Abstract
Circulating proteins are vital in human health and disease and are frequently used as biomarkers for clinical decision-making or as targets for pharmacological intervention. Here, we map and replicate protein quantitative trait loci (pQTL) for 90 cardiovascular proteins in over 30,000 individuals, resulting in 451 pQTLs for 85 proteins. For each protein, we further perform pathway mapping to obtain trans-pQTL gene and regulatory designations. We substantiate these regulatory findings with orthogonal evidence for trans-pQTLs using mouse knockdown experiments (ABCA1 and TRIB1) and clinical trial results (chemokine receptors CCR2 and CCR5), with consistent regulation. Finally, we evaluate known drug targets, and suggest new target candidates or repositioning opportunities using Mendelian randomization. This identifies 11 proteins with causal evidence of involvement in human disease that have not previously been targeted, including EGF, IL-16, PAPPA, SPON1, F3, ADM, CASP-8, CHI3L1, CXCL16, GDF15 and MMP-12. Taken together, these findings demonstrate the utility of large-scale mapping of the genetics of the proteome and provide a resource for future precision studies of circulating proteins in human health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The full summary statistics of the Olink CVD-I protein GWAS have been deposited at the SCALLOP CVD-I online resource (www.scallop-consortium.com), allowing access to interactive SCALLOP CVD-I tools and unrestricted download access for secondary analyses. Additionally, a full copy has been deposited with https://doi.org/10.5281/zenodo.2615265 for long-term retention, as well as with the GWAS Catalog. A copy of the polygenic scores have been deposited at the Polygenic Score Catalog (PGS) Catalog.
References
Chames, P., Van Regenmortel, M., Weiss, E. & Baty, D. Therapeutic antibodies: successes, limitations and hopes for the future. Br. J. Pharmacol. 157, 220–233 (2009).
Holmes, M. V., Ala-Korpela, M. & Smith, G. D. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat. Rev. Cardiol. 14, 577–590 (2017).
Folkersen, L. et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 13, e1006706 (2017).
Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018).
Williams, S. A. et al. Plasma protein patterns as comprehensive indicators of health. Nat. Med. 25, 1851–1857 (2019).
Lehallier, B. et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med. 25, 1843–1850 (2019).
Enroth, S., Johansson, A., Enroth, S. B. & Gyllensten, U. Strong effects of genetic and lifestyle factors on biomarker variation and use of personalized cutoffs. Nat. Commun. 5, 4684 (2014).
Emilsson, V. et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 361, 769–773 (2018).
Melzer, D. et al. A genome-wide association study identifies pQTLs. PLoS Genet. 4, e1000072 (2008).
Assarsson, E. et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity and excellent scalability. PLoS ONE 9, e95192 (2014).
Gamazon, E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016).
Sun, W. et al. Common genetic polymorphisms influence blood biomarker measurements in COPD. PLoS Genet. 12, e1006011 (2016).
Chick, J. M. et al. Defining the consequences of genetic variation on a proteome-wide scale. Nature 534, 500–505 (2016).
Zhernakova, D. V. et al. Individual variations in cardiovascular-disease-related protein levels are driven by genetics and gut microbiome. Nat. Genet. 50, 1524–1532 (2018).
Solomon, T. et al. Identification of common and rare genetic variation associated with plasma protein levels using whole-exome sequencing and mass spectrometry. Circ. Genom. Precis. Med. 11, e002170 (2018).
Cabre, A. et al. Fatty acid binding protein 4 is increased in metabolic syndrome and with thiazolidinedione treatment in diabetic patients. Atherosclerosis 195, e150–e158 (2007).
Nishimoto, N. et al. Mechanisms and pathologic significances in increase in serum IL-6 and soluble IL-6 receptor after administration of an anti-IL-6 receptor antibody, tocilizumab, in patients with rheumatoid arthritis and Castleman disease. Blood 112, 3959–3964 (2008).
Gustot, T. et al. Profile of soluble cytokine receptors in Crohn’s disease. Gut 54, 488–495 (2005).
