Abstract
Trace elements are important for human health but may exert toxic or adverse effects. Mechanisms of uptake, distribution, metabolism, and excretion are partly under genetic control but have not yet been extensively mapped. Here we report a comprehensive multi-element genome-wide association study of 57 essential and non-essential trace elements. We perform genome-wide association meta-analyses of 14 trace elements in up to 6564 Scandinavian whole blood samples, and genome-wide association studies of 43 trace elements in up to 2819 samples measured only in the Trøndelag Health Study (HUNT). We identify 11 novel genetic loci associated with blood concentrations of arsenic, cadmium, manganese, selenium, and zinc in genome-wide association meta-analyses. In HUNT, several genome-wide significant loci are also indicated for other trace elements. Using two-sample Mendelian randomization, we find several indications of weak to moderate effects on health outcomes, the most precise being a weak harmful effect of increased zinc on prostate cancer. However, independent validation is needed. Our current understanding of trace element-associated genetic variants may help establish consequences of trace elements on human health.
Similar content being viewed by others
Introduction
Trace elements are present in living organisms at low concentrations, often defined as concentrations below 100 parts per million or less than 100 μg/g1. Essential trace elements are vital for growth, development and normal physiology and biochemistry, and external sources are necessary as the body is unable to synthesize them. Other trace elements are classified as non-essential or toxic2. The major sources of exposure to trace elements in the general population are food, water, and air. Trace elements therefore often show considerable geographical variations3. Essential trace elements may also be toxic at elevated concentrations, and their uptake and metabolism are generally tightly regulated in the body4,5,6.
The mechanisms underlying variation in trace element concentrations between individuals are far from fully understood. Variations in trace element concentrations have previously been considered to be governed mainly by dietary intakes of food items with elevated concentrations7 and environmental exposure (e.g., geochemical variations, work-related exposure and anthropogenic pollution)8,9, but genetic variation may also be an important factor10,11,12,13,14,15,16. A well-known example is body iron, where protein-altering mutations in the HFE gene cause a severe iron overload condition, hemochromatosis17. Knowledge about genetic factors controlling trace element concentrations may shed light on the biological pathways of trace elements in the body.
Few genome-wide association studies (GWASs) have investigated trace element concentrations and no previous studies have examined a comprehensive multielement panel of trace elements. Early twin studies have shown evidence for heritable variation in humans for copper, selenium, zinc, arsenic, cadmium, lead, and mercury10,11 and recent GWASs have reported genetic associations with blood concentrations of manganese, copper, selenium, zinc, lead, cadmium, and mercury12,13,14,15,16,18. However, many of the previous studies were limited by sample size or investigated only one or a few trace elements. Among the trace elements we investigated here, no published GWAS was found for 39 of them as of today.
Essential as well as non-essential trace elements have been linked to several health-related outcomes in humans, including neurodegenerative disorders19,20,21,22,23,24,25,26,27,28,29,30,31, autoimmune diseases32,33,34, endocrinological diseases35,36,37,38,39, cancers40,41,42 and bone health43,44,45,46. However, there is inconsistent and sometimes conflicting evidence for a protective or harmful effect of the trace elements on different diseases. Most of these studies have been observational and so the findings may have been influenced by confounding factors. Mendelian randomization (MR) methods have been developed to obtain an additional level of evidence for causal relationships. The potential causality of increased trace element levels on various diseases may be explored using genetic instruments for the individual elements.
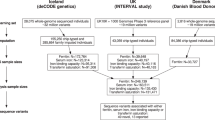
Trace elements have been measured in whole blood in two large Norwegian population-based studies: the Trøndelag Health Study (HUNT)8,47,48,49,50, and the Norwegian Mother, Father, and Child Cohort Study (MoBa)7,51,52,53. Further, genetic variants have been genotyped genome-wide and imputed using the Haplotype Reference Consortium reference panel (v1.1) 54 in both cohorts. GWASs of trace elements have also been reported in other cohorts, including the Swedish Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study15. The main aim of the current study was to identify genetic variants associated with whole blood trace element concentrations, and to explore potential causal associations between trace elements and cancers, neurodegenerative, autoimmune, endocrinological and bone related health outcomes. We, therefore, performed GWASs in HUNT and MoBa followed by a meta-analysis with the PIVUS study15. Further, we used the genetic associations for causal inference with Mendelian randomization, thereby estimating effects of circulating trace elements on health-related outcomes highlighted in previous literature.
Results
Genetic loci associated with trace elements
We identified 20 independent genetic loci (among 21 associations) that reached the genome-wide significance threshold (p < 5 × 10–8) in the meta-analyses of blood concentrations of 14 trace elements measured in the HUNT (14 trace elements, sample size N = 2819), MoBa (11 trace elements, N = 2812) and PIVUS (11 trace elements, N = 949) studies (Supplementary Data 1). The loci were consistently associated (i.e., same direction of the effect) with concentrations of essential (copper [number of variants, n = 1], manganese [n = 10], selenium [n = 2], and zinc [n = 3]) and toxic (arsenic [n = 1], cadmium [n = 3], and lead [n = 1]) trace elements across the meta-analyzed cohorts (Table 1, Supplementary Data 2 and 3, Supplementary Figs. 1–14). To the best of our knowledge, ten of these associations had previously been reported12,13,14,15,16,55,56 (one locus was associated with both manganese and cadmium levels), and 11 had not. Additionally, in HUNT alone we analyzed 43 trace elements that we could not meta-analyze because they had not been measured or the associated genetic variants were not tested in any other cohort. Here, we identified 27 genetic loci for 12 trace elements in the analysis (Supplementary Data 2, Supplementary Figs. 15–38). Among these, we observed a common variant in MORC4 with strong evidence of association with strontium concentrations (minor allele frequency (MAF) = 0.19, p = 2.0 × 10–16), and a common variant in HK1 that was associated with magnesium concentrations (MAF = 0.14, p = 2.2 × 10–8). One locus in SLC18A2 was associated with selenium in HUNT (MAF = 0.27, p = 3.8 × 10–8), but the index (lowest p-value) variant was not tested in MoBa, and none of the tested proxy variants (in high linkage disequilibrium (LD) in HUNT, i.e., correlation r2HUNT > 0.8 with the index variant) were associated with selenium concentrations in MoBa. Most of the other variants from HUNT only were low-frequency (MAF < 5%) or rare (MAF < 0.5%) and given that they were only tested for association in one cohort, they were more likely to be false positive findings. Sensitivity analyses correcting for fish intake (arsenic, selenium), smoking status (arsenic, cadmium, copper, lead), weekly alcohol intake (iron, manganese, zinc, lead) and diabetes (magnesium) in HUNT did not substantially change the effect sizes of the index variants (Supplementary Data 4).
Protein-altering variants
Among the 20 top association signals, we identified eight protein-altering single nucleotide polymorphisms (SNPs) associated with trace elements: Four manganese associated index variants (rs13107325 in SLC39A8, rs1800562 in HFE, rs6099115 in FAM210B and rs855791 in TMPRSS6) and one selenium associated index variant (rs1047891 in CPS1) were nonsynonymous SNPs. Among these, only rs13107325 had previously been reported for manganese in a GWAS15. Although the hemochromatosis variant rs1800562 was not associated with manganese at the GWAS significance level in a previous GWAS15, women with HFE variants were reported to have 12% lower blood manganese concentrations in another study56. Further, three nonsynonymous SNPs were in strong LD (r2 > 0.8) with index variants: rs17279437 in SLC6A20 (p = 9.8 × 10−16, r2HUNT = 0.88 with arsenic index variant rs7306032), rs269868 in DUOX2 (p = 5.7 × 10−10, r2HUNT = 0.96 with cadmium index variant rs953733) and rs57659670 in DUOX2 (p = 3.2 × 10−10, r2HUNT = 0.99 with cadmium index variant rs953733) (Supplementary Data 5).
SNP heritability estimates and genetic and phenotypic correlations of trace elements
For the concentrations of 10 trace elements that had sample sizes above 5000 (cadmium, cobalt, copper, lead, manganese, mercury, molybdenum, selenium, thallium, and zinc), the estimated narrow-sense SNP heritability h2 ranged from h2 = 0.01 ± 0.09 (thallium) to h2 = 0.29 ± 0.10 (manganese). The estimates were higher for essential trace elements (h2 between 0.16 and 0.29) than for non-essential trace elements (h2 between 0.01 and 0.11) (Supplementary Data 6).
We estimated the genetic correlation between all pairs of these 10 trace elements and found absolute values of the genetic correlations to range from 0.01 (cobalt against lead and cobalt against mercury) to 0.97 (molybdenum against cadmium), although the estimates were imprecise and therefore mostly uninformative (Supplementary Fig. 39a, Supplementary Table 1). For comparison, we also estimated the phenotypic correlation (Supplementary Fig. 39b, Supplementary Table 1), where the absolute values ranged from around 0 (manganese against lead and manganese against mercury) to 0.45 (mercury against selenium). We did not observe patterns among the genetic or the phenotypic correlations related to the known ion binding preferences of the trace elements57, which we chose to group as class A (oxygen-seeking) (molybdenum, manganese, zinc), intermediate (cobalt, cadmium, copper), and class B (sulfur/nitrogen-seeking) (lead, mercury, selenium).
