Abstract
Secreted laccases are important enzymes on a broad ecological scale for their role in mediating plant-microbe interactions, but within ascomycete fungi these enzymes have been primarily associated with melanin biosynthesis. In this study, a putatively secreted laccase, Sslac2, was characterized from the broad-host-range plant pathogen Sclerotinia sclerotiorum, which is largely unpigmented and is not dependent on melanogenesis for plant infection. Gene knockouts of Sslac2 demonstrate wide ranging developmental phenotypes and are functionally non-pathogenic. These mutants also displayed indiscriminate growth behaviors and enhanced biomass formation, seemingly as a result of their inability to respond to canonical environmental growth cues, a phenomenon further confirmed through chemical stress, physiological, and transcriptomic analyses. Transmission and scanning electron microscopy demonstrate apparent differences in extracellular matrix structure between WT and mutant strains that likely explain the inability of the mutants to respond to their environment. Targeting Sslac2 using host-induced gene silencing significantly improved resistance to S. sclerotiorum, suggesting that fungal laccases could be a valuable target of disease control. Collectively, we identified a laccase critical to the development and virulence of the broad-host-range pathogen S. sclerotiorum and propose a potentially novel role for fungal laccases in modulating environmental sensing.
Similar content being viewed by others
Introduction
Laccases are a broadly conserved class of multicopper oxidase (MCO), which are known for their capacity to oxidize otherwise recalcitrant phenolic compounds, both directly and indirectly through the activity of mediators1. While these enzymes have garnered industrial interest in recent years as drivers of xenobiotic bioremediation, they play a pivotal role in development across a range of species and can be found in the genomes of plants, animals, fungi, oomycetes, and bacteria1,2,3. One of their most important roles in natural ecosystems is to mediate the interplay between lignin production and degradation in interactions between plants and fungi. Higher plants utilize laccases in the biosynthesis and polymerization of lignin, which acts as a structural compound facilitating plant development and helps to defend plant tissue in response to pathogenesis3. Intriguingly, many wood rot fungi similarly use secreted laccases in the degradation of lignin, in which terminal phenolic lignin is directly oxidized by laccases and non-phenolic lignin is degraded through the activity of mediators, which are oxidized by the enzyme4. This interplay between production and degradation is critical to carbon cycling in the environment and the maintenance of healthy soils.
Unlike wood rot fungi, which are almost exclusively basidiomycetes, laccase activity within ascomycete fungi is typically associated with melanin/pigment deposition on fungal tissue, with most laccase knockout mutants within the phylum demonstrating reduced pigmentation5,6,7,8,9,10,11. The best studied examples of such laccases are Abr2 and ya, from Aspergillus fumigatus and Aspergillus nidulans, respectively, which function in melanization through the polymerization of phenolic monomers into mature melanin in fungal cell walls5. Intriguingly, the effect on fungal development and growth within these various knockouts is highly variable, likely due to the expanded repertoire of laccases and possible functional redundancy2. This redundancy has been suggested by the broad substrate overlap observed within individual species’ laccase repertoires and demonstrated in mutants of the Cochliobolus heterostrophus laccase ChMCO1, in which mutant pigmentation defects could be complemented through the chemical induction of other laccases in vitro11,12. In the plant pathogen Fusarium oxysporum f. sp. lycopersici, multiple distinct laccases were knocked out, and while some differences in sensitivity to oxidative/chemical stress could be measured, no clear changes to development or virulence were observed13.
In some cases, however, roles for specific laccase genes have been noted, such as in Colletotrichum gloeosporioides, Colletotrichum orbiculare, and Setosphaeria turcica where individual laccase gene knockouts result in reduced virulence on host plants6,7,8. These laccases and the laccase Mlac1 from the entomopathogenic fungus Metarhizium anisopliae are also deficient in appressorium formation6,7,8,10. Given that these laccases are almost always associated with pigmentation in ascomycetes, it has broadly been assumed that a lack of melanin/pigment deposition is driving the wide variety of phenotypes observed in knockouts, although a causal relationship has never been confirmed7,8,9. It is known, however, that laccase-mediated melanization is not a general requirement for pathogenicity given the unpigmented nature of the entomopathogenic fungus Beauveria bassiana. Laccases studied within this species appear highly involved in virulence, through either scavenging insect immune response-generated reactive oxygen species or the biosynthesis of the secondary metabolite oosporein14,15,16.
Sclerotinia sclerotiorum is a broad-host-range pathogen of dicotyledonous plants and is distinct from many of the previously mentioned fungi in that its melanin biosynthesis pathway has been relatively well studied. Three genes, encoding a scytalone dehydratase (SCD1), a trihydroxynaphthalene reductase (THR1), and a polyketide synthase (PKS13), with putative upstream roles in melanin biosynthesis have been knocked out and characterized17,18. Although mutants were deficient in melanization of either sclerotia (SCD1 and THR1) or compound appressoria (PKS13), no changes in lesion size were observed, suggesting that melanin deposition does not significantly alter S. sclerotiorum virulence17,18. This has a parallel in the closely related species Botrytis cinerea in which melanogenic genes are dispensable for proper growth and virulence19. Of note, the B. cinerea laccase Bclcc2 is broadly believed to be involved in the oxidation of environmental antimicrobials20,21. In this study we sought to characterize a S. sclerotiorum laccase gene, Sslac2, which is the putative ortholog of the B. cinerea laccase Bclcc2, and elucidate the role of this gene in fungal development and host-microbe interaction. Unlike Bclcc2, which lacks a clear role in development or virulence, our data shows that Sslac2 is crucial for the pathogen’s environmental sensing, as ΔSslac2 strains exhibit significant alterations in development, infection, and response to environmental cues21. This is unlike other fungal systems, such as C. orbiculare, where orthologous laccases between related species have been shown to be functionally interchangeable8. We also discuss the developmental and virulence phenotypes seen in ΔSslac2 mutants, demonstrate the potential for targeting fungal laccases to achieve increased resistance in host plants, and consider the broader context of laccases within fungal and pathogen biology.
