study and obtained funding. All of the authors read, edited and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Metabolism thanks Ping-Chih Ho-and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary handling editor: George Caputa
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
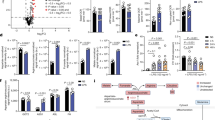
Extended Data Fig. 1 GLS1 is dispensable for macrophage homeostasis but supports macrophage effector functions.
(a) Comparative analysis of Gls1 mRNA expression in different macrophage populations from a publicly available dataset (Immgen). Red indicates up- and blue down-regulated expression (b) Macrophage population gating strategy (left) and numbers (right) measured by flow cytometry in multiple mouse tissues (bone marrow, peritoneal cavity, spleen, liver and brain). Both sexes were analyzed. (c) [3H]-Thymidine incorporation (left) and apoptosis percentage (right) in Gls1fl/fl control or Mac∆Gls1 macrophages at steady state or after an overnight stimulation with IL-4. (d) Phospho-S6 (left) and c-myc (right) expression measured by flow cytometry in these cells. (e) RNAseq of control or Mac∆Gls1 cell sorted PCMs at steady state or after IL-4 stimulation. (f) RNAseq analysis with focus on alternatively activated genes in control or Mac∆Gls1 PCMs stimulated overnight or not with IL-4. (g) CD206 and CD301 expression by flow cytometry in Cre+ control or Mac∆Gls1 mice injected i.p. with PBS or IL-4−complex. (h) mRNA expression of key efferocytic receptors such as MertK, Timd4 or Gas6 or immunoregulatory cytokines such as Tgfβ or Il10 (left panel) or IL-10 secretion (right panel) in reparative Gls1fl/fl control and MacΔGls1 PCMs. (i) Phagocytosis assays of heat-killed, fluorescently pre-labeled E.coli (calculated after quenching the extracellular probe by trypan blue) in these cells. All values are mean ± SEM and are representative of at least one independent experiment (n = 4 to 8 independent animals for b-d, n = 3 to 5 for g-i). P values were determined by a two-tailed Student’s t-test. Source data are provided as a Source Data file (b-d, g-i).
Extended Data Fig. 2 Atherosclerosis development relies on GLS1-dependent glutaminolyis.
(a) Metabolic pathway (left) and RNAseq analysis (right) of Apoe−/− versus WT mouse aortas (6 weeks on WD) performed with Phantasus software (GSE10000). Red indicates up- and blue down-regulated expression. (b) Oil red O staining (left) or 14C glutamine accumulation after i.v. injection (right) in descending aortas extracted from female WT and Apoe−/− mice maintained on a WD for 6 weeks. (c) 14C glutamine incorporation in aortas obtained from female Apoe−/− and Apoe−/− Mac∆Gls1 mice fed for 6 weeks on WD. (d) Cholesterol (left) and triglyceride (right) content in plasma of Apoe−/− and Apoe−/− Mac∆Gls1 mice. (e) Representative sections (left) and quantification (right) of aortic plaques from Apoe−/− or Apoe−/− HSC∆Gls1 mice (12 weeks WD) stained for Oil Red O and Hematoxylin Eosin. Scale bar: 200 μm. (f) Representative sections (top) and quantification (bottom) of aortic plaques from Apoe−/− or Apoe−/− Mac∆Gls1 mice (12 weeks WD) stained for CD68 and Ki67. (g) Gating strategy (left) and quantification (right) of PCM efferocytic index in Apoe−/− and Apoe−/− MacΔGls1 mice 1-hour after labeled ACs i.p. injection. Both sexes were analyzed. All values are mean ± SEM and are representative of at least one independent experiment (n = 4 independent animals for c, n = 7 to 9 for e-g). P values were determined by a two-tailed Student’s t-test. Source data are provided as a Source Data file (c-g).
Extended Data Fig. 3 Glutaminolysis supports mitochondrial OXPHOS and ATP generation to promote efficient efferocytosis.