Phenome-wide associations of trace element loci
We investigated the genetic relationship between trace element concentrations and other complex traits by examining associations of the index variants identified by the meta-analyses with 1326 phenotype codes (‘phecodes’58), 30 blood biomarkers and 167 other continuous traits and measurements of the UK Biobank. In total, 17 index variants from trace element GWA meta-analyses were associated (p < 9.7 × 10−7, threshold Bonferroni corrected for 51,782 tests) with additional phenotypes (Fig. 1, Supplementary Fig. 40, Supplementary Data 7–9):
Each triangle represents a statistically significant (p < 9.7 × 10–7) association between an index variant from trace element GWA meta-analysis (x-axis, grouped by trace element: cadmium, manganese, selenium and zinc), and an outcome in the UK Biobank (y-axis, grouped and colored according to biological domain: anthropometry [light blue], circulatory [dark blue], digestive [light green], endocrine/metabolic [dark green], genitourinary [pink], hematopoietic [red], musculoskeletal [yellow], neoplasms [orange], respiratory [dark purple] and other [light purple]). Larger triangles represent smaller p-values, and the direction indicates if the direction of effect for the indicated allele is the same (up) or opposite (down) as the association with the trace element concentration. This figure only presents a selection of the associated outcomes in UK Biobank. The full list of associations with meta-analysis index variants are visualized in Supplementary Fig. 40.
Three nonsynonymous index variants for manganese (rs13107325 [SLC39A8], rs1800562 [HFE], rs855791 [TMPRSS6]) and one index variant for cadmium in high LD with nonsynonymous SNPs (rs953733 [DUOX2]) were associated with phecodes (Supplementary Data 7, Fig. 1): The manganese decreasing variant in the metal ion transporter gene SLC39A8 (rs13107325) was positively associated with diseases of the esophagus and with musculoskeletal conditions. The manganese decreasing, but iron increasing, hemochromatosis variant (rs1800562) was positively associated with disorders of mineral and iron metabolism, but also with several diseases in other biological domains. The manganese decreasing and iron increasing59 variant rs855791 was positively associated with other anemias. The cadmium increasing variant, rs953733, found in a locus that has also been associated with iron status biomarkers60 (rs73060324), was positively associated with iron deficiency anemias.
Further, eight index variants, representing three trace elements (cadmium, manganese, and selenium), were associated with blood biomarkers in the UK Biobank (Supplementary Data 8, Fig. 1, Supplementary Fig. 40): Five manganese variants (rs13107325 [SLC39A8], rs1800562 [HFE], rs7176565 [DENND4A], rs5997397 [HSCB] and rs855791 [TMPRSS6]) were associated primarily with endocrine/metabolic biomarkers, but also with digestive, genitourinary, musculoskeletal, and inflammatory biomarkers. The cadmium increasing variant rs953733 (DUOX2) was positively associated with levels of creatinine, a marker for kidney function61, while the cadmium decreasing rs6987313 (LACTB2-AS1) was associated with phosphate and triglyceride levels. The selenium increasing nonsynonymous SNP rs1047891 (CPS1) was associated with endocrine/metabolic, genitourinary, and digestive biomarkers. Finally, the selenium increasing rs17823744 (DMGDH) was negatively associated with insulin-like growth factor 1 (IGF-1).
Among the GWAS results for continuous traits in the UK Biobank, we observed associations for 17 meta-analysis index variants (for cadmium, manganese, selenium, and zinc) (Supplementary Data 9, Fig. 1, Supplementary Fig. 40). The majority of the associations were with different blood cell indices, but cadmium, manganese and selenium variants were also associated with continuous traits from several other biological domains, including musculoskeletal and anthropometric measures (cadmium, manganese, selenium), blood pressure traits (cadmium and manganese), biomarkers in urine and variables derived from endocrine/metabolic blood biomarkers (manganese and selenium), measures of cognitive ability (manganese) and alcohol habits (manganese).
In addition to the associations with the meta-analysis index variants, we also assessed the phenome-wide associations with common and low-frequency (MAF > 0.5%) variants identified only in HUNT. Here, four common variants (strontium [rs17326228], cesium [rs7785293], magnesium [rs16926246] and silicon [rs62228297]) were associated (p < 9.7 × 10−7) with blood biomarkers and/or continuous traits in the UK Biobank (Supplementary Data 10–11, Supplementary Fig. 41): The cesium and magnesium variants were associated with a range of blood cell related traits, the cesium and strontium variants with genitourinary biomarkers and measures, the magnesium variant with endocrine/metabolic biomarkers and measures, and the silicon variant with standing height.
Mendelian randomization
We used two-sample MR to perform an exploratory examination of potential causal effects of trace elements on health outcomes within neurodegenerative, autoimmune, and endocrinological diseases, cancers, and bone related domains as highlighted in the literature19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46. Here, we used the index SNPs from the meta-analyses (manganese, selenium, zinc, copper, lead, arsenic, and cadmium) or from the GWASs in HUNT (strontium) as instruments for the selected trace elements (F-statistics > 10 for all trace elements except cadmium [F-statistic = 9]: Supplementary Data 12). For the SNP-outcome associations, we used summary-level data from large GWASs of selected neurodegenerative disorders (Alzheimer’s disease, Parkinson’s disease, and multiple sclerosis), autoimmune diseases (rheumatoid arthritis and autoimmune thyroid disorder), endocrinological diseases (hypothyroidism and type 2 diabetes), bone related traits and disorders (bone mineral density, bone fractures, osteoporosis) and cancers (prostate cancer and colorectal cancer). The most precise association indicated a weak causal effect of zinc on prostate cancer (odds ratio (OR) = 1.06, 95% CI = 1.01–1.12) (Supplementary Data 12). There were also stronger but less precise associations for arsenic and copper on Parkinson’s disease (OR = 1.24, 95% CI = 0.98–1.57 [arsenic], OR = 1.17, 95% CI = 0.89–1.54 [copper]), selenium on colorectal cancer (OR = 0.89, 95% CI = 0.75–1.07) and selenium, cadmium and lead on multiple sclerosis (OR = 0.86, 95% CI = 0.64–1.15 [selenium], OR = 0.86, 95% CI = 0.72–1.01 [cadmium], OR = 0.70, 95% CI = 0.52–0.95 [lead]) which warrant follow-up (Supplementary Data 12). The results should, however, be interpreted with caution due to limitations such as few instrument SNPs per trace element and known correlations and common biological pathways of trace elements. Otherwise, the MR estimates were generally imprecise, and generally they did not give convincing evidence for causal effects of trace elements in the remaining outcomes (Supplementary Data 12).
Discussion
In the present study, we investigated genetic variants associated with whole blood trace element concentrations, combining data from up to 6564 individuals in HUNT, MoBa and PIVUS. Our GWA meta-analyses identified genetic contributions to whole blood concentrations of essential (copper, manganese, selenium, and zinc) and non-essential or toxic (arsenic, cadmium, and lead) trace elements. In total, 20 loci were associated with trace element concentrations across multiple cohorts, confirming 9 loci from previous studies and identifying 11 novel loci. Seven of the novel loci were associated with manganese concentrations, including two nonsynonymous index variants. Four novel genetic loci were associated with selenium, zinc, arsenic, and cadmium, among which we also identified nonsynonymous variants in high LD (r2 > 0.8) with the index variants. The genetic variants that we identified in the present study had small to moderate effect sizes. They could, however, still contribute to individual differences in trace element concentrations in combination with many other factors. Identification of these loci expands our knowledge about the genetic contribution to trace element concentrations and indicates proteins that could aid in establishing mechanisms for absorption, distribution, metabolism, and excretion.
Because essential trace elements are not produced in the body, but are necessary for normal physiology, we would expect genes associated with essential trace elements to encode proteins that include these elements or are involved in their respective regulatory processes. This is in line with our observations in the meta-analyses of copper, zinc, and manganese, where we replicated previously reported loci with index variants near or in genes encoding the copper-binding metalloprotein ceruloplasmin (CP)62, the metalloenzyme carbonic anhydrase 1 (CA1) involved in the zinc balance63, and two divalent metal ion transporters SLC30A1064 and SLC39A865 (nonsynonymous variant), where deficiency in the latter is known to cause severe manganese deficiency66. Other nonsynonymous variants we observed had, to our knowledge, little or no known underlying biology related to the associated trace elements. For example, we observed a novel association between selenium and a nonsynonymous variant in the urea cycle gene CPS1. Although the variant is associated with vitamin D67, which has in turn been hypothesized to interact with selenium68, and the gene has been observed to be upregulated in mice fed excessive amounts of selenium69, the underlying biology of the association with selenium in humans is not established. Because some trace elements interact with each other, or have common uptake mechanisms and co-transport, an imbalance in the concentration of one trace element might also change the concentration of others7,49,70,71: For example, manganese is partly transported by both zinc and iron transporters72, and low iron stores have been associated with higher blood concentrations of other trace elements7,49. Further, functional variants in iron metabolism genes have been associated with lower blood manganese concentrations56, including rs1800562 in HFE that we replicated here. The novel loci we observed to be associated with manganese, as well as the CPS1 locus for selenium, are in line with this pattern: One manganese associated nonsynonymous SNP is known in iron deficiency anemia (rs855791 in TMPRSS6)73. Further, the gene that is closest to the index variant in another locus is the interleukin-6 regulator gene TUT774, which stimulates the iron regulator hepcidin75. The index variants in other novel loci are close to genes involved in mitochondrial iron uptake (FAM210B76, where we identified the nonsynonymous manganese index variant rs6099115) or genes associated with hematological traits or involved in the synthesis of iron containing compounds (LYRM4, FAM210B, DENND4A, FECH, HSCB)76,77,78,79. The CPS1 index variant (rs1047891) was in high LD (r2HUNT = 0.92) with an index variant (rs715) for total iron binding capacity in our previous study60. These loci were not associated (p > 5 × 10−8) with iron in the current study; however, the analysis of iron had limited statistical power with less than half the sample size of manganese.