Results
Sslac2 is the primary laccase expressed during pathogenesis
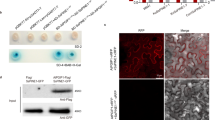
Some fungal laccases play a role in plant pathogenicity, but fungi typically maintain multiple laccases within their genomes, so an evaluation of the S. sclerotiorum laccase repertoire was performed2. Genomic analysis identified 7 putative laccases within the S. sclerotiorum genome, all of which contained predicted secretion signal domains, but which varied in length and cupredoxin domain architecture (Fig. 1A). While enzyme secretion is typically mediated by the presence of a signal peptide on the N-terminus of a protein, the C-termini of fungal laccases is additionally processed during secretion and is considered critical to laccase activity22. The canonical motif mediating this activity is a C-terminal DSGL, and Sslac2-6 contain a conserved DSGx motif in their C-termini, although this is apparently missing in Sslac1 and Sslac7 (Supplementary Fig. 1)22. To determine the laccases most important during S. sclerotiorum pathogenesis, a transcriptomic dataset generated from S. sclerotiorum infection of soybean at 24, 48, and 96 h post inoculation (HPI) was analyzed to identify which homologs were upregulated in-planta compared to an in vitro culture control23. Surprisingly, only Sslac2 appeared to be highly upregulated during infection, particularly in the early stages of disease development (Fig. 1B). Because the onset of infection requires the pathogen to interact with plant surfaces, it was considered whether any S. sclerotiorum laccases were distinctly upregulated on solid surfaces. To examine this, a separate transcriptomic dataset was analyzed from S. sclerotiorum grown on potato dextrose agar (PDA) or in potato dextrose broth (PDB), as these two media are nearly identical outside of surface rigidity produced by the agar24. Similar to pathogenesis, Sslac2 appeared to be the primary laccase upregulated (Supplementary Fig. 2). This upregulation was additionally validated through RT-PCR (Fig. 1C). In toto, these data indicate that Sslac2 is likely upregulated upon deposition of the pathogen on plant material or other inducive surfaces.
A Length and domain architecture of the seven putative laccases in the S. sclerotiorum genome. SP refers to a predicted secretion signal peptide. CuRo 1, 2, and 3 correspond to the three cupredoxin domains characterized in Melanocarpus albomyces (cd13854, cd13901, and cd13880, respectively). B Relative transcript fold change of the seven laccases identified in the S. sclerotiorum genome during infection of soybean when compared to in vitro culture growth. Fold change values are from the transcriptomic analysis performed in Westrick et al.23. C RT-PCR of Sslac2 and Histone 3 (H3) expression when grown on Potato Dextrose Agar (PDA) or in Potato Dextrose Broth (PDB).
Laccase and sclerotial production are abolished in ΔSslac2 knockout mutants
To evaluate the role of Sslac2 in disease and development, a CRISPR-Cas9 assisted method was used to generate two independent Sslac2 gene knockouts. Surprisingly, ΔSslac2 strains completely lost the ability to produce sclerotia, and this was accompanied by an increase in the formation of aerial hyphae (Fig. 2A). Laccase production of these mutants was also evaluated by growing WT (1980) and mutant strains on media containing 0.2 mM 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid (ABTS) or tannic acid and evaluating pigment production, as oxidation by laccases is known to generate a violet/blue pigment from ABTS and a brown pigment from tannic acid6,21. These pigments were clearly observed in plates growing the WT (1980) strain but were completely absent in the mutants (Fig. 2B). All strains were additionally grown on ABTS and copper sulfate (CuSO4) in an attempt to complement this laccase deficiency through the activation of other laccase homologs, as CuSO4 has been shown to induce laccase production in related species11,25. The addition of CuSO4 did not appear to increase laccase production in either of the knockout mutants, suggesting that Sslac2 is the primary secreted laccase utilized by S. sclerotiorum during growth and development on solid surfaces (Fig. 2B).
Sslac2 expression affects fungal tropism
Initial observations of ΔSslac2 strains indicated a growth defect, as reduced radial growth could be observed on PDA (Fig. 3A). In contrast, when grown in liquid culture on PDB ΔSslac2 mutants grew to a significantly higher mass than WT (1980), suggesting that the initial growth defect was specific to radial growth (Fig. 3B). A clear increase in aerial hyphae was noted in older cultures of ΔSslac2 (Fig. 2A), so ΔSslac2 and WT (1980) strains were grown for 2 weeks to assess whether the mutants were growing downward into the agar as well. At 2 weeks post inoculation (WPI), WT hyphae rested in a thin layer atop the agar, whereas ΔSslac2 strains visibly penetrated the agar surface (Fig. 3C). Remarkably, when grown on split plates, ΔSslac2 strains grew over the high barrier separating the plate sections and continued to grow into the neighboring chamber (Supplementary Fig. 3). When confronted with such barriers, the WT strain typically enters dormancy and produces survival structures (Supplementary Fig. 3). Thus, ΔSslac2 may be unable to sense the environmental triggers leading to dormancy and the production of sclerotia.
A WT and ΔSslac2-1 one day after inoculation on 60 mm PDA plates. B Hyphal dry weight of WT and mutant strains grown in PDB for 48 h. C Cross section of PDA colonized with WT and mutant strains 2 weeks post inoculation (WPI). Statistical analysis utilized a Student’s t-test on three biological replicates of each strain (***<0.001). Scale bars represent 1 mm. The source data underlying this figure can be found in Supplementary Data 1.
To assess if the agar penetration and slow radial growth phenotypes were caused by defects in directional growth or a failure to respond to dormancy triggers, all strains were grown on PDA with and without a top layer of sterile cellophane to halt agar penetration. While a significant defect in radial growth was measured on PDA, this defect disappeared when grown on cellophane (Fig. 4). These data and the noted sclerotial phenotype in the mutants suggest that the mutant grows in a random, indiscriminate manner, whereas the WT prioritizes radial growth and is able to respond to environmental cues. This suggests that Sslac2 plays a role in hyphal thigmotropism and the recognition of environmental triggers leading to the differentiation of morphological features.
A WT and ΔSslac2-1 strains 48 h after inoculation. B Quantification of colony area on PDA. C Quantification of colony area on PDA covered in cellophane. Statistical analysis utilized a Student’s t-test on three biological replicates of each strain (***<0.001). The source data underlying this figure can be found in Supplementary Data 1.
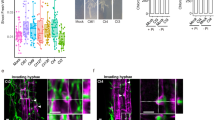
Sslac2 is required for penetration structure formation and oxalic acid (OA) production
Because Sslac2 is highly upregulated during the early stages of infection (Fig. 1B), we assessed the ability of ΔSslac2 mutants to generate canonical compound appressoria (the penetration structures formed by S. sclerotiorum to infiltrate plant tissue). Strikingly, ΔSslac2 mutants generated severely malformed and far fewer compound appressoria when grown on inductive surfaces such as glass slides, suggesting that Sslac2 may be required for proper signaling and differentiation of penetration structures on host surfaces (Fig. 5A, B). Exogenously applied cAMP has been shown to rescue appressorium defects in G-protein-coupled receptor mutants;26 however, the addition of cAMP to the glass slides used for compound appressorium production did not rescue this defect in the ΔSslac2 mutants.
A Comparison of compound appressorium formation between WT (1980) and ΔSslac2-1 strains. B Quantification of canonical compound appressoria and attempted compound appressoria generated by the WT and mutant strains, respectively. C Growth of the WT and mutant strains on bromophenol blue plates to qualitatively assess OA production. Statistical analysis utilized a Student’s t-test on three biological replicates of each strain (***<0.001). Scale bars represent 100 μm. The source data underlying this figure can be found in Supplementary Data 1.
