Abstract
The translocated intimin receptor (Tir) is an essential type III secretion system (T3SS) effector of attaching and effacing pathogens contributing to the global foodborne disease burden. Tir acts as a cell-surface receptor in host cells, rewiring intracellular processes by targeting multiple host proteins. We investigated the molecular basis for Tir’s binding diversity in signalling, finding that Tir is a disordered protein with host-like binding motifs. Unexpectedly, also are several other T3SS effectors. By an integrative approach, we reveal that Tir dimerises via an antiparallel OB-fold within a highly disordered N-terminal cytosolic domain. Also, it has a long disordered C-terminal cytosolic domain partially structured at host-like motifs that bind lipids. Membrane affinity depends on lipid composition and phosphorylation, highlighting a previously unrecognised host interaction impacting Tir-induced actin polymerisation and cell death. Furthermore, multi-site tyrosine phosphorylation enables Tir to engage host SH2 domains in a multivalent fuzzy complex, consistent with Tir’s scaffolding role and binding promiscuity. Our findings provide insights into the intracellular Tir domains, highlighting the ability of T3SS effectors to exploit host-like protein disorder as a strategy for host evasion.
Similar content being viewed by others
Introduction
Intrinsically disordered proteins and regions (IDPs and IDRs, respectively) are widespread across all kingdoms of life1. They do not adopt well-defined structures but are dynamic conformational ensembles that display unique properties complementary to globular proteins2,3. IDPs/IDRs play essential roles in signalling pathways4, including cell cycle5, circadian circuits6, post-transcriptional regulation7, and protein degradation8. Given their ubiquitous relevance in cellular signalling, the onset of several human diseases, including cancer and neurodegeneration, are linked to dysfunctional disordered proteins9. A hallmark of IDPs/IDRs is the high occurrence of short linear motifs, also known as eukaryotic linear motifs (ELMs)10. ELMs are stretches of 3–10 contiguous residues mediating transient protein-protein interactions (e.g., post-translational modification sites, targeting signals)11. For instance, phosphorylation sites are often found in protein IDRs, modifying charge and hydrophobicity and modulating partner interactions7. The prevalence of such functional elements in IDRs provides versatility to cell interaction hubs by enabling flexibility and adaptability to multiple interaction interfaces12,13. Moreover, IDR sequence composition often has low complexity and charged residues favouring electrostatic interactions at lipid bilayer surfaces14, with lipid-binding proteins accounting for 15% of all disordered proteins15. As a result, IDRs are common in membrane signalling receptors, particularly in their intracellular domains16.
Disordered proteins comprise 30-50% of eukaryotes’ proteins17,18. In contrast, they are rare in prokaryotes. However, increasing experimental evidence in the literature shows several effector proteins secreted by pathogenic bacteria containing functionally relevant IDRs19. An example is the Helicobacter pylori type IV secretion system effector CagA, which recruits host proteins to potentiate oncogenic signalling via a long IDR20. Another illustrative example is the effector CyaA of Bordetella pertussis, the causative agent of whooping cough that interacts with host calmodulin via a disordered stretch of 75 amino acid residues21. Other less-ordered effectors are also secreted by Enteropathogenic and Enterohemorrhagic Escherichia coli strains (EPEC and EHEC, respectively), such as EspF(U)/TccP22 and EspB23.
