Abstract
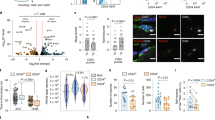
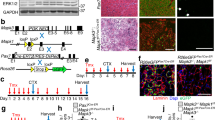
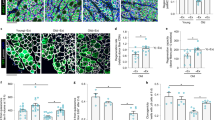
Muscle undergoes progressive weakening and regenerative dysfunction with age due in part to the functional decline of skeletal muscle stem cells (MuSCs). MuSCs are heterogeneous, but whether their gene expression changes with age and the implication of such changes are unclear. Here we show that in mice, growth arrest-specific gene 1 (Gas1) is expressed in a small subset of young MuSCs, with its expression progressively increasing in larger fractions of MuSCs later in life. Overexpression of Gas1 in young MuSCs and inactivation of Gas1 in aged MuSCs support that Gas1 reduces the quiescence and self-renewal capacity of MuSCs. GAS1 reduces RET signalling, which is required for MuSC quiescence and self-renewal. Indeed, we show that the RET ligand, glial-cell-line-derived neurotrophic factor can counteract GAS1 by stimulating RET signalling and enhancing MuSC self-renewal and regeneration, thus improving muscle function. We propose that strategies aimed at targeting this pathway can be exploited to improve the regenerative decline of MuSCs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data that support the findings of this study are available from the corresponding author upon reasonable request. All sequencing data has been deposited with the NCBI Sequence Read Archive database under accession no. PRJNA494728.
Code availability
The custom code used during this study is available from the corresponding author upon reasonable request.
References
Merkle, F. T., Mirzadeh, Z. & Alvarez-Buylla, A. Mosaic organization of neural stem cells in the adult brain. Science 317, 381–384 (2007).
Lo Celso, C. et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457, 92–96 (2009).
Hayashi, K., de Sousa Lopes, S. M. C., Tang, F., Lao, K. & Surani, M. A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3, 391–401 (2008).
Lepper, C., Conway, S. J. & Fan, C. M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 460, 627–631 (2009).
Liu, N. et al. A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat. Cell Biol. 19, 202–213 (2017).
Lepper, C., Partridge, T. A. & Fan, C. M. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646 (2011).
Murphy, M. M., Lawson, J. A., Mathew, S. J., Hutcheson, D. A. & Kardon, G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637 (2011).
McCarthy, J. J. et al. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development 138, 3657–3666 (2011).
Sambasivan, R. et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656 (2011).
Sherwood, R. I. et al. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell 119, 543–554 (2004).
Gilbert, P. M. et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329, 1078–1081 (2010).
Cheung, T. H. et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482, 524–528 (2012).
Rocheteau, P., Gayraud-Morel, B., Siegl-Cachedenier, I., Blasco, M. A. & Tajbakhsh, S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148, 112–125 (2012).
Sousa-Victor, P., García-Prat, L., Serrano, A. L., Perdiguero, E. & Muñoz-Cánoves, P. Muscle stem cell aging: regulation and rejuvenation. Trends Endocrinol. Metab. 26, 287–296 (2015).
Brack, A. S. & Muñoz-Cánoves, P. The ins and outs of muscle stem cell aging. Skelet. Muscle 6, 1 (2016).
Chakkalakal, J. V., Jones, K. M., Basson, M. A. & Brack, A. S. The aged niche disrupts muscle stem cell quiescence. Nature 490, 355–360 (2012).
Cosgrove, B. D. et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 20, 255–264 (2014).
Liu, L. et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 4, 189–204 (2013).
Del Sal, G., Ruaro, M. E., Philipson, L. & Schneider, C. The growth arrest-specific gene, gas1, is involved in growth suppression. Cell 70, 595–607 (1992).
Martinelli, D. C. & Fan, C. M. The role of Gas1 in embryonic development and its implications for human disease. Cell Cycle 6, 2650–2655 (2007).
Fukada, S. et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 25, 2448–2459 (2007).
Martinelli, D. C. & Fan, C. M. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 21, 1231–1243 (2007).
Zammit, P. S. et al. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 166, 347–357 (2004).
Leem, Y. E. et al. Gas1 cooperates with Cdo and promotes myogenic differentiation via activation of p38MAPK. Cell. Signal. 23, 2021–2029 (2011).
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Holzenberger, M. et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421, 182–187 (2003).
Lukjanenko, L. et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 22, 897–905 (2016).
Airaksinen, M. S. & Saarma, M. The GDNF family: signalling, biological functions and therapeutic value. Nat. Rev. Neurosci. 3, 383–394 (2002).