After penetrating into host tissue, S. sclerotiorum secretes copious amounts of OA, an organic acid that facilitates host tissue acidification as well as directly induces cell death and subverts defenses in the host27,28,29. As OA is the dominant acid secreted by S. sclerotiorum, its production can be assessed using bromophenol blue plates which shift from blue to yellow as the medium is acidified. A failure to acidify bromophenol blue plates has been broadly used as a hallmark of OA deficiency in S. sclerotiorum, and while ΔSslac2 mutants were capable of acidifying the media to a limited degree, they were clearly deficient relative to WT (1980) (Fig. 5C)18,30,31.
Given the importance of both penetration and OA production in S. sclerotiorum pathogenic development, it is unsurprising that these mutants were essentially non-pathogenic (Fig. 6A). To assess whether this defect in pathogenicity was due only to the loss of host penetration, WT and ΔSslac2 strains were inoculated onto damaged soybean leaves, which provide direct access to the hosts tissues and have been shown in other systems to complement penetration structure defects (Fig. 6B). Additionally, in order to assess the role that OA may be playing in the ΔSslac2 virulence, mutant strains were compared with a previously described ΔSsoah1 strain deficient in OA production due to the deletion of oxaloacetate acetylhydrolase, which converts oxaloacetic acid to OA31. As expected, WT (1980) was capable of infecting both damaged and undamaged soybean leaves and ΔSsoah1 colonization was limited to the area around the plant vasculature as previously reported30. In contrast, even with damaged tissue, ΔSslac2 strains were incapable of inducing lesions on soybean leaves, indicating that the previously observed loss of pathogenicity is not solely the result of defects in penetration and acidification (Fig. 6B). Because soybean is a moderately resistant host of S. sclerotiorum, we additionally tested ΔSslac2 infection of the more susceptible host Nicotiana benthamiana. No lesions were observed on undamaged leaves, but limited lesions could be seen on damaged leaves, albeit to a far lesser extent than either WT (1980) or ΔSsOah1 (Fig. 6C). While the ΔSslac2 strain is able to marginally colonize wounded N. benthamiana leaves, overall, it is clear that this mutant’s virulence defect extends beyond penetration structures and OA production and likely involves the loss of other virulence factors or an inability to adequately respond to a hostile host environment.
A Comparison of WT (1980), ΔSslac2-1, and ΔSslac2-2 during infection of soybean leaves. B Infection of damaged and undamaged soybean tissue by WT (1980), ΔSslac2-1, ΔSslac2-2, and ΔSs-oah1. C Infection of damaged N. benthamiana at 48 and 96 h post infection (HPI) by WT (1980), ΔSslac2-1, ΔSslac2-2, and ΔSs-oah1.
Transcriptional activity of ΔSslac2 is less responsive to environmental factors
Many of the defects observed in ΔSslac2 mutants suggest a failure to respond to environmental factors, but it’s unknown if this failure is specific to features such as pH and surface recognition or more generalized. To address this question, an RNA sequencing analysis was conducted to compare WT (1980) and ΔSslac2-1 strains in both defined glucose minimal media (GMM) and GMM with the addition of soybean green stem extract (GrSt) to simulate the presence of plant material. A principal component analysis was conducted to examine the variation in gene expression profiles, and unsurprisingly, a large transcriptional shift was observed in the WT strain with the addition of GrSt, reflecting the pathogen’s response to additional proteins, carbohydrates, and plant chemical signals (Fig. 7A). Remarkably, the ΔSslac2-1 is largely non-responsive to the addition of GrSt (Fig. 7A). This is further visualized by the quantification of differentially expressed genes in ΔSslac2-1 and WT strains, in which a drastically increased number of genes demonstrate both up- and down-regulation in the WT when exposed to GrSt (Fig. 7B, Supplementary File 1). To further validate the noted deficiency in OA production by ΔSslac2 mutants, we analyzed the expression of the critical enzyme Ssoah1 (Sscle_10g075560), revealing ~160x lower expression than the WT (Fig. 5C; Supplementary Data 2).
A PCA plot comparing expression profiles of WT (1980) and ΔSslac2-1 grown in GMM or GMM + GrSt. B Number of genes found to be differentially regulated between GMM and GMM + GrSt for each strain. Colors within the bars represent genes categorized by their log2 fold-change (FC) (FDR < 0.05; log2 FC > 1 or <−1). Up = upregulated in GMM + GrSt vs GMM. Down = downregulated in GMM + GrSt vs GMM. The source data underlying Fig. 7 can be found in Supplementary Data 2.
ΔSslac2 environmental sensing defect is likely due to alterations in fungal cell wall structure
Laccases within ascomycete fungi have primarily been associated with detoxification of exogenous compounds and cell wall modification, so the sensitivity of Sslac2 to antifungal plant defense compounds and cell wall stressors was assessed5,6,8,9,10,11,12,16,21. WT and ΔSslac2 mutants were grown on PDA supplemented with different upstream and downstream components of the plant phenylpropanoid pathway21,32. ΔSslac2 mutants were significantly more susceptible to all tested compounds (Supplementary Fig. 4A). This result is somewhat surprising, as the ΔBclcc2 strain is known to show greater resistance to the phytoalexin resveratrol than WT B. cinerea.
To assess the susceptibility of ΔSslac2 mutants to cell wall stressors, all strains were additionally grown on plates amended with poacic acid, calcofluor white (CFW), or Congo red (Supplementary Fig. 4B)33. Both mutants demonstrated significantly greater susceptibility to poacic acid and CFW and moderately increased susceptibility to Congo red (Supplementary Fig. 4B). As this difference in susceptibility could be due to either alterations in hyphal cell wall architecture or a general difficulty in responding to chemical stresses, as is suggested by Supplementary Fig. 4A, we attempted to form protoplasts of both strains using a typical cocktail of cell wall degrading enzymes from Trichoderma harzianum. While protoplasts could be efficiently generated from WT (1980) hyphae, ΔSslac2 hyphae remained largely intact, suggesting that alterations to the hyphal cell wall may be reducing the efficiency of the enzyme cocktail (Supplementary Fig. 4C).
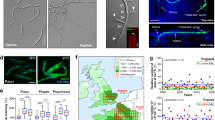
Additional evidence for alterations to cell wall structure can be seen in the assessment of surface hydrophobicity through hyphal wetting, which show dramatically decreased hyphal hydrophobicity in the ΔSslac2 mutants (Supplementary Fig. 5). This alteration may help to additionally explain the accelerated growth of the mutants in liquid culture, as nutrient exchange through liquid media may be more efficient through a more hydrophilic hyphal cell wall, as was noted in hydrophobin mutants of Trichoderma spp34. An assessment of the hyphal surface was made with scanning electron microscopy, and while clear differences in the cell surface were difficult to discern, obvious changes to hyphae and hyphal growth patterns were observed (Fig. 8A). WT growth typically consists of thin hyphae branching to evenly disperse across an environment. In contrast, ΔSslac2 mutant hyphae were noticeably thicker in diameter and often found to grow in bundles (Fig. 8A). This may be due to increasingly hydrophilic hypha adhering to one another and may be either a cause or an effect of the previously noted defect in growth directionality.