EPEC is a leading cause of child diarrhoea worldwide24, and EHEC causes hemolytic-uremic syndrome (HUS), defined by acute kidney failure, low platelet count, and destruction of red blood cells25. These bacteria attach to the gut mucosa, causing attaching and effacing (A/E) lesions, characterised by the formation of actin-rich pedestal-like structures and the brush border microvilli effacement. To establish a replicative niche on host cells, EPEC and EHEC deliver an array of T3SS effectors26. Here, we aimed to identify disordered effectors expressed by A/E pathogens. To this end, we ran a structural disorder analysis on the collection of effectors and searched for ELMs evidence. Among the disordered effectors, we found the prominent translocated intimin receptor (Tir) with a relatively high density of host-like ELMs, particularly in its intracellular domains. Tir is the first effector secreted during infection27. Once inside the host, it migrates to the plasma membrane to act as a receptor for the outer membrane bacterial adhesin intimin28, anchoring A/E pathogens to the plasma membrane of the host cells. Moreover, Tir promotes actin polymerisation29,30, suppresses autophagy31, and immune response32 to allow bacterial survival on the host cell surface. It also triggers pyroptotic cell death in macrophages33 and intestinal epithelial cells34. As a double-pass membrane receptor, Tir adopts a hairpin topology with the external intimin‐binding domain (IBD)35 connecting two transmembrane domains and both N- and C-terminus in the host cytoplasm (N-Tir and C-Tir, respectively). Those intracellular regions enable Tir to interact with at least 25 host targets36, such as host tyrosine phosphatases SHP-1/2 to suppress pro-inflammatory cytokines signalling32,37 and TAK1-mediated immune response38.
How Tir hijacks intracellular signalling by interacting with multiple host proteins to assist infection is poorly understood. To gain insights into Tir’s detailed biophysics and biological function, we devised a structural biophysical study to assess its intrinsic disorder and interactions using Nuclear Magnetic Resonance (NMR) and Small-angle X-ray scattering (SAXS) combined with AlphaFold-239. We focus on stoichiometry, accessibility to multi-site phosphorylation, host protein- and membrane-binding of Tir intracellular regions. We found that N-Tir is a flexible dimer with a hybrid architecture of ordered and disordered parts brimmed with host-like ELMs. Moreover, we show that C-Tir is an IDR able to form phosphorylation-mediated fuzzy complexes with SH2 host domains that might facilitate binding promiscuity. C-Tir has non-random structures around host-like phosphorylation sites with lipid-binding ability pre-phosphorylation, suggesting a regulation based on the interplay between membrane association and phosphorylation. We uncover that Tir-induced actin polymerization and cell death depend on lipid-binding sites within disordered C-Tir.
Our findings highlight a molecular mechanism through which Tir can interact with host components by mimicking disordered host proteins, suggesting that pathogenic bacteria may share this strategy to assist infection.
Results
Structural disorder and short linear motifs are common features of A/E pathogen effectors
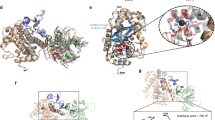
EPEC, EHEC and the mouse-specific pathogen Citrobacter rodentium (CR) are A/E pathogens. To quantify the prevalence of disorder propensity in these pathogens, we estimated the disorder content of their T3SS effectors using DISOPRED340 (Fig. 1) and IUPred 1.041 (Supplementary Figure 1). We observed that effector proteins have a higher disorder propensity than their proteome counterparts in EHEC O157:H7, the closely related EPEC O127:H6, and CR strain ICC168 (Fig. 1a, Supplementary Table 1). Subsequently, we classified the effectors into five structural categories based on disorder content (Supplementary Table 2). Our analysis shows that A/E pathogens have a structurally diverse repertoire of effectors, ranging from fully unstructured to ordered proteins (Fig. 1b, Supplementary Table 3). While most proteins are folded in all prokaryotic collections (Supplementary Figure 2), IDP effectors are predicted to be two to four-fold more frequent than the whole bacterial proteome (1.9% vs. 7.7% on average). Partially Disordered Proteins (PDR) with long disordered regions occur in EPEC O127:H6 effectors at a similar frequency as the human proteome. For instance, we classified as PDR the EspB effector, involved in cytoskeleton rearrangement and previously identified as an inherently less-ordered effector23. An additional example of PDR is the NleH effector, a protein kinase with a disordered N-terminal domain that binds the ribosomal protein 3 (RPS3) to manipulate the NF-κB pathway42. In the order-to-disorder continuum, the effectors EspF and EspF(U)/TccP are IDPs (Fig. 1c). Both proteins contain multiple consecutive repeats of linear motif pairs critical to trigger actin assembly, with EspF having three to five repetitions depending on the strain43 and EspF(U)/TccP, only secreted by EHEC, five-and-a-half repeats44 (Fig. 1d). Our analysis further highlighted Tir as a disordered effector (Fig. 1c). Tir displays a high disorder propensity within its IBD and intracellular regions (Fig. 1d, e).