Non-essential trace elements typically do not have their own transport proteins or specific mechanisms for metabolism and are generally taken up into the body using the routes of macronutrients80 or essential trace elements with similar chemical properties70. This is illustrated by one of the known loci we replicate for the toxic metal cadmium, where the index variant is nearest to the zinc/manganese transporter gene SLC39A8, which was also associated with manganese. For cadmium, we also report a novel locus with two nonsynonymous variants in DUOX2, both in high LD with the index variant. The gene codes for a thyroid hormone synthesis related protein81 with a heme binding site82 and was associated with iron status in previous GWASs60,83. Similarly, the nonsynonymous variant rs17279437 in the glycine transporter gene SLC6A20 has been associated with excessive glycine excretion84. In this study, the variant was in high LD with the index variant for arsenic, which could potentially indicate that the protein can also transport arsenic. Glycine levels have also been associated with arsenic exposure in mouse models85.
The genetic associations observed only in HUNT warrant further replication. One interesting result identified in HUNT was a selenium locus within a solute carrier gene (SLC18A2). Although the index variant was not tested in MoBa, variants in high LD (r2 > 0.8) with the index variant were tested, but not associated with selenium in that data. The non-association in MoBa could however potentially be a result of sex-specific or pregnancy-specific exposure patterns. Further, index variants in gold and magnesium loci were in or nearest to genes associated with hemoglobin and iron regulation (CD163 and HK1)86,87. The strong association between hypomagnesemia and type 2 diabetes88, combined with a previously reported association between the magnesium HK1 index variant with levels of the type 2 diabetes biomarker glycated hemoglobin (HbA1c)89 could support this finding. However, associations between HK1 and HbA1c could also reflect the erythrocyte lifetime regardless of diabetes status90. We also observed a strong association between strontium concentrations and a locus in MORC4. Strontium is chemically very similar to calcium. It has effects on bone balance91, and could potentially induce skeletal abnormalities in very high doses91. A rare genetic variant in MORC4 has been associated with a 3.4 times increased risk of osteoarthritis (Open Targets Platform92, accessed 25.05.2022), and another MORC family member, MORC3, is involved in calcium homeostasis and maintenance of bone remodeling93. We therefore speculate that MORC4 could have similar functions or be involved in similar pathways.
In the PheWAS analyses, we observed the well-known association of functional variants in iron metabolism genes (the hemochromatosis HFE variant, rs1800562, and the TMPRSS6 variant, rs855791) with disorders of iron and mineral metabolism and anemias. As expected, and previously demonstrated in the UK Biobank60, rs1800562 was also associated with several known clinical manifestations of HFE hemochromatosis94,95. Likewise, the index variant in the DUOX2 (cadmium) locus known from GWASs of iron status biomarkers60,83,96, was associated with iron deficiency anemia. The many associations between trace element index variants and blood cell indices could also potentially reflect their correlation with iron status. The associations between digestive and musculoskeletal disorders and rs13107325 (manganese), which alters the metal ion transporter ZIP8 (encoded by SLC39A8), could potentially highlight the role of manganese or other divalent trace metals in these conditions. Associations with basal metabolic rate and metabolic biomarkers could possibly be related to the processing of trace elements in the body, and associations with biomarkers for liver and kidney function could be related to the clearance of trace elements that are toxic or in high abundance. However, population stratification could also have caused false associations with some of the variants.
We estimated the heritability of concentrations of 10 trace elements with meta-analysis sample size above 5000, ranging from low to moderately high heritability (0.01 ± 0.09 [thallium] to 0.29 ± 0.10 [manganese]). The lower heritability for non-essential trace elements compared to essential trace elements could reflect that humans have not evolved genes and biological pathways to handle non-essential trace elements. The very low heritability of thallium could also be because most of the samples from MoBa were below the detection limit. Further, many of the heritability estimates could be low because LD Score regression underestimates the heritability of traits that are not highly polygenic97.
The genetic and phenotypic correlations between nine trace elements did generally not correspond well to each other, however, the genetic correlations were highly imprecise and therefore mostly uninformative. Mercury and selenium had the strongest phenotypic correlation, which is in line with the known mercury-selenium antagonistic relationship98. In general, the genetic correlations were higher than the phenotypic correlations, although the range of the different phenotypic correlation estimates were similar to those previously reported in MoBa7. This could potentially reflect a relatively low polygenicity of trace element concentrations, where few genetic loci might influence many different trace elements, while there are a variety of different factors influencing the phenotypic variations.
Using two-sample MR, we observed indications of a weak harmful effect of circulating zinc on prostate cancer. The effect of zinc on prostate cancer is debated, but a harmful effect has been found in some previous studies99. There was also a weak positive association for arsenic and copper on Parkinson’s disease, and weak negative associations for selenium on colorectal cancer and selenium, cadmium, and lead on multiple sclerosis. Otherwise, we observed little evidence for causal roles of trace elements in the remaining selected health outcomes. The apparently protective effect of lead on multiple sclerosis is not in line with the known inhibitory effect of lead on a heme biosynthesis catalyst, aminolevulinate dehydratase (ALAD)100, and (lead induced) impairment of heme synthesis as a suggested potential trigger for multiple sclerosis101. Generally, true direct protective effects of non-essential or toxic trace elements seem implausible, although their effect could potentially be indirect, for example, if the toxic trace elements influence essential trace elements in the body. Based on the PheWAS results and the known correlations and common biological pathways between several trace elements, the estimates were particularly vulnerable to misspecification of the primary phenotype and/or horizontal pleiotropy. Further, blood concentrations might not indicate other tissue or organ specific trace element concentrations. MR analyses can also be influenced by selection bias and competing risks, which might also explain apparent protective effects of toxic trace elements on late-life health outcomes. Few instrument SNPs per trace element provided limited opportunities for reliable sensitivity analyses. Some estimates were precise around the null, but in general, the confidence intervals were wide and compatible with both moderate protection and harmful effects and were therefore uninformative. The low precision could be due to low case sample sizes in the outcome data and/or genetic instruments that had few SNPs with small to moderate effects on the exposures, potentially introducing weak instrument bias102. This was quantified by the low variance explained and low F-statistics of some of the instruments. We chose not to include any correction for multiple testing, since this was a first exploratory analysis with low power. The findings were based on the analyses of variation in trace element concentrations in a general population, and we were therefore unable to estimate any effects of extreme trace element concentrations. The current results should be interpreted with caution, and future well-powered multivariable MR analyses might be helpful to discriminate between the effects of different trace elements. Further, as we only analyzed genetic associations with trace element concentrations measured in adults, it was not possible to evaluate any effects of trace elements specific to growth and development stages in newborns to adolescents103. Low sample sizes and lack of cohorts with both trace elements and health outcomes available also prevented us from exploring non-linear associations with MR.
This study has several limitations. Neither the HUNT nor the MoBa populations have generally been exposed to high levels of toxic trace elements, which makes these excellent populations for detecting genetic factors, but on the other hand many trace elements had a high proportion of measurements below the detection limit, thereby lowering the effective sample size. Accordingly, expanding the analyses to additional populations and ancestry groups could have been highly informative, both due to differences in LD structure and variation in exposure to trace elements from the diet and the environment. Further, trace elements are distributed differently in different organs, tissues, and body fluids104,105,106, and whole blood is therefore not the preferred tissue for monitoring the status of all trace elements. Some trace elements could also be influenced by evaporation, contamination from syringes or leakage of trace elements from glass vials. Differences between studies and populations also created known and potential limitations. Without access to individual level data in PIVUS, we were unable to harmonize all covariates. For example, the PIVUS association model included triglycerides and cholesterol as covariates, while the models used in HUNT and MoBa did not. Although HUNT, PIVUS and MoBa are all Scandinavian cohorts, the populations from which they have been sampled have clear differences, particularly in terms of birth year, age, and sex, and although the directions of effect were consistent across all three studies, the differences observed in effect size across these studies for a few SNPs could potentially be related to population-specific exposure patterns or factors related to pregnancy (MoBa). For example, fish intake might influence Swedes more than Norwegians, because there are indications that some toxic trace elements have accumulated more in fish caught in the shallow Bothnian Bay than along the deep Norwegian coastline107. Further, pregnant women might change their diet and different generations could have been exposed to different levels of environmental pollution throughout their lives. The main limitation was that most of the trace elements were only measured in HUNT, and both low sample sizes and the lack of replication cohorts were general limitations. Further, trace element concentrations are associated with many different factors, and while we have controlled for some of these, there might still be unmeasured confounding by population stratification related to factors we have not controlled for, especially for rare variants. A particular limitation was the lack of detailed data on dietary habits. The heritability and genetic correlation estimates found using LD Score regression were limited by the potentially low polygenicity of the trace elements and genomic control (GC) correction of summary statistics, both which may contribute to heritability being underestimated. Additionally, using summary statistics from mixed models may contribute to overestimation. Finally, small sample sizes (and for some trace elements many measurements below the detection limit) resulted in highly imprecise estimates. The imprecision was especially large for the genetic correlations, which were further limited by heritability z-scores that were all below 4108.