A Scanning electron micrographs of WT and mutant strains of S. sclerotiorum. White scale bars correspond to 10 microns. B Transmission electron microscopy (TEM) photos of the cell walls and extracellular matrices of WT (1980), ΔSslac2-1, and ΔSslac2-2. R Resin, FC fungal cell, CW cell wall, arrows denote the fungal extracellular matrix. Black scale bars correspond to 200 nm.
A higher resolution analysis of cell well structure was conducted through transmission electron microscopy (TEM) to assess potential changes in cell wall architecture in the mutant strains. While no structural modifications could be confirmed in the ΔSslac2 strains given the inherent variability in S. sclerotiorum cell walls relative to hyphal age, a clear textural difference in the extracellular matrix coating the cells was seen (Fig. 8B; Supplementary Fig. 6). This apparent failure by the laccase mutants to properly assemble their exterior cell wall ultrastructure may contribute to the previously noted changes in their physiochemical properties and altered responses to external environmental signals (Fig. 7; Supplementary Fig. 5).
Host-induced gene silencing (HIGS) targeting Sslac2 induces resistance to S. sclerotiorum in soybean
Given the clear importance that Sslac2 plays in both the virulence and development of S. sclerotiorum, we considered the value that this gene may have as a target of HIGS-mediated disease control. Targeted gene silencing using stable transgenic lines has been shown to provide robust disease control against S. sclerotinia in some pathosystems, so Sslac2 gene silencing was first assessed using a viral vector. To achieve this, a segment of Sslac2 was cloned into a modified Bean pod mottle virus (BPMV) vector which was then used to biolistically inoculate soybean seedlings35. Infected material was used to inoculate new seedlings with either empty vector (EV) or Sslac2-targetting BPMV, which were subsequently infected with WT S. sclerotiorum. Stem lesions were measured over a week of infection, and significantly smaller lesions were observed on plants in which Sslac2 was being silenced (Fig. 9). The reddening seen on BPMV-Sslac2 stems in response to infection indicates a successfully induced resistance response by soybeans against S. sclerotiorum invasion. This finding indicates that the gene silencing of Sslac2 likely serves to limit the virulence of the pathogen, while allowing the host time to mount a more successful defense (Fig. 9)32.
A Visual appearance of lesions from S. sclerotiorum infection soybeans containing an empty vector (EV) strain of Bean pod mottle virus (BPMV) and soybeans containing BPMV targeting Sslac2. B Quantification of lesion lengths from infection of BPMV-EV and BPMV-Sslac2 soybeans. Statistical analysis utilized a Student’s t-test on six biological replicates of each construct. Experiment was repeated twice (*<0.05, **<0.01, ***<0.001). The source data underlying this figure can be found in Supplementary Data 1.
Discussion
In this study a novel laccase, Sslac2, was identified within the broad-host-range fungal pathogen S. sclerotiorum and found to be critical for the proper regulation of an array of developmental and virulence traits. The phenotypes observed from ΔSslac2 mutants appear surprisingly expansive, in contrast, some of these phenotypes have only been noted in individual laccase mutants of other ascomycetes (Fig. 10). The mutants most closely resembling ΔSslac2 are that of ΔStLAC2, from the northern corn leaf blight pathogen S. turcica, and Lac1, from the anthracnose pathogen C. gloeosporioides (Fig. 10). In both cases, laccase knockout mutants were similar to ΔSslac2 in that they had severe defects in appressorial production, leading to a loss of pathogenicity on intact plant tissue and drastically reduced virulence on wounded tissue6,7. ΔStLAC2 additionally displays a similar increased susceptibility to cell wall stressors and altered hydrophobicity, with an identical hyphal wetting phenotype6. A primary difficulty in comparing the spectrum of biological roles mediated by individual laccases is that many of these functions have not been assessed across all species. In the mulberry pathogen Scleromitrula shiraiana, knockdown mutants of the laccase Sh-lac generated significantly less OA than WT strains, similar to ΔSslac29. Unfortunately, environmental acidification has not been assessed outside of these two systems, so it is unknown whether this is a common feature of laccase mutants (Fig. 10).
Many of the assessed laccase mutants have some defect in melanin/pigment formation, and it is typically suggested that subsequent phenotypic alterations are due to melanogenic defects, but we argue that this explanation is unlikely in S. sclerotiorum (Fig. 10)6,8,9. S. sclerotiorum is known to generate melanin through the DHN melanin pathway and multiple components of this pathway have been deleted and characterized17,18. Although a loss of melanization was seen in sclerotia and compound appressoria, there was no effect on fungal virulence. This agrees with expansive studies on the melanogenic genes from the closely related fungus B. cinerea, none of which was found to play a clear role in infection19.
S. sclerotiorum has seven putative laccases in its genome, all of which have predicted secretion signal peptides on their N-termini, but only five of which (Sslac2-6) contain the canonical C-terminal motif DSGx (Fig. 1A; Supplementary Fig. 2)22. While Sslac2 is the only of these laccases with clear induction in-planta from our transcriptomic analysis, existing S. sclerotiorum expressed sequence tag libraries suggest that other laccases are expressed at distinct developmental stages (sclerotial development, carpogenic germination apothecial formation), possibly playing a more classic role in melanin deposition (Fig. 1B)36. Given the overlapping substrate ranges of many laccases, some amount of redundancy is expected and has been demonstrated in other systems11,12. Additionally, it was demonstrated in C. orbiculare that ΔLac2 mutants could be functionally complemented with orthologous laccases from related species. The unexpected severe phenotype in ΔSslac2 mutants is surprising given that the closest characterized ortholog, B. cinerea laccase Bclcc2, does not significantly contribute to development or virulence. Knockout mutants of Bclcc2 displayed no alterations in virulence and appear to be primarily involved in oxidation of environmental phenolics, although both ΔSslac2 and ΔBclcc2 strains show abolished tannic acid oxidation activity20,21. Laccases are a class of multicopper oxidase requiring a core of copper to catalyze oxidation, and while no phenotypes similar to ΔSslac2 have been observed in B. cinerea laccase mutants, a nearly identical phenotype has been observed in mutants of the copper transporter BcCcc237. Knockouts of BcCcc2 show reduced melanization, malformed and reduced compound appressoria, no pathogenicity on unwounded tissue, and reduced virulence on wounded tissue, all of which we observed in ΔSslac2 strains. These deficiencies were attributed to the failure of the ΔBcCcc2 mutant to provide copper to copper-containing enzymes, of which laccases were major likely recipients, and suggests that B. cinerea may utilize laccases other than Bclcc2 to perform similar functions to Sslac237.