a Distribution of disorder fraction displayed as box plots for effectors (orange) and full-proteomes (blue) of three representative A/E pathogens. Statistical analysis with Mann-Whitney U-test. b Accumulated fractions of the structural categories in terms of structural disorder. IDP: Intrinsically disordered proteins; PDR: Proteins with intrinsically disordered regions; FRAG: Proteins with fragmented disorder; NDR: Not disordered proteins; ORD: Ordered Proteins. See definitions in Supplementary Table 2. c Fraction of ELMs vs. disorder fraction in A/E effectors. Tir, EspF, and EspF(U) display a high motif and disorder content. IDP and PDR effectors are labelled. See all data in Supplementary Table 3. d Disorder propensity of EspF (top), Espf(U)/Tccp (middle), and Tir (bottom) from EHEC O157:H7 (black line), EPEC O127:H6 (blue line), and CR strain ICC168 (red line). Tir’s disorder propensity is conserved among A/E pathogens. Experimental verified GBD-, SH3- and SH2-binding motifs are indicated in light red, blue, and green bars, respectively. The 14-3-3 motif within N-Tir first residues is indicated by a black bar. The transmembrane domains (TMDs) of Tir are in purple. e EPEC O127:H6 Tir in hairpin topology as determined using TopGraph124, with the IBD flanked by the two TMDs and cytosolic termini with high content in disorder-promoting residues (T, A, G, R, D, H, Q, K, S, E, P).
Pathogens can exploit host-like motifs as a strategy to interact with host components to subvert host cellular functions45,46. On this basis, we searched the A/E effectors for eukaryotic linear motifs (ELMs) and found that aside from EspF and EspF(U), other A/E effectors bear multiple putative ELM instances (Fig. 1c). Tir emerged again as having several predicted ELMs in its intracellular regions, including experimentally verified SH3- and SH2-domain binding motifs (Fig. 1d)47. Structural disorder and high ELM density support Tir’s ability to interact with several host proteins36. There is a correlation between disorder content and ELM density, being Tir, EspF(U), and EspF clustered with the highest disorder fraction and overall motif density (Fig. 1c), similar to eukaryotic IDPs11,48. This correlation highlights ELMs within disordered segments of bacterial effectors as a potential strategy to disrupt host networks.
Tir’s N-terminal tail is a dimer with order-disorder duality
Tir shares substantial sequence conservation (Supplementary Fig. 3) and a high frequency of disorder-promoting residues49 at the N- and C-terminal regions (N-Tir and C-Tir, respectively), both localised in the host cytosol (Fig. 1e). We assessed the structural disorder propensity of EPEC O127:H6 N-Tir and C-Tir by multiple biophysical methods, including small-angle X-ray scattering (SAXS), circular dichroism (CD), and nuclear magnetic resonance (NMR). For this integrative study, we used the protein constructs encompassing the residues 1-233 and 388-550 from the N-terminal and C-terminal cytosolic regions of EPEC O127:H6 Tir, respectively (Fig. 1e, Supplementary Fig. 4, Supplementary Table 4). Based on size-exclusion chromatography (SEC)-SAXS data (Supplementary Fig. 5), the N-Tir is a non-globular protein with a radius of gyration (Rg) of 37.72 ± 0.50 Å and a maximum distance (Dmax) of 140.0 ± 10.0 Å (Fig. 2a, Supplementary Table 5), and a molecular weight compatible with a 52 kDa dimer, in line with the observed gel filtration retention time (Supplementary Figs. 4c and 5b). The respective P(r) curve is highly asymmetric and bimodal, with a peak at 21.9 Å and a prominent shoulder at 35.9 Å, suggesting that the dimer is elongated with spatially separated lobes. Its SAXS-derived Kratky plot is also not bell-shaped as expected for a non-globular and flexible arrangement (Fig. 2b). The corresponding SAXS-driven ab initio reconstruction highlights an elongated S-shaped core for the N-Tir dimer (Supplementary Fig. 6). This extended dimer is largely disordered but contains stable secondary structural elements. Its far-UV CD profile reveals a partially folded protein, marked by a negative maximum at 205 nm due to high disorder content. The positive signal at 190 nm and the negative shoulder at 220 nm reflect ordered elements (Fig. 2c). So, SAXS and CD data indicated that N-Tir is a partially folded dimer.