In summary, we have identified novel genetic loci and replicated previously indicated loci for essential and non-essential trace elements. These highlighted interesting genes that may help establish biological pathways and mechanisms for uptake, distribution, metabolism, and excretion of trace elements in humans. MR analyses provided several indications of weak to moderate associations between trace elements and health outcomes, the most precise being a weak harmful effect of genetically determined circulating zinc on prostate cancer. However, generally imprecise MR estimates gave no convincing evidence for causal roles of genetically determined levels of other trace elements on health outcomes. Studies of populations with higher trace element exposure burdens or with larger samples are needed to investigate moderate and weak effects.
Methods
Cohort descriptions
HUNT
The HUNT Study47,48 is a longitudinal population-based health study conducted in the county of Trøndelag, Norway. Data and samples have been collected through four cross-sectional surveys: HUNT1 (1984–86), HUNT2 (1995–97), HUNT3 (2006–08) and HUNT4 (2017–2019). Approximately 123,000 individuals (aged ≥ 20 years) have participated in one or more HUNT surveys. Approximately 88,000 individuals have been genotyped using one of three Illumina HumanCoreExome arrays: 12 v.1.0, 12 v.1.1 and 24 with custom content (UM HUNT Biobank v1.0). Sample and variant quality control (QC) was performed using standard practices as described elsewhere109: Samples were excluded if they had a call rate < 99%, estimated contamination > 2.5%, large chromosomal copy number variants, lower call rate of a technical duplicate pair or twin, gonosomal constellations other than XX or XY, or a discrepancy between the inferred sex and reported gender. After genotyping, variants were excluded if they had a call rate < 99%, deviation from the Hardy–Weinberg equilibrium (p < 10−4 in unrelated samples of European ancestry), Gentrain score < 0.15, cluster separation < 0.3, probe sequences that could not be mapped perfectly to the reference genome, or if another assay with higher call rate had genotyped the same variant. All variants were imputed from the HRC v1.1 reference panel110 merged with 2201 sequenced samples from HUNT, using Minimac3111.
Trace elements have been measured in whole blood samples collected in HUNT2 and HUNT3 using high-resolution inductively coupled plasma mass spectrometry (HR-ICP-MS) (Thermo Finnigan Element 2, Thermo Finnigan, Bremen, Germany) at three laboratories as part of previous studies. Here, we combined measurements of nine trace elements (As, Cd, Co, Cu, Pb, Mn, Hg, Se, Zn) in 930 samples (HUNT2) that have been analyzed by the National Institute of Occupational Health in Norway (STAMI), 53 trace elements (Al, Sb, As, Ba, Be, B, Br, Cd, Ca, Ce, Cs, Cl, Cr, Co, Cu, Ga, Ge, Au, Ho, In, Fe, La, Pb, Li, Mg, Mn, Hg, Mo, Nd, Ni, Nb, Pa, P, Pt, Pr, Re, Rh, Rb, Sm, Se, Si, Sr, S, Ta, Tb, Tl, Sn, W, U, V, Y, Zn, Zr) that have been measured in 757 samples (HUNT3) by one laboratory at NTNU (hereafter named NTNU1), and 30 trace elements (As, Be, Bi, B, Br, Cd, Ca, Cs, Cr, Cu, Ga, Au, In, Ir, Fe, Pb, Mg, Mn, Hg, Mo, Ni, Rb, Se, Ag, Sr, Tl, Th, Sn, W, Zn) that have been measured in 1539 samples (HUNT3) by a second laboratory at NTNU (hereafter named NTNU2). For 23 trace elements in HUNT3, some samples (maximum 30) had missing information on lab assignment, and we assigned these to NTNU2. There were no individuals with multiple measurements. Trace element measurements were returned to the HUNT Databank after sample and trace element QC at the respective laboratories. The samples analyzed by the STAMI laboratory were collected as part of a sub-study of iron status in women (selection criteria: female, age between 20 and 55 years old, non-pregnant, not blood donor in the past 2 years), the samples analyzed by HUNT1 were collected as part of a neuroimaging sub-study (selection criteria: age between 50 and 65, participation in previous HUNT surveys, exclusion criteria: MRI contraindications (pacemaker of the heart, clipped cerebral aneurysm, cochlear implants, severe claustrophobia, or body weight above 150 kg)), and 267 of the 1539 samples analyzed by NTNU2 were selected among type 2 diabetes cases as part of a diabetes related sub-study. Samples from the NTNU1 laboratory that were either below the detection limit or more than 10 times the median value had been removed and were not included in the current analysis. The specific reason for removal of these measurements was not available, therefore we excluded these measurements for our analysis. The proportion of unavailable samples per trace element from NTNU1 ranged from 0 to 69% (average 17%, median 2%). For samples from the NTNU2 laboratory, we replaced measurements below the detection limit with randomly generated numbers between 0 and the element-specific detection limit, because the true measurements were unavailable. The proportion of measurements below the detection limit from NTNU2 ranged from 0% to 97% (average 11%, median 0%). Details of sample collection, storage of samples, sample processing, quality control and ICP-MS analyses have been reported in detail elsewhere8,49,50.
MoBa
The Norwegian Mother, Father and Child Cohort Study (MoBa) is a population-based pregnancy cohort study conducted by the Norwegian Institute of Public Health. Participants were recruited from all over Norway from 1999–2008. The women consented to participation in 41% of the pregnancies. The cohort includes approximately 114,500 children, 95,200 mothers and 75,200 fathers53. The current study is based on version 12 of the quality-assured data files released for research in January 2019. The establishment of MoBa and initial data collection was based on a license from the Norwegian Data Protection Agency and approval from The Regional Committees for Medical and Health Research Ethics.
In short, pregnant women were recruited in their first trimester and invited to fill in three questionnaires during pregnancy, and to donate blood and urine samples at the time of ultrasound screening around gestational weeks 17–19 (mean 18.5). Blood samples were obtained from both parents during pregnancy and from mothers and children (umbilical cord) at birth. Follow-up is conducted through questionnaires and linkage to national health registries53. A total of 98,000 MoBa participants have been genotyped using one of three arrays: Illumina HumanCoreExome, Illumina Global Screening Array, or Illumina OmniExpress. In the current study, we used genetic data collected as part of the Better Health by Harvesting Biobanks (HARVEST) study released by MoBa Genetics v.1.0 (https://github.com/folkehelseinstituttet/mobagen/wiki/MoBaGenetics1.0). The sample and variant quality control in MoBa has been described elsewhere112. All variants were imputed from the HRC reference panel v1.1 at the Sanger Imputation Service.
The Norwegian Environmental Biobank is a sub-study within MoBa established with the aim of biomonitoring nutrients and environmental contaminants in MoBa participants. The study included 2999 pregnant women with available genetic data who had donated blood and urine samples and had responded to questionnaires 1–6 in MoBa7,52. A total of 11 trace elements (As, Cd, Co, Cu, Mn, Mo, Pb, Se, Tl, Zn, Hg), were measured in whole blood donated by the women in gestational week 18 at the department of laboratory medicine at Lund University (Sweden). All trace elements except Hg were analyzed using an ICP-MS (iCAP Q, Thermo Fisher Scientific, Bremen, Germany). Total Hg was determined in acid-digested samples by cold vapor atomic fluorescence spectrophotometry (Sandborgh-Englund 1998). During the analysis campaign, the laboratory participated in the German External Quality Assessment Scheme (G-EQAS), with good agreement between obtained element concentrations in quality control samples used and expected values. Details of sample collection, storage of samples, sample processing and ICP-MS analyses have been reported in detail elsewhere7.
Association analyses
We performed genome-wide association analyses of 59 trace elements measured in up to 2819 individuals in HUNT and 11 trace elements measured in 2812 individuals in MoBa. For 48 trace elements in HUNT (Al, Sb, As, Ba, Be, Bi, B, Br, Cd, Ca, Ce, Cs, Cl, Co, Cu, Ga, Ge, Au, Ho, In, Ir, Fe, La, Pb, Li, Mg, Mn, Hg, Mo, Nd, Nb, Pa, P, Rh, Rb, Sm, Se, Ag, Sr, S, Ta, Tb, Tl, Sn, W, U, Y, Zn) and 9 trace elements in MoBa (Cd, Co, Cu, Hg, Mn, Mo, Pb, Se, Tl, Zn), we used a linear mixed model regression under an additive genetic model for each variant as implemented in BOLT-LMM v.2.3.4113, thereby controlling for relatedness between study participants. We performed association analyses of 9 trace elements in HUNT (Cr, Ni, Pt, Pr, Re, Si, Th, V, Zr) and 2 trace elements in MoBa (As, Mo) in unrelated individuals using PLINK 2.0114, because BOLT-LMM was unable to estimate the trace element heritability. In total, human GWASs had not been previously published for 39 trace elements (Sb, Ba, Be, B, Br, Bi, Ce, Cs, Ga, Ge, Au, Ho, In, Ir, La, Li, Nd, Nb, Pd, Pt, Pr, Re, Rh, Rb, Sm, Si, Ag, Sr, S, Ta, Tb, Tl, Th, Sn, W, U, V, Y, Zr). Distributions, sample size and proportion of measurements below the detection limit per study are given for each trace element in Supplementary Data 1.