Our data show that ΔSslac2 mutants are reduced in hydrophobicity relative to WT strains, as observed through enhanced hyphal wetting (Supplementary Fig. 5). Typically, surface hydrophobicity is largely mediated by the presence of surface hydrophobins, the presence of which are both ubiquitous among and unique to filamentous fungi38. It’s surprising that surface hydrophobicity would be affected after the deletion of a single laccase because the S. sclerotiorum hydrophobins should still be intact; however, TEM analysis of the WT and ΔSslac2 strains suggests that the extracellular matrix (ECM) of mutant strains may be more intrinsically disordered than the WT (Fig. 8B; Supplementary Fig. 6). Such a change in the ECM, the component of hyphae that interacts most directly with the environment, may help to explain why the mutant hyphae were observed to “stick” to one another and why hydrophobins may by incapable of maintaining hydrophobicity. This loss of hydrophobicity may also explain the enhanced growth of the mutants in liquid culture, as a similar phenotype has been observed in more hydrophilic strains of Trichoderma34. Additionally, modifications to the ECM could undermine fungal cell receptor activity, including G-protein-coupled receptors (GPCRs) or ion channels. This could help to explain the aberrant environmental sensing phenotypes observed in this study. The defect in environmental sensing might also explain many of the developmental phenotypes we observed, including the observed reduction in OA secretion and canonical compound appressoria formation (Fig. 5). This study is the first to provide evidence for roles of fungal laccases in ECM formation and environmental sensing.
Other Sslac2-driven modifications to cell wall composition are likely, as we show that ΔSslac2 is very resistant to protoplasting by a cocktail of cell wall degrading enzymes (Supplementary Fig. 4C). Such a phenotype was additionally observed in knockout mutants of Rho1, a GTPase, from F. oxysporum, which displayed strikingly similar developmental defects to ΔSslac2, including a growth defect specific to solid surfaces, attenuated virulence on plants, increased susceptibility to cell wall stressors, and resistance to protoplasting39. Broadly, Rho1 orthologues in yeast are known to play a role in polarized cell growth through regulation of the actin cytoskeleton and can directly interact with the β-1,3-glucan synthase in fungal cell walls39. This activity likely extends to filamentous fungi as well, given the growth morphology and cell wall alterations observed in ΔRho1 from F. oxysporum) and ΔRhoA (A. nidulans), and supports a connection between cell wall composition and the thigmotropic phenotypes observed in ΔSslac2 (Figs. 7 and 8; Supplementary Fig. 3-6)39,40. The precise interplay between cell wall biosynthesis and thigmotropism is currently unclear, but such alterations may additionally play a role in the attenuated virulence of ΔSslac2 strains, as plants are known to activate defenses in response to fungal cell wall components (Fig. 6)41. If the cell wall of ΔSslac2 mutants were modified in a way that increases the release of such components, as was predicted in ΔRho1, then a reduction in virulence would be expected. Moreover, compromised virulence in the mutants could be the result of heightened susceptibility and/or impaired transcriptional response to plant-produced antifungal compounds, as evidenced by in vitro plate assays and transcriptomic analysis (Supplementary Fig. 4A; Fig. 7).
Given the clear and pivotal role that Sslac2 plays in virulence, we chose to evaluate it as a target of host induced gene silencing (HIGS) to achieve disease resistance in soybeans. An initial screen using a viral vector confirmed that silencing Sslac2 significantly increases plant resistance to S. sclerotiorum (Fig. 9). A drawback of such an approach is that viral vectors are often only capable of partial gene silencing, given the relatively low viral titer in plants used for this assay32. We are currently generating stable transgenic soybean lines expressing hairpin dsRNA targeting Sslac2 and will be evaluated for resistance to S. sclerotiorum infection.
In summary, this study characterizes a single fungal laccase (Sslac2) critical for proper development, virulence, and environmental sensing in the broad-host-range fungal plant pathogen S. sclerotiorum. Future work will focus on elucidating the chemical substrates of Sslac2 and precise mechanisms by which this protein mediates fungal thigmotropism and responses to environmental stimuli. Efforts will also focus on evaluating Sslac2 and other fungal laccases as targets of gene silencing for disease control.
Materials and methods
Plant and fungal growth
All soybean and N. benthamiana plants were maintained in the greenhouse or growth chamber at 24 ± 2 °C with 16-h light/8-h dark photoperiod cycle. Plants were watered daily and supplemented with fertilizer (Miracle-Gro) every week.
All S. sclerotiorum cultures were maintained on potato dextrose agar (PDA) plates or PDA supplemented with 50 μg/ml hygromycin in the case of knockout strains. Liquid cultures were grown in potato dextrose broth. Cellophane assays were conducted by autoclaving pre-cut rings of cellophane prior to being placed on 100 × 15 mm petri dishes containing PDA.
Fungal transformation
The wild-type S. sclerotiorum strain used in the generation of all mutants was the type strain referred to as 1980 (ATCC Product Name: 18683), as this strain was used to generate the genome sequence used for genomic and RNA-Seq analysis (NCBI BioProject Acc. PRJNA15530). The ΔSsoah1 mutant was additionally generated in a WT (1980) background31. Gene knockouts were generated in S. sclerotiorum using a CRISPR-Cas9 method in combination with a modified form of the protocols described in Rollins et al.42. and Westrick et al.43 Split markers targeting Sslac2 were generated using polymerase chain reaction (PCR) by amplifying 5–600 bp regions upstream (Sslac2-LF-F and Sslac2-LF-R) and downstream (Sslac2-RF-F and Sslac2-RF-R) regions of Sslac2. These amplicons were designed to contain 20 bp sequences with homology to the 5′ and 3′ regions, respectively, of the hygromycin resistance cassette (HygR; 1.8 kb) found in pCRISPR-Cas9-TrpC-Hyg18. The hygromycin resistance cassette was amplified using the two flanking regions and the HygR were connected through fusion PCR as described in Szewczyk et al.44. for a product of ~3kb44. Split markers were generated from this product by using primers internal to HygR (Hyg Split F and Hyg Split R) in conjunction with Sslac2-RF-F and Sslac2-RF-R, yielding two amplicons with an overlapping region of ~400 bp.
Two small guide RNAs (sgRNAs) targeting Sslac2 were designed using the E-CRISP Design Tool (http://www.e-crisp.org/E-CRISP/index.html), generated using the GenCrispr sgRNA Screening Kit (L00689; Genscript Biotech Corp.), and diluted to a concentration of 4 μM. Alt-R S.p. Cas9 nuclease 3NLS (1081058; IDT) was diluted to a concentration of 4 μM and combined with sgRNA at a 1.2-to-1 ratio (3.6 μl of sgRNA to 3 μl of Cas9 protein) and incubated at room temperature for 5 min to assemble the RNP complex. These complexes were combined with 1 μg of each split-marker and transfected into S. sclerotiorum protoplasts using the polyethylene glycol (PEG) transformation described in Rollins et al.42.