Gale, J. D. et al. Effect of PF-04634817, an oral CCR2/5 chemokine receptor antagonist, on albuminuria in adults with overt diabetic nephropathy. Kidney Int. Rep. 3, 1316–1327 (2018).
Bashore, A. C. et al. Targeted deletion of hepatocyte Abca1 increases plasma HDL (high-density lipoprotein) reverse cholesterol transport via the LDL (low-density lipoprotein) receptor. Arterioscler. Thromb. Vasc. Biol. 39, 1747–1761 (2019).
Burkhardt, R. et al. Trib1 is a lipid- and myocardial infarction-associated gene that regulates hepatic lipogenesis and VLDL production in mice. J. Clin. Invest. 120, 4410–4414 (2010).
Rosa, M. et al. A Mendelian randomization study of IL-6 signaling in cardiovascular diseases, immune-related disorders and longevity. NPJ Genom. Med 4, 23 (2019).
Interleukin 1 Genetics Consortium. Cardiometabolic effects of genetic upregulation of the interleukin 1 receptor antagonist: a Mendelian randomisation analysis. Lancet Diabetes Endocrinol. 3, 243–253 (2015).
Mahdessian, H. et al. Integrative studies implicate matrix metalloproteinase-12 as a culprit gene for large-artery atherosclerotic stroke. J. Intern. Med. 282, 429–444 (2017).
Kaplanski, G. Interleukin-18: biological properties and role in disease pathogenesis. Immunol. Rev. 281, 138–153 (2018).
Heilig, R. et al. The Gasdermin-D pore acts as a conduit for IL-1β secretion in mice. Eur. J. Immunol. 48, 584–592 (2018).
Autiero, M. et al. Role of PlGF in the intra- and inter-molecular cross talk between the VEGF receptors Flt1 and Flk1. Nat. Med. 9, 936–943 (2003).
Dri, P. et al. TNF-induced shedding of TNF receptors in human polymorphonuclear leukocytes: role of the 55-kDa TNF receptor and involvement of a membrane-bound and non-matrix metalloproteinase. J. Immunol. 165, 2165–2172 (2000).
Tenenhouse, H. S. & Sabbagh, Y. Novel phosphate-regulating genes in the pathogenesis of renal phosphate wasting disorders. Pflug. Arch. 444, 317–326 (2002).
Xie, J. H. et al. Engineering of a novel anti-CD40L domain antibody for treatment of autoimmune diseases. J. Immunol. 192, 4083–4092 (2014).
de Miguel, D., Lemke, J., Anel, A., Walczak, H. & Martinez-Lostao, L. Onto better TRAILs for cancer treatment. Cell Death Differ. 23, 733–747 (2016).
Holmes, M. V. & Davey Smith, G. Can Mendelian randomization shift into reverse gear? Clin. Chem. 65, 363–366 (2019).
McCarthy, C. P. & Januzzi, J. L. Jr Soluble ST2 in heart failure. Heart Fail. Clin. 14, 41–48 (2018).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics 26, 2190–2191 (2010).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, S361–S363 (2012).
Tigchelaar, E. F. et al. Cohort profile: LifeLines-DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open 5, e006772 (2015).
GTEx Consortium Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Urmo V, et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. Preprint at bioRxiv https://doi.org/10.1101/447367 (2018).
Westra, H. J. et al. Systematic identification of trans-eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013).
Lloyd-Jones, L. R. et al. The genetic architecture of gene expression in peripheral blood. Am. J. Hum. Genet. 100, 371 (2017).