Prior to analysis, we applied rank-based inverse normal transformation of the trace element concentrations after adjusting for age and sex (HUNT) using linear regression. Age, and the first ten genetic principal components (PCs) of ancestry were included as covariates in all the analyses. Sex, genotyping batch, geographical region (coast, town/fjord, or inland/mountain) and analysis laboratory (NTNU1, NTNU2, STAMI) were included as additional covariates in HUNT where appropriate. We performed genomic control correction of all GWAS results with an inflation factor λ > 1 (calculated from variants with MAF ≥ 0.01).
Variants with a minor allele count <10 or an imputation R2 < 0.3 were excluded from the analyses. After visual inspection of the Manhattan and quantile-quantile plots, we excluded the full set of results for Bi and Th due to excessive inflation of the p-values (Supplementary Figs. 42–45).
We used METAL v.2011-03-25115 to perform fixed-effect inverse variance weighted GWA meta-analysis of 14 trace elements (Al, As, Cd, Cr, Co, Cu, Pb, Mn, Hg, Mo, Se, Tl, Zn) analyzed in at least two of three studies: the HUNT, MoBa and/or publicly available summary statistics from the PIVUS study15, where 11 trace elements (Al, Cd, Co, Cu, Cr, Hg, Mn, Mo, Ni, Pb, Zn) have been measured in 949 seniors from Uppsala, Sweden, and rank inverse normalized trace element concentrations had been tested for association using a linear regression under an additive model, adjusting for triglycerides, cholesterol, gender and two principal components of ancestry. Age was not included as covariate because the PIVUS participants were of the same age.
Definition of associated variants, associated loci, and locus novelty
We considered genetic variants with p < 5 × 10−8 to be statistically significant at a genome-wide significance level and defined these as associated with the given trace element. Genetic loci were defined as 500 kilobase pairs to each side of genome-wide significant variants in the same region. A locus was classified as novel for a given trace element if it had not been reported for the respective trace element before. Previously published loci were identified with a literature search and a look-up in the NHGRI-EBI GWAS catalog116.
Index SNPs were identified as the genetic variant with the lowest p-value in each locus. We used PLINK v.1.9114 to identify variants in high LD (correlation r2 > 0.8) with the index variants, based on a reference panel of 5000 unrelated individuals in HUNT. The index variants and variants in high LD with the index variants were annotated using ANNOVAR v.2019Oct24117.
Sensitivity analyses
For trace elements where tobacco smoke (Cd, As, Pb, Cu)118, wine (Pb)119 or fish (As, Se)120 is a major source of human intake, or where alcohol is thought to regulate uptake or metabolism (Fe, Mn, Zn)121,122,123, we repeated the association analyses of the associated loci in HUNT, including smoking status (self-reported, never versus ever smokers (including ex-smokers, occasionally and daily smokers)), frequency of fat fish intake (self-reported) and units of alcohol per week (self-reported) as covariates, respectively (Supplementary Data 4). Further, since some of the analyzed samples were selected from type 2 diabetes cases, and type 2 diabetes is associated with hypomagnesemia88, we repeated the association analyses of magnesium including diabetes status (excluding type 1 diabetes) as covariate. Diabetes cases were defined as either any non-type 1 diabetes (self-reported) and/or fasting serum glucose ≥7.0 mmol/liter and/or serum glucose ≥ 11.0 mmol/liter 2 hours after first having fasted overnight and then consumed 75 g of glucose dissolved in ~3 dl water.
Heritability estimation and genetic and phenotypic correlation between trace elements
We used LD Score regression108,124 to estimate the narrow-sense SNP heritability (Vg/Vp ± 1SE, where Vg is the variance explained by the SNPs and Vp is the total phenotypic variance) of 11 trace elements from the GWA meta-analysis summary level SNP results, using LD scores estimated from individuals of European ancestry in the HUNT population. The analysis was performed in trace elements with a meta-analysis sample size > 5000, as recommended for the LD Score regression software. Each set of summary statistics were restricted to well imputed SNPs in HapMap3. Further, we used LD Score regression to estimate the genetic correlation between all pairs of 10 trace elements with an estimated SNP heritability > 0 and a sample size > 5000 in the GWA meta-analysis. The cadmium-mercury and molybdenum-cobalt correlations were excluded because the genetic estimates were out of bounds (higher than 1.0 or lower than −1.0). Additionally, we estimated the phenotypic correlation between the same pairs of trace elements in HUNT using Spearman rank correlation. Prior to the phenotypic correlations, the trace elements were corrected for median concentration per lab and log2 transformed.
Phenome-wide association tests (PheWAS)
We tested for associations of 34 common and low-frequency (MAF > 0.5%) trace element index variants (21 from meta-analysis and 13 from GWAS in HUNT) with 1326 phecodes, 167 continuous traits and 30 biomarkers in participants of white British ancestry in the UK Biobank, using publicly available summary statistics (https://pan.ukbb.broadinstitute.org/)125. Four variants (rs146233512 [gold], rs763064690 [indium], rs78394934 [iron], rs927502065 [tungsten]) were excluded because they were not tested in the UK Biobank. To correct for the total number of tests (n = 51,782), we used a Bonferroni corrected p-value significance threshold of 9.7 × 10−7.
Mendelian randomization (MR) of individual trace elements on selected health related outcomes
To explore potential causal associations of trace elements on selected outcomes, we used two-sample MR: We applied the inverse-variance weighted (IVW) method for trace elements with multiple index SNPs, and the Wald ratio method for trace elements with only one index SNP, as implemented in the TwoSampleMR126 and MRInstruments127 packages in R v3.6.3 and R v4.0.5. The exposures were selected based on robust genetic associations with trace elements in the current study, i.e., meta-analyzed index variants and the common index variant in the locus strongly associated with Sr in HUNT. The outcomes were selected based the availability of instrument summary statistics for a priori outcomes of interest highlighted in previous literature: Alzheimer’s disease (Mn, Pb, As, Cd, Cu, Se and Zn)19,20,24,25,26,27,128, Parkinson’s disease (Pb, As, Cd, Cu, Mn, Se, and Zn)19,28,29,30, multiple sclerosis (Zn, Mn, Se, Pb, As and Cd)21,22,23, autoimmune thyroid disease (Se)32, hypothyroidism (Se)32, osteoporosis (Cu)43,45, bone mineral density (Cu, Cd, Mn, Zn, and Sr)44,45,46, bone fracture (Sr)46, rheumatoid arthritis (Cd and Zn)33,34, type 2 diabetes (Cu, Mn, Se and Zn)37,38,39, colorectal cancer (Zn and Se)41,42 and prostate cancer (Se, Zn)40,42. We used SNP-exposure associations from the GWA meta-analysis (Cu, Zn, Mn, Se, Pb, As, Cd) or GWAS in HUNT (Sr) of each trace element (Supplementary Data 2), and SNP-outcome associations were collected from independent genome-wide summary level data129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146 (Supplementary Data 12). For the SNP associations with multiple sclerosis, we calculated the beta coefficient as the natural logarithm of the OR, and the standard error as \({SE}={|beta}/Q(\frac{p}{2})|\), where | | denotes absolute values, p denotes the p-value and \(Q\) denotes the inverse standard normal distribution. Outcomes obtained from https://pan.ukbb.broadinstitute.org125 were defined as: hypothyroidism (phecode: 244), and osteoporosis (phecode: 743.1). We estimated the variance explained and F-statistic for the instruments from a linear regression of the trace element concentrations versus the sum of estimated allele counts (dosages) for the trace element increasing alleles in HUNT (Supplementary Data 12). Except for manganese, the trace element instruments consisted of fewer than 4 SNPs, which provided limited opportunities for reliable sensitivity analyses.
Statistics and reproducibility
Unless otherwise specified, the statistical analyses were performed in R v.3.6.4. The sample size per trace element, proportion of measurements below the detection limit, and the distributions of trace element concentrations, age, and sex in HUNT, MoBa and PIVUS are reported in Supplementary Data 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Summary level data supporting the findings are available in the Supplementary materials and from https://dataverse.no/dataverse/ntnu/ (https://doi.org/10.18710/UYPCL0). Numerical source data underlying Fig. 1 can be found in Supplementary Data 7–9. The consent given by the participants in HUNT and MoBa does not open for storage of individual level data in repositories or journals. Researchers associated with Norwegian research institutes can apply for the use of HUNT data. Researchers from other countries may apply if collaborating with a Norwegian Principal Investigator. Information for data access can be found at https://www.ntnu.edu/hunt/data. Researchers who want access to MoBa data sets for replication should apply to https://helsedata.no. Access to data from either HUNT or MoBa requires approval from The Regional Committees for Medical and Health Research Ethics in Norway.