Transformants capable of surviving on PDA containing 50 μg/ml hygromycin were subjected to 5 rounds of hyphal tipping before undergoing DNA extraction to confirm the replacement of Sslac2 with the HygR marker. Primers internal and external to Sslac2 and HygR were used to confirm deletion and primers targeting Histone 3 (H3 F and H3 R) were used as a control (Supplementary Fig. 7: Supplementary Table 1). DNA was extracted using the cetyl trimethyl ammonium bromide (CTAB) method described in Talbot et al.45.
Virus-induced gene silencing (VIGS) assay and construct generation
A modified Bean pod mottle virus (BPMV) vector was used to assess Sslac2 as a potential target of VIGS for disease control35. In order to silence Sslac2 (Sscle_03g023030; SS1G_00974; XM_001598835), a 267 base pair sequence was selected within the mRNA of S. sclerotiorum strain 1980 (GenBank Accession XM_001590428). Total RNA was extracted from S. sclerotiorum using the Maxwell® RSC Plant RNA Kit, and cDNA was generated using an AMV first strand cDNA synthesis kit (New England Biolabs, Catalog # E6550). The segment was amplified through PCR with PstI and BamHI restriction sites incorporated onto the double-stranded cDNA using specific primers (PstI-VIGS-F and BamHI-VIGS-R) (Supplementary Table 1). The amplicon underwent gel purification (QIAquick Gel Extraction Kit®, QIAGEN), then both the amplicon and the viral vector RNA2 plasmid (pBPMV-IA-V1) were subjected to restriction digestion with PstI/BamHI before being ligated to form BPMV-Sslac235. The vector plasmids were then transformed into DH5α competent cells using 5 μl of the purified ligation product per 50 μl of competent cells, a 30 min ice incubation, 45 s heat shock in a 42 °C water bath, and incubation in 500 μl of Luria broth (LB) for 1 h at 37 °C. Using glycerol stocks, midi preparations were conducted (Fast Ion Plasmid Midi Kit®, IBI Scientific) for subsequent biolistic inoculations. Biolistic inoculations were performed as described in McCaghey et al. 46.
Plant disease assays
Soybeans: For detached leaf assays, leaves were taken from the first trifoliate of 5–6-week-old plants (cv. Williams 82) and placed in petri dishes containing two layers of filter paper/paper towel and 7 milliliters of sterile water. Leaves were inoculated near the center with agar plugs of actively growing wild-type (1980) or mutant S. sclerotiorum. Damaged leaves were scored four times in a crisscross pattern using a sterile scalpel directly under the agar plug. Petri dishes were wrapped in parafilm, placed at room temperature, and photographed every twenty-four hours. All disease assays were conducted in triplicate.
For VIGS assays, 10–14 day old plants (cv. Traff) were rub-inoculated with lyophilized leaves infected with BPMV-Sslac2 as described in McCaghey et al. 46. Plants were then allowed to grow an additional 5 weeks prior to being inoculated with WT (1980) S. sclerotiorum through cut petioles. Briefly, deep-well plates (100 mm × 55 mm) containing 75 ml of PDA were inoculated with WT (1980) and allowed to grow for two days. The petiole of the first trifoliate was cut ~2 cm from the main stem with a razor and a plug from the leading edge of mycelia was punctured using an inverted one-ml pipette tip. The inverted pipette tip with agar plug, was then slid onto the excised petiole. Lesions were measured with digital calipers 4–7 Days post inoculation (DPI). Three plants were inoculated per 1 L pot and 5 pots were tested for both the empty vector and BPMV-Sslac2 infected plants. The experiment was replicated twice.
N. benthamiana: Leaves were taken from 6–7 week-old plants and placed in petri dishes containing two layers of filter paper/paper towel and 7 ml of sterile water. Leaves were inoculated near the center with agar plugs of actively growing wild type (1980) or mutant S. sclerotiorum. Damaged leaves were scored four times in a crisscross pattern using a sterile scalpel directly under the agar plug. Petri dishes were wrapped in parafilm, placed at room temperature, and photographed every twenty-four hours.
Stress Testing
All stress test assays were performed in 60 mm × 15 mm petri dishes containing PDA supplemented with benzoic acid (150 μg/ml), cinnamic acid (150 μg/ml), ferulic acid (500 μg/ml), resveratrol (200 μg/ml), congo red (50 μg/ml), calcofluor white (250 μg/ml), or poacic acid (50 μg/ml). Cultures were allowed to grow for 24–48 h prior to being photographed. Colony areas were quantified in ImageJ47. All comparisons were tested in triplicate and the statistical significance of colony area differences was assessed with a Student’s t-test.
Protoplasting assay
Protoplasting was done using a modified protocol from Rollins et al.42 Briefly, three agar plugs of each strain were grown for two days in petri dishes containing PDB at room temperature. Agar plugs were excised with tweezers and a scalpel, and each sample was washed with water and then protoplast buffer (0.8 M MgSO4·7H2O, 0.2 M Sodium citrate·2H2O, pH 5.5). Samples were roughly chopped with a sterile razor blade and placed in 17 ml of protoplast buffer. For each sample, 100 mg of lysing enzyme from Trichoderma harzianum (Sigma Aldrich, L1412) was dissolved in 3 ml of Novozyme buffer (1 M Sorbitol, 50 mM Sodium citrate·2H2O) and then filtered through a 0.45 μm filter directly into to protoplast buffer containing the sample. All samples were incubated in a 28 °C shaker at 120 RPM for 3 hours before being filtered through four layers of Miracloth to collect protoplasts. Samples were centrifuged at 3000 × g for 10 min to pellet protoplasts, which were subsequently reconstituted in 1 ml protoplast buffer before quantification with a hemocytometer. All samples were run in triplicate. The statistical significance of protoplast count differences was assessed with a Student’s t-test (*<0.05, **<0.01, ***<0.001). The experiment was replicated twice.
Assay of laccase production
Laccase production was assessed using either 0.2 mM 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid (ABTS), 0.2 mM ABTS + 0.6 mM CuSO4, or 2.5 mg/ml tannic acid amended to PDA plates (100 mm × 15 mm)6,21. Agar plugs of actively growing hyphae from each strain was used to inoculate the center of plates and were allowed to grow for 24 h prior to being photographed. Laccase production was associated with the accumulation of bluish/purple pigments in the case of ABTS and brown pigment in the case of tannic acid. CuSO4 amendment was utilized as a known inducer or laccase production25.
Hydrophobicity assay
Three-week-old cultures of all strains were topped with 100 μl of either H2O or H2O + 0.01% Triton X-100. Photos were taken 2 minutes after treatment to observe mycelial soaking.