McRae, A. F. et al. Identification of 55,000 replicated DNA methylation QTL. Sci. Rep. 8, 17605 (2018).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Vilhjalmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Acknowledgements
Secure computing was supported by NeIC Tryggve, which is the Nordic collaboration for sensitive data funded by NeIC and ELIXIR nodes of participating countries. Sources of funding for SMCC, part of the national research infrastructure SIMPLER: We acknowledge the national research infrastructure SIMPLER (the Swedish Infrastructure for Medical Population-based Life-course and Environmental Research) for provisioning of facilities and support. SIMPLER receives funding through the Swedish Research Council under grant 2017-00644. This study was also supported by additional grants from the Swedish Research Council (grants 2017-06100, 2015-05997 and 2015-03257), the Swedish Research Council for Health, Working Life and Welfare (FORTE grant 2017-00721) and Stiftelsen Olle Engkvist Byggmästare (grant 2017/49). S.L was supported by NIH grant 1R01HL139731 and American Heart Association grant 18SFRN34250007. The Orkney Complex Disease Study (ORCADES) was supported by the Chief Scientist Office of the Scottish Government (CZB/4/276 and CZB/4/710), a Royal Society URF to J.F.W., the MRC Human Genetics Unit quinquennial programme ‘QTL in Health and Disease', Arthritis Research UK and the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947). DNA extractions were performed at the Edinburgh Clinical Research Facility. G.D.S. works in the Medical Research Council Integrative Epidemiology Unit at the University of Bristol (MC_UU_00011/1). We would like to acknowledge the invaluable contributions of the research nurses in Orkney, the administrative team in Edinburgh and the people of Orkney. M.A-.K. is supported by a Senior Research Fellowship from the National Health and Medical Research Council (NHMRC) of Australia (APP1158958). He also has a research grant from the Sigrid Juselius Foundation, Finland. A.D.B. was supported by a Wellcome PhD training fellowship for clinicians (204979/Z/16/Z), the Edinburgh Clinical Academic Track (ECAT) programme. J.G.S. and the genotyping of MPP-RES was supported by grants from the Swedish Heart-Lung Foundation (2016-0134 and 2016-0315), the Swedish Research Council (2017-02554), the European Research Council (ERC-STG-2015-679242), the Crafoord Foundation, Skåne University Hospital, Scania County, governmental funding of clinical research within the Swedish National Health Service, a generous donation from the Knut and Alice Wallenberg foundation to the Wallenberg Center for Molecular Medicine in Lund, and funding from the Swedish Research Council (Linnaeus grant Dnr 349-2006-237, Strategic Research Area Exodiab Dnr 2009-1039) and the Swedish Foundation for Strategic Research (Dnr IRC15-0067) to the Lund University Diabetes Center. The study of the LifeLines-DEEP cohort is supported by the Netherlands Heart Foundation CVON grant 2018-27 to J.F. and A.Z., Netherlands Organization for Scientific Research (NWO-VIDI grant 864.13.013 to J.F., 016.178.056 to A.Z., 917.14.374 to L. Franke, VENI grant 194.006 to D.V.Z., Gravitation grant ExposomeNL 024.004.017 to A.Z. and gravitation 024.003.001 to J.F.), European Research Council (ERC starting grant 715772 to A.Z. and 637640 to L. Franke). L. Franke also receives financial support from Oncode Institute. The CROATIA_Vis study was funded by grants from the Medical Research Council (UK), European Commission Framework 6 project EUROSPAN (contract no. LSHG-CT-2006-018947) and the Republic of Croatia Ministry of Science, Education and Sports research grants (108-1080315-0302). We would like to acknowledge the staff of several institutions in Croatia that supported the field work, including but not limited to The University of Split and Zagreb Medical Schools, Institute for Anthropological Research in Zagreb and the Croatian Institute for Public Health. The SNP genotyping for the Vis cohort was performed in the core genotyping laboratory of the Clinical