Code availability
We used the following publicly available software (URLs) to generate and analyze the data: R v3.6.3 and R v.4.0.5, BOLT-LMM v.2.3.4 (https://alkesgroup.broadinstitute.org/BOLT-LMM/downloads), PLINK 2.0 (https://www.cog-genomics.org/plink/2.0/), METAL v.2011-03-25 (https://genome.sph.umich.edu/wiki/METAL), ANNOVAR v.2019Oct24 (https://annovar.openbioinformatics.org) and LD Score Regression (https://github.com/bulic/ldsc).
References
Morrison, G. H., Cheng, K. L. & Grasserbauer, M. General aspects of trace analytical methods-IV. Recommendations for nomenclature, standard procedures and reporting of experimental data for surface analysis techniques. Pure. Appl. Chem. 51, 2243–2250 (1979).
Nielsen, F. H. Should bioactive trace elements not recognized as essential, but with beneficial health effects, have intake recommendations. J. Trace Elem. Med. Biol. 28, 406–408 (2014).
Elder, A., Nordberg, G. F. & Kleinman, M. Chapter 3—Routes of exposure, dose, and toxicokinetics of metals. in Handbook on the Toxicology of Metals 4th edn (eds Nordberg, G. F., Fowler, B. A. & Nordberg, M.) 45–74 (Academic Press, 2015).
Dev, S. & Babitt, J. L. Overview of iron metabolism in health and disease. Hemodial. Int. 21, S6–S20 (2017).
Li, Y. Copper homeostasis: emerging target for cancer treatment. IUBMB Life 72, 1900–1908 (2020).
Chen, P., Bornhorst, J. & Aschner, M. Manganese metabolism in humans. Front Biosci. (Landmark Ed.) 23, 1655–1679 (2018).
Caspersen, I. H. et al. Patterns and dietary determinants of essential and toxic elements in blood measured in mid-pregnancy: The Norwegian Environmental Biobank. Sci. Total Environ. 671, 299–308 (2019).
Simić, A. et al. Trace elements in whole blood in the general population in Trøndelag County, Norway: The HUNT3 Survey. Sci. Total Environ. 806, 150875 (2022).
Birgisdottir, B. E. et al. Essential and toxic element concentrations in blood and urine and their associations with diet: Results from a Norwegian population study including high-consumers of seafood and game. Sci. Total Environ. 463–464, 836–844 (2013).
Whitfield, J. B. et al. Genetic effects on toxic and essential elements in humans: arsenic, cadmium, copper, lead, mercury, selenium, and zinc in erythrocytes. Environ. Health Perspect. 118, 776–782 (2010).
Whitfield, J. B. et al. Evidence of genetic effects on blood lead concentration. Environ. Health Perspect. 115, 1224–1230 (2007).
Evans, D. M. et al. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum. Mol. Genet 22, 3998–4006 (2013).
Warrington, N. M. et al. Genome-wide association study of blood lead shows multiple associations near ALAD. Hum. Mol. Genet 24, 3871–3879 (2015).
Jäger, S. et al. Blood copper and risk of cardiometabolic diseases: a Mendelian randomization study. Hum. Mol. Genet. 31, 783–791 (2022).
Ng, E. et al. Genome-wide association study of toxic metals and trace elements reveals novel associations. Hum. Mol. Genet. 24, 4739–4745 (2015).
Borné, Y. et al. Genome wide association study identifies two loci associated with cadmium in erythrocytes among never-smokers. Hum. Mol. Genet. 25, 2342–2348 (2016).
Girelli, D. et al. Hemochromatosis classification: update and recommendations by the BIOIRON Society. Blood 139, 4–7 (2022).
Yang, W. et al. Genome-wide association and Mendelian randomization study of blood copper levels and 213 deep phenotypes in humans. Commun. Biol. 5, 405 (2022).
Chin-Chan, M., Navarro-Yepes, J. & Quintanilla-Vega, B. Environmental pollutants as risk factors for neurodegenerative disorders: Alzheimer and Parkinson diseases. Front. Cell Neurosci. 9, 124 (2015).
Rahman, M. A. et al. Exposure to environmental arsenic and emerging risk of Alzheimer’s disease: perspective mechanisms, management strategy, and future directions. Toxics 9, 188 (2021).
Sheykhansari, S. et al. Redox metals homeostasis in multiple sclerosis and amyotrophic lateral sclerosis: a review. Cell Death Dis. 9, 348 (2018).
Nirooei, E. et al. Blood trace element status in multiple sclerosis: a systematic review and meta-analysis. Biol. Trace Elem. Res. 200, 13–26 (2022).
Tsai, C. P. & Lee, C. T. C. Multiple sclerosis incidence associated with the soil lead and arsenic concentrations in Taiwan. PLoS One 8, e65911 (2013).
Branca, J. J. V., Morucci, G. & Pacini, A. Cadmium-induced neurotoxicity: Still much ado. Neural Regen. Res. 13, 1879–1882 (2018).
Lopes de Andrade, V., Marreilha dos Santos, A. P. & Aschner, M. Chapter Eleven—Neurotoxicity of metal mixtures. in Advances in Neurotoxicology 5 (eds. Aschner, M. & Costa, L. G.) 329–364 (Academic Press, 2021).
Lei, P., Ayton, S. & Bush, A. I. The essential elements of Alzheimer’s disease. J. Biol. Chem. 296, 100105 (2021).
Xie, Z., Wu, H. & Zhao, J. Multifunctional roles of zinc in Alzheimer’s disease. Neurotoxicology 80, 112–123 (2020).
Lee, C.-P., Zhu, C.-H. & Su, C.-C. Increased prevalence of Parkinson’s disease in soils with high arsenic levels. Parkinsonism Relat. Disord. 88, 19–23 (2021).
Pamphlett, R. et al. Concentrations of toxic metals and essential trace elements vary among individual neurons in the human locus ceruleus. PLoS One 15, e0233300 (2020).
Adani, G., Filippini, T., Michalke, B. & Vinceti, M. Selenium and other trace elements in the etiology of Parkinson’s disease: a systematic review and meta-analysis of case-control studies. Neuroepidemiology 54, 1–23 (2020).
Parmalee, N. L. & Aschner, M. Manganese and aging. Neurotoxicology 56, 262–268 (2016).
Rayman, M. P. Multiple nutritional factors and thyroid disease, with particular reference to autoimmune thyroid disease. Proc. Nutr. Soc. 78, 34–44 (2019).
Reyes-Hinojosa, D. et al. Toxicity of cadmium in musculoskeletal diseases. Environ. Toxicol. Pharmacol. 72, 103219 (2019).
Bonaventura, P., Benedetti, G., Albarède, F. & Miossec, P. Zinc and its role in immunity and inflammation. Autoimmun. Rev. 14, 277–285 (2015).
Marín Martínez, L., Molino Pagán, D. & López Jornet, P. Trace elements in saliva as markers of type 2 diabetes mellitus. Biol. Trace Elem. Res. 186, 354–360 (2018).
Fernández-Cao, J. C. et al. Zinc intake and status and risk of type 2 diabetes mellitus: a systematic review and meta-analysis. Nutrients 11, 1027 (2019).
Sanjeevi, N., Freeland-Graves, J., Beretvas, N. S. & Sachdev, P. K. Trace element status in type 2 diabetes: a meta-analysis. J. Clin. Diagn. Res. 12, OE01–OE08 (2018).
Eshak, E. S. et al. Manganese intake from foods and beverages is associated with a reduced risk of type 2 diabetes. Maturitas 143, 127–131 (2021).
Vinceti, M., Filippini, T. & Rothman, K. J. Selenium exposure and the risk of type 2 diabetes: a systematic review and meta-analysis. Eur. J. Epidemiol. 33, 789–810 (2018).
Costello, L. C. & Franklin, R. B. Zinc is decreased in prostate cancer: an established relationship of prostate cancer! J. Biol. Inorg. Chem. 16, 3–8 (2011).
Prasad, A. S., Beck, F. W. J., Snell, D. C. & Kucuk, O. Zinc in cancer prevention. Nutr. Cancer 61, 879–887 (2009).
Vinceti, M. et al. Selenium for preventing cancer. Cochrane Database Syst. Rev. 1, CD005195 (2018).
Zheng, J., Mao, X., Ling, J., He, Q. & Quan, J. Low serum levels of zinc, copper, and iron as risk factors for osteoporosis: a meta-analysis. Biol. Trace Elem. Res. 160, 15–23 (2014).
Alghadir, A. H., Gabr, S. A., Al-Eisa, E. S. & Alghadir, M. H. Correlation between bone mineral density and serum trace elements in response to supervised aerobic training in older adults. Clin. Interv. Aging 11, 265–273 (2016).
Aaseth, J., Boivin, G. & Andersen, O. Osteoporosis and trace elements—an overview. J. Trace Elem. Med. Biol. 26, 149–152 (2012).
Kołodziejska, B., Stępień, N. & Kolmas, J. The influence of strontium on bone tissue metabolism and its application in osteoporosis treatment. Int. J. Mol. Sci. 22, 6564 (2021).