Compound appressoria observation/quantification
Agar plugs of actively growing mycelium were collected and placed face down on glass slides and then incubated in the dark at room temperature in a sealed container overnight (~16 h). A scalpel was used to cut the agar plugs away from hyphae which had grown onto the glass slide and the plugs were removed. Mycelia were stained with 0.05% trypan blue for 1 h before being rinsed with water to remove the dye. As the mutant strains are substantially less hydrophobic than the wild-type and therefore attach poorly to the glass slide, the water rinse was done by carefully removing the dye and replacing it with water. This process was repeated 10× for each agar plug. Stained mycelia were then observed under a compound microscope to observe the production of compound appressoria. The statistical significance of compound appressorium count differences was assessed with a Student’s t-test.
RT-PCR of Sslac2 on PDA and PDB
WT (1980) S. sclerotiorum was grown for 48 h at room temperature either in a 125 ml Erlenmeyer flask containing PDB on a rotary shaker (120 RPM) or on a plate of PDA (100 mm × 15 mm). Samples in PDB were then removed from the broth and flash frozen in liquid nitrogen and ground into a powder with a mortar and pestle. Samples on PDA had liquid nitrogen poured directly onto plates to flash freeze mycelium and the underlying PDA, then a thin layer of mycelia was scraped off using a pre-chilled scalpel. Total RNA was extracted from frozen mycelia using the Maxwell® RSC Plant RNA Kit, and cDNA was amplified using an AMV first strand cDNA synthesis kit and was normalized to 50 ng/μl for each sample (New England Biolabs, Catalog # E6550). Sslac2 was amplified using specific detection primers (Sslac2 Det F and R) for 30 cycles. The raw gel is included as Supplementary Fig. 8.
Scanning and transmission electron microscopy (SEM and TEM)
For SEM samples were grown on PDA plates embedded with 10 mm Whatman filters shortly after plates were poured, allowing for a thin layer of agar to cover the filters. Samples were grown for 2 days until they had completely covered the filter paper, before being submerged in a chemical fixative (78% - Ultrapure ddH2O, 10% - 10× PBS, 10–37% formaldehyde, 2–50% glutaraldehyde) overnight. Samples were treated with 1% osmium tetroxide for 30 min at 22 °C. Samples were subsequently washed with a series of increasing ethanol concentrations (30–100% [vol/vol]), followed by critical point drying and sputter coating with platinum. Scanning electron microscopy (SEM) of samples was performed using a LEO 1530 microscope. TEM samples were grown on PDA overlayed with cellophane for two days before 10 mm circles were peeled off and placed in the above fixative overnight. Sample preparation, sectioning, and imaging was conducted by the UW Madison Medical School Electron Microscope Facility on a Philips CM120 STEM.
Genomic and RNA-Seq analysis
For the genomic analysis, known laccase proteins from Cucurbita maxima, Melanocarpus albomyces, Myrothecium verrucaria, and Saccharomyces cerevisiae were used as queries in a BLASTp search of S. sclerotiorum proteins in the National Center for Biotechnology Information (NCBI) Genbank repository, as described in Feng et al.2. All identified genes were assessed for a secretion signal peptide using SignalP 6.048. Protein features were described using the NCBI Conserved Domain database after querying through BLASTp.
For the RNA-Seq analysis, WT (1980) and Sslac2 mutant strains were grown in 1% GMM liquid culture for 3 days before cultures were moved to either fresh media alone or fresh media containing 1% soybean SE and allowed to grow for 4 h prior to flash freezing in liquid nitrogen. RNA was extracted with a Maxwell RSC Plant RNA Kit (AS1500). Differential gene expression values were generated using the bioinformatic pipeline described in Westrick et al.43.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA sequencing data was deposited in NCBI’s Gene Expression Omnibus (GEO) under project GSE246683. Source data underlying the figures can be found in Supplementary Data Set 1. Strains generated in this manuscript are available upon request.
References
Morozova, O. V., Shumakovich, G. P., Shleev, S. V. & Yaropolov, Y. I. Laccase – mediator systems and their applications: a review. Appl. Biochem. Microbiol. 43, 523–535 (2014).
Feng, B. Z., Li, P. Q., Fu, L. & Yu, X. M. Exploring laccase genes from plant pathogen genomes: a bioinformatic approach. Genet. Mol. Res. 14, 14019–14036 (2015).
Arregui, L. et al. Laccases: structure, function, and potential application in water bioremediation. Microb. Cell Factories 18, 1–33 (2019).
Ten Have, R. & Teunissen, P. J. M. Oxidative mechanisms involved in lignin degradation by white-rot fungi. Chem. Rev. 101, 3397–3413 (2001).
Upadhyay, S. et al. Subcellular compartmentalization and trafficking of the biosynthetic machinery for fungal melanin report subcellular compartmentalization and trafficking of the biosynthetic machinery for fungal melanin. Cell Rep. 1–8, https://doi.org/10.1016/j.celrep.2016.02.059 (2016).
Ma, S. et al. The StLAC2 gene is required for cell wall integrity, DHN-melanin synthesis and the pathogenicity of Setosphaeria turcica. Fungal Biol. 121, 589–601 (2017).
Wei, Y. et al. The laccase gene (LAC1) is essential for Colletotrichum gloeosporioides development and virulence on mango leaves and fruits. Physiol. Mol. Plant Pathol. 99, 55–64 (2017).
Lin, S. Y., Okuda, S., Ikeda, K., Okuno, T. & Takano, Y. LAC2 encoding a secreted laccase is involved in appressorial melanization and conidial pigmentation in Colletotrichum orbiculare. Mol. Plant Microbe Interact. 25, 1552–1561 (2012).
Lu, Z. Laccase gene Sh-lac is involved in the growth and melanin biosynthesis of Scleromitrula shiraiana. Phytopathology 107, 353–361 (2017).
Fang, W., Fernandes, É. K. K., Roberts, D. W., Bidochka, M. J. & St, R. J. A laccase exclusively expressed by Metarhizium anisopliae during isotropic growth is involved in pigmentation, tolerance to abiotic stresses and virulence. Fungal Genet. Biol. 47, 602–607 (2010).
Saitoh, Y., Izumitsu, K., Morita, A., Shimizu, K. & Tanaka, C. ChMCO1 of cochliobolus heterostrophus is a new class of metallo-oxidase, playing an important role in DHN-melanization. Mycoscience 51, 327–336 (2010).
Baldrian, P. Fungal laccases-occurrence and properties. FEMS Microbiol. Rev. 30, 215–242 (2006).
Cordoba Cañero, D. & Roncero, M. I. G. Functional analyses of laccase genes from Fusarium oxysporum. Phytopathology 98, 509–518 (2008).
Fan, Y. et al. Regulatory cascade and biological activity of Beauveria bassiana oosporein that limits bacterial growth after host death. Proc. Natl Acad. Sci. USA 114, E1578–E1586 (2017).
Feng, P., Shang, Y., Cen, K. & Wang, C. Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc. Natl Acad. Sci. USA 112, 11365–11370 (2015).