(a) Publicly available gene expression datasets analysis of macrophages ingesting apoptotic cells (GSE98169). Red indicates up- and blue down-regulated expression. (b) Gls1 mRNA expression and (c) Glutamine/Glutamate ratio during efferocytosis in control BMDMs in a time course experiment. (d) Quantification of leucine and valine levels and (e) ornithine, arginine, proline and putrescine levels in control or Mac∆Gls1 BMDMs at steady state or after IL-4 stimulation. (f) Efferocytic index in control or Mac∆Gls1 BMDMs in basal conditions or stimulated overnight with IL-4 and + /− Gabapentin or Bcat inhibitor (left panel) or treated with IL-4 and ornithine (right panel). (g) OCR measurements in reparative control and MacΔGls1 PCMs or (h) in resting and reparative control or Mac∆Gls1 BMDMs. (i) ECAR measured by Seahorse in these BMDMs. (j) Mitochondrial complex I (i.e, NADH-ubiquinone oxidoreductase) (left panel) and complex II (i.e, succinate dehydrogenase (SDH)) (right panel) activities in reparative MacΔGls1 BMDMs. (k) OCR measurements after one (45 min) and two (1-hour rest + 45 min) incubations with ACs in control or Mac∆Gls1 BMDMs (resolving condition) (left panel) or after a second incubation with ACs in control or Mac∆Gls1 BMDMs following a standard ‘Seahorse’ procedure (right panel). (l) OCR measurements after empty or Gls1 lentivirus overexpression in resting control macrophages or reparative control and Mac∆Gls1 BMDMs. (m) ATP production measurements in these cells. (n) Mitochondrial ROS quantification using Mitosox probe in resting and reparative control or Mac∆Gls1 BMDMs and (o) in control and Mac∆Gls1 BMDMs after empty or Gls1 lentivirus overexpression. (p) ATP production measurements in reparative control or Mac∆Gls1 BMDMs after lentiviral vector-mediated overexpression of empty or mitochondrial alternative oxidase (AOX). (q) Efferocytic index in resolving control or Mac∆Gls1 BMDMs + /− 3NPA or antimicyn A between the two rounds of efferocytosis. (r) Efferocytic index after one round efferocytosis (left) or after two round efferocytosis (right) in control or Risp−/− macrophages. All values are mean ± SEM and are representative of at least one independent experiment (n = 2 to 3 independent animals for b-e, n = 3 to 9 for f-r). P values were determined by ordinary two-tailed Student’s t-test (d, e, g, j-m, o, p, r) or one-way ANOVA with Tukey post hoc test for multiple comparisons (f, h, i, n, q). Source data are provided as a Source Data file (d-r).
Extended Data Fig. 4 Gls1 deficiency metabolically reprograms macrophages.
(a) CoMBI-T profiling analysis from RNAseq data of reparative IL-4 treated Gls1-deficient or sufficient PCMs. (b) KEGG mapping with DreamBio from the same RNAseq data. Red indicates up- and green down-regulated expression.
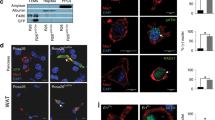
Extended Data Fig. 5 Gls1 deficiency limits non-canonical transaminase metabolism to modulate OXPHOS and redox balance.
(a) ATP production measured by Seahorse in control or Mac∆Gls1 BMDMs at steady state or after overnight IL-4 stimulation + /− EGCG, AOA or NAC. (b) KDM6 activity assay (left) and Tet2 activity assay (right) in resting and reparative control or Mac∆Gls1 BMDMs. (c) OCR quantification control or Mac∆Gls1 BMDMs stimulated overnight with IL-4 in presence or absence of αKetoglutarate. (d) GLUD1 protein expression assessed by Western blotting in control or HSC∆Glud1 PCMs. (e) Effect of Gls1 deficiency on PCA (pyroglutamate), a glutathione precursor, as a read out of cellular GSH replenishment. (f) Efferocytic index in resolving control or Mac∆Gls1 BMDMs + /− AOA treatment between the two rounds of efferocytosis. (g) Efferocytic index in control or Mac∆Gls1 BMDMs stimulated overnight or not with IL-4 and transfected with scramble or Got1/Got2 siRNA. (h) BMDMs analyzed by liquid chromatography-mass spectrometry. (i) ROS quantification using flow cytometry in resting and reparative control or Mac∆Gls1 BMDMs. (j) OCR quantification and ATP production measured by Seahorse. (k) NAD(P)H levels and (l) efferocytic index determined by flow cytometry in control or Mac∆Gls1 BMDMs in basal conditions or overnight IL-4 stimulation + /− GSH. All values are mean ± SEM and are representative of at least one independent experiment (n = 3 to 9 independent animals for a, n = 3 for b, e, g, k, n = 3 to 8 for c, f, h, i, l, n = 4 to 12 for j). P values were determined by ordinary two-tailed Student’s t-test (b, e, g-i) or one-way ANOVA with Tukey post hoc test for multiple comparisons (a, f, j-l). Each statistical bar color-coded represents an independent one-way ANOVA test against IL-4 conditions. Source data are provided as a Source Data file (a-c, e-l).