Krokstad, S. et al. Cohort profile: the HUNT study, Norway. Int. J. Epidemiol. 42, 968–977 (2013).
Åsvold, B. O. et al. Cohort profile update: the HUNT study, Norway. Int. J. Epidemiol. 52, e80–e91 (2022).
Meltzer, H. M. et al. Low iron stores are related to higher blood concentrations of manganese, cobalt and cadmium in non-smoking, Norwegian women in the HUNT 2 study. Environ. Res. 110, 497–504 (2010).
Syversen, T. et al. Trace elements in the large population-based HUNT3 survey. Biol. Trace Elem. Res. 199, 2467–2474 (2021).
Rønningen, K. S. et al. The biobank of the Norwegian mother and child cohort study: a resource for the next 100 years. Eur. J. Epidemiol. 21, 619–625 (2006).
Paltiel, L. et al. The biobank in the Norwegian Mother and Child Cohort Study—present status. Nor. Epidemiol. 24, 29–35 (2014).
Magnus, P. et al. Cohort Profile Update: The Norwegian Mother and Child Cohort Study (MoBa). Int. J. Epidemiol. 45, 382–388 (2016).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
Rentschler, G. et al. Cadmium concentrations in human blood and urine are associated with polymorphisms in zinc transporter genes. Metallomics 6, 885–891 (2014).
Claus Henn, B. et al. Associations of iron metabolism genes with blood manganese levels: a population-based study with validation data from animal models. Environ. Health 10, 97 (2011).
Nieboer, E. & Richardson, D. H. S. The replacement of the nondescript term ‘heavy metals’ by a biologically and chemically significant classification of metal ions. Environ. Pollut. B 1, 3–26 (1980).
Wei, W. Q. et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One 12, e0175508 (2017).
Wang, C.-Y., Meynard, D. & Lin, H. Y. The role of TMPRSS6/matriptase-2 in iron regulation and anemia. Front Pharm. 5, 114 (2014).
Moksnes, M. R. et al. Genome-wide meta-analysis of iron status biomarkers and the effect of iron on all-cause mortality in HUNT. Commun. Biol. 5, 591 (2022).
Waikar, S. S. & Bonventre, J. V. Creatinine kinetics and the definition of acute kidney injury. J. Am. Soc. Nephrol. 20, 672–679 (2009).
Yang, F. et al. Characterization, mapping, and expression of the human ceruloplasmin gene. Proc. Natl. Acad. Sci. USA 83, 3257–3261 (1986).
Pinter, T. B. J. & Stillman, M. J. The zinc balance: competitive zinc metalation of carbonic anhydrase and metallothionein 1A. Biochemistry 53, 6276–6285 (2014).
Chen, P., Bowman, A. B., Mukhopadhyay, S. & Aschner, M. SLC30A10: a novel manganese transporter. Worm 4, e1042648 (2015).
Nebert, D. W. & Liu, Z. SLC39A8 gene encoding a metal ion transporter: discovery and bench to bedside. Hum. Genom. 13, 51 (2019).
Park, J. H. et al. SLC39A8 deficiency: biochemical correction and major clinical improvement by manganese therapy. Genet. Med. 20, 259–268 (2018).
Manousaki, D. et al. Genome-wide association study for vitamin D levels reveals 69 independent loci. Am. J. Hum. Genet. 106, 327–337 (2020).
Mathew, A., Bashir, S., de Roos, B. & Sneddon, A. A. Interaction of selenium and vitamin D and its relevance to atherosclerosis. Proc. Nutr. Soc. 78, E41 (2019).
Hu, L. et al. Mitochondrial protein profile in mice with low or excessive selenium diets. Int. J. Mol. Sci. 17, 1137 (2016).
Bárány, E. et al. Iron status influences trace element levels in human blood and serum. Environ. Res. 98, 215–223 (2005).
Tapiero, H., Townsend, D. M. & Tew, K. D. Trace elements in human physiology and pathology. Copper. Biomed. Pharmacother. 57, 386–398 (2003).
Ye, Q. et al. Influence of iron metabolism on manganese transport and toxicity. Metallomics 9, 1028–1046 (2017).
An, P. et al. TMPRSS6, but not TF, TFR2 or BMP2 variants are associated with increased risk of iron-deficiency anemia. Hum. Mol. Genet. 21, 2124–2131 (2012).
Lin, C. C. et al. Terminal uridyltransferase 7 regulates TLR4-triggered inflammation by controlling Regnase-1 mRNA uridylation and degradation. Nat. Commun. 12, 3878 (2021).
Camaschella, C., Nai, A. & Silvestri, L. Iron metabolism and iron disorders revisited in the hepcidin era. Haematologica 105, 260–272 (2020).
Yien, Y. Y. et al. FAM210B is an erythropoietin target and regulates erythroid heme synthesis by controlling mitochondrial iron import and ferrochelatase activity. J. Biol. Chem. 293, 19797–19811 (2018).
Tong, W. H. et al. TLR-activated repression of Fe-S cluster biogenesis drives a metabolic shift and alters histone and tubulin acetylation. Blood Adv. 2, 1146–1156 (2018).
Ast, T. et al. Hypoxia rescues frataxin loss by restoring iron sulfur cluster biogenesis. Cell 177, 1507–1521.e16 (2019).
Chen, M. H. et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182, 1198–1213.e14 (2020).
Roggenbeck, B. A., Banerjee, M. & Leslie, E. M. Cellular arsenic transport pathways in mammals. J. Environ. Sci. 49, 38–58 (2016).
Rigutto, S. et al. Activation of dual oxidases Duox1 and Duox2: differential regulation mediated by cAMP-dependent protein kinase and protein kinase C-dependent phosphorylation. J. Biol. Chem. 284, 6725–6734 (2009).
Meitzler, J. L. & Ortiz De Montellano, P. R. Structural stability and heme binding potential of the truncated human dual oxidase 2 (DUOX2) peroxidase domain. Arch. Biochem. Biophys. 512, 197–203 (2011).
Bell, S. et al. A genome-wide meta-analysis yields 46 new loci associating with biomarkers of iron homeostasis. Commun. Biol. 4, 156 (2021).
Suhre, K. et al. A genome-wide association study of metabolic traits in human urine. Nat. Genet. 43, 565–569 (2011).
Shi, X. et al. Metabolomic analysis of the effects of chronic arsenic exposure in a mouse model of diet-induced fatty liver disease. J. Proteome Res. 13, 547–554 (2014).
Madsen, M., Graversen, J. H. & Moestrup, S. K. Haptoglobin and CD163: Captor and receptor gating hemoglobin to macrophage lysosomes. Redox Rep. 6, 386–388 (2001).
Soranzo, N. et al. Common variants at 10 genomic loci influence hemoglobin A1C levels via glycemic and nonglycemic pathways. Diabetes 59, 3229–3239 (2010).
Gommers, L. M. M., Hoenderop, J. G. J., Bindels, R. J. M. & De Baaij, J. H. F. Hypomagnesemia in type 2 diabetes: a vicious circle? Diabetes 65, 3–13 (2016).
Mohlke, K. L. & Boehnke, M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum. Mol. Genet. 24, R85–R92 (2015).
Wang, D., Wang, Y., Madhu, S., Liang, H. & Bray, C. L. Total hemoglobin count has significant impact on A1C—Data from National Health and Nutrition Examination Survey 1999–2014. Prim. Care Diabetes 13, 316–323 (2019).
Marie, P. J., Ammann, P., Boivin, G. & Rey, C. Mechanisms of action and therapeutic potential of strontium in bone. Calcif. Tissue Int. 69, 121–129 (2001).
Ochoa, D. et al. Open targets platform: supporting systematic drug-target identification and prioritisation. Nucleic Acids Res. 49, D1302–D1310 (2021).
Hong, G. et al. The emerging role of MORC family proteins in cancer development and bone homeostasis. J. Cell Physiol. 232, 928–934 (2017).
Brissot, P. et al. Haemochromatosis. Nat. Rev. Dis. Prim. 4, 18016 (2018).
Pietrangelo, A. Hereditary hemochromatosis—a new look at an old disease. N. Engl. J. Med. 350, 2383–2397 (2004).
Benyamin, B. et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat. Commun. 5, 4926 (2014).
Evans, L. M. et al. Comparison of methods that use whole genome data to estimate the heritability and genetic architecture of complex traits. Nat. Genet. 50, 737–745 (2018).
Khan, M. A. K. & Wang, F. Mercury-selenium compounds and their toxicological significance: toward a molecular understanding of the mercury-selenium antagonism. Environ. Toxicol. Chem. 28, 1567–1577 (2009).
Oczkowski, M., Dziendzikowska, K., Pasternak-Winiarska, A., Włodarek, D. & Gromadzka-Ostrowska, J. Dietary factors and prostate cancer development, progression, and reduction. Nutrients 13, 496 (2021).
Bernard, A. & Lauwers, R. Metal‐induced alterations of δ‐aminolevulinic acid dehydratase. Ann. N. Y. Acad. Sci. 514, 41–47 (1987).