Research Facility at the Western General Hospital, Edinburgh, Scotland. C.H. is supported by MRC University Unit Programme Grant MC_UU_00007/10 (QTL in Health and Disease). P.W.F. was supported by a grant from the European Research Council (CoG-2015_681742_NASCENT). P.S. is supported by a Rutherford Fund Fellowship from the Medical Research Council, grant MR/S003746/1. J.D. holds a British Heart Foundation Professorship and a National Institute for Health Research Senior Investigator Award. Participants in the INTERVAL randomized controlled trial were recruited with active collaboration and funding from NHS Blood and Transplant England (www.nhsbt.nhs.uk), the National Institute for Health Research (NIHR), the NIHR BioResource (http://bioresource.nihr.ac.uk) and the NIHR Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust, NIHR Blood and Transplant Research Unit in Donor Health and Genomics (NIHR BTRU-2014-10024), UK Medical Research Council (MR/L003120/1) and British Heart Foundation (SP/09/002, RG/13/13/30194 and RG/18/13/33946). We would like to thank J. Parks at Wake Forest School of Medicine, Winston-Salem, NC, USA, and D. Rader at Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA, for their kind donations of samples from transgenic mice and controls. Estonian Biobank analyses were funded by EU H2020 grant 692145, Estonian Research Council grant PUT1660, European Union through the European Regional Development Fund Project nos. 2014-2020.4.01.15-0012 and 2014-2020.4.01.16-0125, ERA-CVD grant Detectin-HF. Data analyses were carried out in part in the High Performance Computing Center of the University of Tartu. M.V.H. works in a unit that receives funding from the UK Medical Research Council and is supported by a British Heart Foundation Intermediate Clinical Research Fellowship (FS/18/23/33512) and the National Institute for Health Research Oxford Biomedical Research Centre. This research has been conducted using the UK Biobank Resource under Application no. 13721. Finally, we would like to thank Moving Science for beautiful work on the front-page art proposal (https://movingscience.dk). This work was carried out on behalf of the SCALLOP consortium.
Author information
Authors and Affiliations
Contributions
L. Folkersen, S.G., Q.W., D.H.H., Å.K.H., D.V.Z., E.F., E.M.D., E.I. and A.M. contributed to meta-analysis. L. Folkersen, Å.K.H., D.V.Z., Y.W., J.R.G., Y.C., A.C., F.M., E.F., L. Franke, T.Q., R.W., H.-J.W., J.Y. and A.M. contributed to functional analysis. L. Folkersen, S.G., Q.W., G.D.S., T.P., T.Q., J.Y., L.W., A.S.B., M.V.H., E.I. and A.M. contributed to MR analysis. S.G., J.P., N.E., S.E.B., T.S.B., A.D.B., St.E., A.K., M.A-.K., S.H.C., J.D., Sö.E, C.F., L. Franke, P.W.F., V.G., C. Haley, A.H., Å.J., P.K.J., L.L., C.M.L., S.L., E.M.D., M.M., A.P.M., R.M., M.W.N., O.P., B.P., E.P., J.S., P.S., U.V., H.-J.W., A.Z., J.Ä., J.F., J.G.S., T.E., C. Hayward, U.G., M.L., A. Siegbahn, J.F.W., L.W., A.S.B., E.I. and A.M. contributed to cohort-level analysis. B.E.S., L.M. and A.M. contributed to mouse experiments. K.P., J.D.G., J.L., W.Z., A.Q. and A.M. contributed to clinical trials. L. Folkersen, Å.K.H., A. Schork, J.R.G., F.M., E.F., A.I., T.W. and A.M. contributed to other downstream analysis. L. Folkersen, S.G., Å.K.H., G.E., C.F., O.M., K.M., P.M.N., J.N., M.O.M., M.S. and A.M. contributed to replication analysis. L. Folkersen, S.G., Q.W., M.V.H., E.I. and A.M. contributed to writing. L. Folkersen, S.G., Q.W., A.S.B., M.V.H., E.I. and A.M. contributed to project planning. All authors gave final approval to publish.
Corresponding author
Ethics declarations
Competing interests
J.P. received travel support from Olink AB. R.W., F.M. and D.H.H. are employees of Intomics A/S. A.M., A.Q., E.F., J.D.G., J.L., K.P., M.W.N. and W.Z. are employees of Pfizer Inc. E.I., after this work was completed, became an employee of GSK Inc.
Additional information
Peer review information Primary Handling Editor: Pooja Jha.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
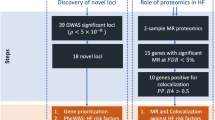
Extended Data Fig. 1 MR-selected loci.