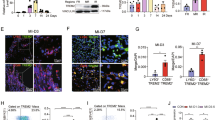
Extended Data Fig. 6 Glutaminolysis is essential for post-transcriptional regulation of actin dynamics and efferocytosis.
(a) Schematic representation of efferocytosis steps and RNAseq analysis of “find-me”, “eat-me” and “tolerate-me” signals in reparative IL-4 treated control and MacΔGls1 PCMs. (b) Schematic representation of actin polymerization and depolarization (left panel) and RNAseq analysis with focus on F-actin dynamic regulators in reparative control or Mac∆Gls1 PCMs (right panel). (c) Dbl expression quantified by flow cytometry in control or IL-4-treated macrophages that were incubated ± ACs for 45 min. (d) RNAseq analysis with focus on GTP-converting ezymes. (e) qPCR quantification (left) and efferocytic index (right) in Gls1fl/fl control and Mac∆Gls1 BMDMs stimulated overnight with IL-4 after Nme1/6 or Sucgl1 lentivirus overexpression. (f) Cdc42 and (g) Rac1 activity assays in control or Mac∆Gls1 BMDMs in basal condition or stimulated overnight with IL-4 + /− AOA. (h) Efferocytic index in reparative control or Mac∆Gls1 BMDMs or (i) in resolving control or Mac∆Gls1 BMDMs + /− NSC23766 (RAC1 inhibitor), ML141 (CDC42 inhibitor) or MDIVI-1 (DRP1-mediated mitochondrial fission inhibitor). All values are mean ± SEM and are representative of at least one independent experiment (n = 2 to 3 independent animals for c-f, n = 5 to 9 for g, n = 3 to 6 for h, i). P values were determined by ordinary one-way ANOVA with Tukey post hoc test for multiple comparisons (c, e-i). Each statistical bar color-coded represents an independent one-way ANOVA test. Source data are provided as a Source Data file (c, e-i).
Extended Data Fig. 7 Graphical abstract illustrating how non-canonical glutamine transamination sustains efferocytosis in atherosclerotic plaques.
Glutaminase (GLS) 1 is upregulated upon reparative (IL-4 stimulation) or resolving (continued clearance of apoptotic cells) to convert glutamine to glutamate. Non-canonical glutamine transamination then acts through the aspartate-arginino-succinate (AAS) shunt to sustain redox buffering (NADPH generation to efficiently supply reduced glutathione GSSG) and fuel energy (i.e, ATP) production through efficient oxidative phosphorylation for cytoskeletal rearrangements.
Supplementary information
Source data
Source Data Fig. 1
Source Data.
Source Data Fig. 2
Source Data.
Source Data Fig. 3
Source Data.
Source Data Fig. 4
Source Data.
Source Data Fig. 5
Source Data.
Source Data Fig. 1a
Uncropped western blot.
Source Data Extended Data Fig. 1
Source Data.
Source Data Extended Data Fig. 2
Source Data.
Source Data Extended Data Fig. 3
Source Data.
Source Data Extended Data Fig. 5
Source Data.
Source Data Extended Data Fig. 5
Uncropped western blot.
Source Data Extended Data Fig. 6
Source Data.
Rights and permissions
About this article
Cite this article
Merlin, J., Ivanov, S., Dumont, A. et al. Non-canonical glutamine transamination sustains efferocytosis by coupling redox buffering to oxidative phosphorylation. Nat Metab 3, 1313–1326 (2021). https://doi.org/10.1038/s42255-021-00471-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-021-00471-y
This article is cited by
-
Dysregulated cellular metabolism in atherosclerosis: mediators and therapeutic opportunities
Nature Metabolism (2024)
-
epHero – a tandem-fluorescent probe to track the fate of apoptotic cells during efferocytosis
Cell Death Discovery (2024)
-
Amino acid metabolism in immune cells: essential regulators of the effector functions, and promising opportunities to enhance cancer immunotherapy
Journal of Hematology & Oncology (2023)
-
Inflamed macrophages sans mitochondrial pyruvate carrier?
Nature Metabolism (2023)
-
Brown adipose tissue-derived Nrg4 alleviates endothelial inflammation and atherosclerosis in male mice
Nature Metabolism (2022)