Morelli, A., Ravera, S., Calzia, D. & Panfoli, I. Impairment of heme synthesis in myelin as potential trigger of multiple sclerosis. Med. Hypotheses 78, 707–710 (2012).
Burgess, S. & Thompson, S. G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 40, 755–764 (2011).
Sanderson, E., Richardson, T. G., Morris, T. T., Tilling, K. & Smith, G. D. Estimation of causal effects of a time-varying exposure at multiple time points through Multivariable Mendelian randomization. PLoS Genet. 18, e1010290 (2022).
Longnecker, M. P. et al. Use of selenium concentration in whole blood, serum, toenails, or urine as a surrogate measure of selenium intake. Epidemiology 7, 384–390 (1996).
Długaszek, M. Studies on relationships between essential and toxic elements in selected body fluids, cells and tissues. Chem. Biol. Interact. 297, 57–66 (2019).
Rahil-Khazen, R., Botann, B. J., Myking, A. & Ulvik, R. Multi-element analysis of trace element levels in human autopsy tissues by using inductively coupled atomic emission spectrometry technique (ICP-AES). J. Trace Elem. Med. Biol. 16, 15–25 (2002).
Leivuori, M. & Niemistö, L. Sedimentation of trace metals in the Gulf of Bothnia. Chemosphere 31, 3839–3856 (1995).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Ferreira, M. A. et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat. Genet. 49, 1752–1757 (2017).
Iglesias, A. I. et al. Haplotype reference consortium panel: Practical implications of imputations with large reference panels. Hum. Mutat. 38, 1025–1032 (2017).
Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284–1287 (2016).
Helgeland, Ø. et al. Characterization of the genetic architecture of infant and early childhood body mass index. Nat. Metab. 4, 344–358 (2022).
Loh, P. R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
U.S. Department of Health and Human Services. Chapter 3–Chemistry and Toxicology of Cigarette Smoke and Biomarkers of Exposure and Harm. in How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease: A Report of the Surgeon General 40–41 (Centers for Disease Control and Prevention (US), 2010).
Towle, K. M., Garnick, L. C. & Monnot, A. D. A human health risk assessment of lead (Pb) ingestion among adult wine consumers. Int. J. Food Contam 4, 7 (2017).
Næss, S. et al. Mercury, lead, arsenic, and cadmium in Norwegian seafood products and consumer exposure. Food Addit. Contam. Part B Surveil 13, 99–106 (2020).
Skalny, A. V., Skalnaya, M. G., Grabeklis, A. R., Skalnaya, A. A. & Tinkov, A. A. Zinc deficiency as a mediator of toxic effects of alcohol abuse. Eur. J. Nutr. 57, 2313–2322 (2018).
Han, M., Böhlke, M., Maher, T. & Kim, J. Alcohol exposure increases manganese accumulation in the brain and exacerbates manganese-induced neurotoxicity in mice. Arch. Toxicol. 95, 3665–3679 (2021).
Dostalikova-Cimburova, M. et al. Role of duodenal iron transporters and hepcidin in patients with alcoholic liver disease. J. Cell Mol. Med. 18, 1840–1850 (2014).
Bulik-Sullivan, B. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Pan-UKBB team [Data set] https://pan.ukbb.broadinstitute.org (2020).
Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018).
Hemani, G. MRInstruments: data sources for genetic instruments to be used in MR. R package version 0.3.2. https://github.com/MRCIEU/MRInstruments/ (2020).
Balachandran, R. C. et al. Brain manganese and the balance between essential roles and neurotoxicity. J. Biol. Chem. 295, 6312–6329 (2020).
Saevarsdottir, S. et al. Multiomics analysis of rheumatoid arthritis yields sequence variants that have large effects on risk of the seropositive subset. Ann. Rheum. Dis. 81, 1085–1095 (2022).
Patsopoulos, N. A. et al. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science (2019) 365, eaav7188 (2019).
Huyghe, J. R. et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat. Genet. 51, 76–87 (2019).
Schumacher, F. R. et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 50, 928–936 (2018).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Morris, J. A. et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 51, 258–266 (2019).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022).
Saevarsdottir, S. et al. FLT3 stop mutation increases FLT3 ligand level and risk of autoimmune thyroid disease. Nature 584, 619–623 (2020).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).
Saevarsdottir, S. et al. Multiomics analysis of rheumatoid arthritis yields sequence variants that have large effects on risk of the seropositive subset. [Data set] https://download.decode.is/form/2022/RA_All.txt.gz (2022).
Patsopoulos, N. A. et al. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. [Data set] https://imsgc.net/ (2019).
Huyghe, J. R. et al. Discovery of common and rare genetic risk variants for colorectal cancer. [Data set] NHGRI-EBI GWAS Catalog https://www.ebi.ac.uk/gwas/studies/GCST012879 (2019).
Schumacher, F. R. et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. [Data set] http://practical.icr.ac.uk/blog/?page_id=8164 (2018).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. [Data set] https://drive.google.com/drive/folders/10bGj6HfAXgl-JslpI9ZJIL_JIgZyktxn (2019).
Morris, J. A. et al. An atlas of genetic influences on osteoporosis in humans and mice. [Data set] http://www.gefos.org/sites/default/files/Morrisetal2018.NatGen.SumStats.tar_0.gz (2019).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. [Data set] NHGRI-EBI GWAS Catalog https://www.ebi.ac.uk/gwas/studies/GCST90027158/ (2022).
Saevarsdottir, S. et al. FLT3 stop mutation increases FLT3 ligand level and risk of autoimmune thyroid disease. [Data set] https://download.decode.is/form/2020/AITD2020.gz (2020).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. [Data set] http://diagram-consortium.org/downloads.html (2018).
Acknowledgements
The Trøndelag Health Study (The HUNT Study) is a collaboration between HUNT Research Center (Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology), Trøndelag County Council, Central Norway Regional Health Authority, and the Norwegian Institute of Public Health. The genotyping in HUNT was financed by the National Institutes of Health (NIH R35 HL135824-03); University of Michigan; the Research Council of Norway; the Liaison Committee for Education, Research and Innovation in Central Norway; and the Joint Research Committee between St Olav’s hospital and the Faculty of Medicine and Health Sciences, NTNU. The genetic investigations of the HUNT Study are a collaboration between researchers from the K.G. Jebsen Center for Genetic Epidemiology, NTNU, and the University of Michigan Medical School and the University of Michigan School of Public Health. The K.G. Jebsen Center for Genetic Epidemiology was financed by Stiftelsen Kristian Gerhard Jebsen (SKGJ-MED-015); Faculty of Medicine and Health Sciences, NTNU, Norway. We thank HUNT participants for donating their time, samples, and information to help others. The Norwegian Mother, Father and Child Cohort Study (MoBa) is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. We are grateful to all the participating families in Norway who take part in this on-going cohort study. We thank the Norwegian Institute of Public Health (NIPH) for generating high-quality genomic data. This research is part of the HARVEST collaboration, supported by the Research Council of Norway (#229624). We also thank the NORMENT Center for providing genotype data, funded by the Research Council of Norway (#223273), South East Norway Health Authorities and Stiftelsen Kristian Gerhard Jebsen. We further thank the Center for Diabetes Research, the University of Bergen for providing genotype data and performing quality control and imputation of the data funded by the ERC AdG project SELECTionPREDISPOSED, Stiftelsen Kristian Gerhard Jebsen, Trond Mohn Foundation, the Research Council of Norway, the Novo Nordisk Foundation, the University of Bergen, and the Western Norway Health Authorities. The NIPH has contributed to funding of the Norwegian Environmental Biobank (NEB). The laboratory measurements have partly been funded by the Research Council of Norway through research projects (#275903 and #268465), and the human biomonitoring project HBM4EU, funded by the European Union’s Horizon 2020 research and innovation program under grant agreement No 733032.
Funding
Open access funding provided by Norwegian University of Science and Technology.
Author information
Authors and Affiliations
Contributions
M.R.M. analyzed the data and wrote the first draft of the manuscript. B.M.B., K.H., and C.J.W. conceived and designed the study. A.F.H., B.N.W., L.F.T., H.R., and L.B. contributed to analyses. A.S., T.S., T.H.N., and T.P.F. gave advice on phenotypes and phenotype definitions. K.H., C.J.W., P.M., P.R.N., O.A.A., A.L.B., T.S., A.S., and T.P.F. contributed to data acquisition. M.R.M., A.F.H., I.S., W.Z., J.Z., L.F., D.M.E., N.M.W., T.H.N., B.O.Å., T.P.F., C.J.W., K.H. and B.M.B. interpreted the results. All authors revised the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
The current study has been approved by the Norwegian Data Protection Authority and approved by the Regional Ethics Committee for Medical and Health Research Ethics in Northern Norway (REK Reference Number: 51604). All MoBa and HUNT participants have signed informed written consent. The MoBa cohort is currently regulated by the Norwegian Health Registry Act.
Peer review
Peer review information
Communications Biology thanks Kaixiong Ye and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Kaoru Ito and George Inglis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Moksnes, M.R., Hansen, A.F., Wolford, B.N. et al. A genome-wide association study provides insights into the genetic etiology of 57 essential and non-essential trace elements in humans. Commun Biol 7, 432 (2024). https://doi.org/10.1038/s42003-024-06101-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06101-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.