Chromosomal location of all primary associations that were selected as instrument variables for Mendelian Randomization, that is those passing Bonferroni corrected GWAS significance P < 5.6 × 10−10 with replication at nominal p < 0.05, or for non-heterogeneous variants (p < 9 × 10−5), surpassing a P-value threshold of P < 5 × 10−8 in the joint discovery and replication meta-analysis.
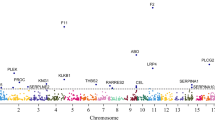
Extended Data Fig. 2 Online tools.
Illustration of the online interactive tools for visualization of genomic loci, regions and plausible networks (www.scallop-consortium.com). a. Illustration of hotspot loci on chromosome 10 (left) and illustration of hotspot loci with independent effects established using COJO analysis (right) b. Circular Manhattan plot for TNF-R2. C. The pathway implicated by trans-pQTLs for plasma TNF-R2. The network shows the likely path from pQTL to TNF-R2.
Extended Data Fig. 3 PrediXcan heatmap.
Heat map showing PrediXcan associations across tissues for any protein with significant associations between protein and predicted mRNA levels (FDR < 0.05) in at least one tissue. In each cell, numeric labels correspond to the uncorrected P-value from the association of protein with predicted expression levels. The colour palette shows the relative expression level of the gene across tissues in the GTeX resource.
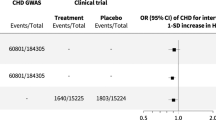
Extended Data Fig. 4 CCR5-2 trial additional results.
Effect of exposure to PF-04634817 on EN-RAGE, FGF-23, KIM-1, myoglobin and TNFR-2. Box plots elements are according to standards for box-and-whisker diagrams.
Extended Data Fig. 5 Work flows 1.
Work flows describing meta analysis.
Extended Data Fig. 6 Work flows 2.
Work flows describing decisions on significance.
Extended Data Fig. 7 Work flows 3.
Work flows describing reasoning behind Mendelian Randomization evidence strength.
Extended Data Fig. 8 Polygenic risk score effects on complex outcomes.
Meta-regression of quantiles of ST2 polygenic risk score and relative risk of asthma (left) and inflammatory bowel disease (right). Values plotted on the x-axis relate to the quantile-specific mean difference in ST2 as compared to the 6th quantile. Values plotted on the y-axis relate to the quantile-specific log odds of disease as compared to the 6th quantile. The red line is the slope derived from the meta-regression across the ST2 quantiles of the PRS on log odds of disease, weighted by the standard error of the log odds.
Extended Data Fig. 9 Cis and trans comparison.
Comparison of absolute effect sizes of all primary cis- and trans loci listed in Supplementary Table 2. Box plots elements are according to standards for box-and-whisker diagrams.
Supplementary information
Supplementary Information
Supplementary Fig. 1: Overview of protein levels showing effects on complex phenotypes using MR. Similar to Fig. 4b, but also showing effects with intermediate evidence strength. Supplementary Fig. 2: Overview of complex phenotypes showing effects on protein levels using MR. Supplementary Note: Detailed overview of systems biology processes.
Rights and permissions
About this article
Cite this article
Folkersen, L., Gustafsson, S., Wang, Q. et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab 2, 1135–1148 (2020). https://doi.org/10.1038/s42255-020-00287-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-00287-2
This article is cited by
-
The genetic etiology of body fluids on chronic obstructive airways disease
Respiratory Research (2024)
-
Peripheral blood indicators and COVID-19: an observational and bidirectional Mendelian randomization study
BMC Medical Genomics (2024)
-
Mendelian randomization identifies circulating proteins as biomarkers for age at menarche and age at natural menopause
Communications Biology (2024)
-
Nanoparticle enrichment mass-spectrometry proteomics identifies protein-altering variants for precise pQTL mapping
Nature Communications (2024)
-
Genetic determinants of plasma protein levels in the Estonian population
Scientific Reports (2024)