Luo, W. et al. A hierarchical NGF signaling cascade controls Ret-dependent and Ret-independent events during development of nonpeptidergic DRG neurons. Neuron 54, 739–754 (2007).
Wilhelm, S. M. et al. Regorafenib (BAY 73-4506): a new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int. J. Cancer 129, 245–255 (2011).
Egner, I. M., Bruusgaard, J. C. & Gundersen, K. Satellite cell depletion prevents fiber hypertrophy in skeletal muscle. Development 143, 2898–2906 (2016).
López-Ramírez, M. A., Domínguez-Monzón, G., Vergara, P. & Segovia, J. Gas1 reduces Ret tyrosine 1062 phosphorylation and alters GDNF-mediated intracellular signaling. Int. J. Dev. Neurosci. 26, 497–503 (2008).
Biau, S., Jin, S. & Fan, C. M. Gastrointestinal defects of the Gas1 mutant involve dysregulated Hedgehog and Ret signaling. Biol. Open 2, 144–155 (2013).
Cabrera, J. R. et al. Gas1 is related to the glial cell-derived neurotrophic factor family receptors α and regulates Ret signaling. J. Biol. Chem. 281, 14330–14339 (2006).
Sousa-Victor, P. et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321 (2014).
Fry, C. S. et al. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regenerative capacity without affecting sarcopenia. Nat. Med. 21, 76–80 (2015).
Brack, A. S. et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317, 807–810 (2007).
Carlson, M. E., Hsu, M. & Conboy, I. M. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature 454, 528–532 (2008).
Fontana, X. et al. c-Jun in Schwann cells promotes axonal regeneration and motoneuron survival via paracrine signaling. J. Cell Biol. 198, 127–141 (2012).
Baudet, C. et al. Retrograde signaling onto Ret during motor nerve terminal maturation. J. Neurosci. 28, 963–975 (2008).
Suzuki, H. et al. Up-regulation of glial cell line-derived neurotrophic factor (GDNF) expression in regenerating muscle fibers in neuromuscular diseases. Neurosci. Lett. 257, 165–167 (1998).
Jin, S., Martinelli, D. C., Zheng, X., Tessier-Lavigne, M. & Fan, C. M. Gas1 is a receptor for sonic hedgehog to repel enteric axons. Proc. Natl Acad. Sci. USA 112, E73–E80 (2015).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Liu, L., Cheung, T. H., Charville, G. W. & Rando, T. A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 10, 1612–1624 (2015).
Hakim, C. H., . & Wasala, N. B. & Duan, D. Evaluation of muscle function of the extensor digitorum longus muscle ex vivo and tibialis anterior muscle in situ in mice. J. Vis. Exp. (72), 50183 (2013).
Rozo, M., Li, L. & Fan, C. M. Targeting β1-integrin signaling enhances regeneration in aged and dystrophic muscle in mice. Nat. Med. 22, 889–896 (2016).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Zheng, X. et al. Low-cell-number epigenome profiling aids the study of lens aging and hematopoiesis. Cell Rep. 13, 1505–1518 (2015).
Acknowledgements
We thank Fan laboratory members and Y. Zheng for comments; E. Dikovsky for the mouse facility team; and S. Satchell for technical assistance. We especially thank T. Cheung for sharing the FACS protocol. L.L. is supported by the Carnegie Institution of Washington. M.R. was supported by a predoctoral fellowship from the NIH (no. HD075345). C.-M.F. is supported by the NIH (grant nos. R01AR060042, R01AR071976 and R01AR072644) and the Carnegie Institution of Washington.
Author information
Authors and Affiliations
Contributions
L.L., M.R. and C.-M.F. conceptualized the projects, designed the experiments and wrote the manuscript. L.L. and C.-M.F. performed the experiments, analysed the data and drew up the conclusions. S.Y. and C.L. assisted with the FACS and ChIP–seq. C.L. provided the mouse strains. X.Z. and F.J.T. performed the bioinformatic analyses.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: Elena Bellafante
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and Table 1
Rights and permissions
About this article
Cite this article
Li, L., Rozo, M., Yue, S. et al. Muscle stem cell renewal suppressed by GAS1 can be reversed by GDNF in mice. Nat Metab 1, 985–995 (2019). https://doi.org/10.1038/s42255-019-0110-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-019-0110-3
This article is cited by
-
Dynamic changes in butyrate levels regulate satellite cell homeostasis by preventing spontaneous activation during aging
Science China Life Sciences (2024)
-
Pro-myogenic small molecules revealed by a chemical screen on primary muscle stem cells
Skeletal Muscle (2020)