Lu, Z. et al. Multifunctional role of a fungal pathogen‐secreted laccase 2 in evasion of insect immune defense. Environ. Microbiol. 23, 1256–1274 (2021).
Liang, Y., Xiong, W., Steinkellner, S. & Feng, J. Deficiency of the melanin biosynthesis genes SCD1 and THR1 affects sclerotial development and vegetative growth, but not pathogenicity, in Sclerotinia sclerotiorum. Mol. Plant Pathol. 19, 1444–1453 (2018).
Li, J. et al. Introduction of large sequence inserts by CRISPR-cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen sclerotinia sclerotiorum. mBio 9, 1–19 (2018).
Schumacher, J. DHN melanin biosynthesis in the plant pathogenic fungus Botrytis cinerea is based on two developmentally regulated key enzyme (PKS)-encoding genes. Mol. Microbiol. 99, 729–748 (2016).
Schouten, A., Maksimova, O., Cuesta-Arenas, Y., Van Den Berg, G. & Raaijmakers, J. M. Involvement of the ABC transporter BcAtrB and the laccase BcLCC2 in defence of Botrytis cinerea against the broad-spectrum antibiotic 2,4-diacetylphloroglucinol. Environ. Microbiol. 10, 1145–1157 (2008).
Schouten, A., Wagemakers, L., Stefanato, F. L., van der Kaaij, R. M. & van Kan, J. A. Resveratrol acts as a natural profungicide and induces self-intoxication by a specific laccase. Mol. Microbiol. 43, 883–894 (2002).
Andberg, M. et al. Essential role of the C-terminus in Melanocarpus albomyces laccase for enzyme production, catalytic properties and structure. FEBS J. 276, 6285–6300 (2009).
Westrick, N. M. et al. Gene regulation of Sclerotinia sclerotiorum during infection of Glycine max: On the road to pathogenesis. BMC Genomics 20, 157 (2019).
Peyraud, R., Mbengue, M., Barbacci, A. & Raffaele, S. Intercellular cooperation in a fungal plant pathogen facilitates host colonization. Proc. Natl Acad. Sci. USA 116, 3193–3201 (2019).
Buddhika, U. V. A., Savocchia, S. & Steel, C. C. Copper induces transcription of BcLCC2 laccase gene in phytopathogenic fungus, Botrytis cinerea. Mycology 12, 48–57 (2021).
Adachi, K. & Hamer, J. E. Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell 10, 1361–1373 (1998).
Mccaghey, M., Willbur, J., Smith, D. L. & Kabbage, M. The complexity of the Sclerotinia sclerotiorum pathosystem in soybean: virulence factors, resistance mechanisms, and their exploitation to control Sclerotinia stem rot. Trop. Plant Pathol. 44, 2-22 (2018).
Williams, B., Kabbage, M., Kim, H.-J., Britt, R. & Dickman, M. B. Tipping the Balance: Sclerotinia sclerotiorum Secreted Oxalic Acid Suppresses Host Defenses by Manipulating the Host Redox Environment. PLoS Pathog. 7, e1002107 (2011).
Kabbage, M., Williams, B. & Dickman, M. B. Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9, e1003287 (2013).
Xu, L., Xiang, M., White, D. & Chen, W. pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 17, 2896–2909 (2015).
Liang, X. et al. Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16, 559–571 (2015).
Ranjan, A. et al. Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 1–15 (2019) https://doi.org/10.1111/pbi.13082.
Piotrowski, J. S. et al. Plant-derived antifungal agent poacic acid targets β-1,3-glucan. Proc. Natl Acad. Sci. USA 112, E1490–E1497 (2015).
Cai, F. et al. Evolutionary compromises in fungal fitness: hydrophobins can hinder the adverse dispersal of conidiospores and challenge their survival. ISME J. 14, 2610–2624 (2020).
Zhang, C., Bradshaw, J. D., Whitham, S. A. & Hill, J. H. The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 153, 52–65 (2010).
Lyu, X. et al. A ‘footprint’ of plant carbon fixation cycle functions during the development of a heterotrophic fungus. Sci. Rep. 5, 1–13 (2015).
Saitoh, Y., Izumitsu, K., Morita, A. & Tanaka, C. A copper-transporting ATPase BcCCC2 is necessary for pathogenicity of Botrytis cinerea. Mol. Genet. Genomics 284, 33–43 (2010).
Linder, M. B., Szilvay, G. R., Nakari-Setälä, T. & Penttilä, M. E. Hydrophobins: The protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 29, 877–896 (2005).
Martínez-Rocha, A. L. et al. Rho1 has distinct functions in morphogenesis, cell wall biosynthesis and virulence of Fusarium oxysporum. Cell. Microbiol. 10, 1339–1351 (2008).
Guest, G. M., Lin, X. & Momany, M. Aspergillus nidulans RhoA is involved in polar growth, branching, and cell wall synthesis. Fungal Genet. Biol. 41, 13–22 (2004).
Pieterse, C. M. J., Van der Does, D., Zamioudis, C., Leon-Reyes, A. & Van Wees, S. C. M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521 (2012).
Rollins, J. A. The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant Microbe Interact. MPMI 16, 785–795 (2003).
Westrick, N. M., Park, S. C., Keller, N. P., Smith, D. L. & Kabbage, M. A broadly conserved fungal alcohol oxidase (AOX) facilitates fungal invasion of plants. Mol. Plant Pathol. 24, 28–43 (2023).
Szewczyk, E. et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111–3120 (2006).
Talbot, N. J., Salch, Y. P., Ma, M. & Hamer, J. E. Karyotypic Variation within Clonal Lineages of the Rice Blast Fungus, Magnaporthe grisea. Appl. Environ. Microbiol. 59, 585–593 (1993).
McCaghey, M. et al. Host-induced gene silencing of a Sclerotinia sclerotiorum oxaloacetate acetylhydrolase using bean pod mottle virus as a vehicle reduces disease on soybean. Front. Plant Sci. 12, 1–13 (2021).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Teufel, F. et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025 (2022).
Acknowledgements
We would like to thank the USDA National Institute of Food and Agriculture (2021-67011-35151 to N.M.W.), the USDA ARS National Sclerotinia Initiative (58-3060-8-023 to M.K. and D.S.), and USDA Hatch (Wis04031 to M.K.) for supporting this research.
Author information
Authors and Affiliations
Contributions
N.M.W. and M.K. conceived the project. N.M.W., M.K., E.D., M.B., C.M.H. and D.L.S. contributed to experimental design. Experiments were conducted by N.M.W. and E.D. All authors contributed to the writing and editing of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks U. V. A. Buddhika, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary handling editors: Tobias Goris.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Westrick, N.M., Dominguez, E.G., Bondy, M. et al. A single laccase acts as a key component of environmental sensing in a broad host range fungal pathogen. Commun Biol 7, 348 (2024). https://doi.org/10.1038/s42003-024-06034-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06034-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.