a Normalised pairwise distance distribution, P(r), computed from experimental SAXS curves of N-Tir (pink) and NS-Tir (purple) consistent with an elongated dimer. Dashed lines indicate the derived Rg and Dmax values. b Kratky plots of N-Tir (pink) and NS-Tir (dark-magenta) highlight a high conformationally flexibility for N-Tir. c Far-CD spectra of N-Tir (pink) and NS-Tir (dark-magenta) and their difference (grey dashed line). d [15N-1HN]-HSQC spectra of N-Tir at 5 °C and e 35 °C reveal two dynamically different regions within the protein.
We have collected NMR data at different temperatures to probe the order-disorder interplay within N-Tir at high resolution. The 52 kDa dimer size is beyond the practical range amenable to traditional NMR spectroscopy in solution. The NMR signals of larger molecules relax faster, leading to line broadening, low spectral sensitivity, and eventually loss of NMR signals, specially at low temperatures. Nevertheless, we found that the 52 kDa dimer has a [15N-1HN]-HSQC NMR spectrum at 5 °C characterised by intense signals and low chemical shift dispersion akin to an IDP (Fig. 2d). We assigned the resonances at 5 °C mostly to the first 80 and last 35 residues of the N-Tir, meaning that those regions are flexible in solution. The missing resonances of the central residues became visible with increasing temperature due to faster tumbling (Supplementary Fig. 7a). At 35 °C, they adopted an NMR fingerprint of a folded-like protein (Fig. 2e), reflecting a more well-structured region. With NMR, we could establish that N-Tir’s central sequence has the broad NMR fingerprint of a well-folded protein flanked by residues with poorly dispersed 1HN backbone resonances pointing to disorder. A closer look at the structural disorder predictions computed for N-Tir also suggests an order-disorder duality, with flanking residues more flexible than those in the central region (Supplementary Fig. 7b). Such disordered segments have linear motifs experimentally identified to interact with host proteins (Fig. 1d)47,50. The partial deletion of these disordered regions reduced the random-coil content reported by CD data, highlighting a more structured central part of N-Tir (N-Tir61-200, hereafter NS-Tir, standing for N-Terminal structured region of Tir). The difference between CD spectra resulted in a pure random-coil CD signature, reinforcing that the flanking regions are disordered. NS-Tir’s CD profile displays positive values below 200 nm and two negative bands at 210 and 222 nm, commonly associated with structured secondary conformations (Fig. 2c). We have employed SAXS to probe further the overall structure of NS-Tir. Our synchrotron SEC − SAXS data confirmed that NS-Tir is still a stable dimer in solution. It has a slightly less broadened Kratky profile that reflects the absence of the disordered flanking regions (Fig. 2c) but is compatible with an elongated dimer retaining a similar overall conformation as supported by NMR. We acquired [15N-1HN]-HSQC data for NS-Tir and superimposed them to N-Tir spectra (Supplementary Figure 7c). Over 75% of the visible resonances overlapped between the two constructs, with a low dispersion for the 1H dimension at 5 °C and a broader [15N-1HN]-fingerprint at 35 °C, indicating that NS-Tir maintained a similar folded isolated and in the full-length N-terminal region context. The nearly reversible thermal denaturation of NS-Tir monitored by far-UV CD shows the dimer unfolding in two discrete steps, capturing both folding and dimerization events. It reveals a stable dimer that unfolds and dissociates by increasing temperature51 (Supplementary Figure 8). Our data firmly supports that N-Tir is partially disordered with a stable dimer core.
N-Tir encodes a central antiparallel OB-like fold dimer
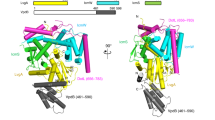
To gain insight into the structure of N-Tir’s dimer core, we turned to AlphaFold-2 (AF-2)52 and Flexible Meccano (FM)53 to create a structural ensemble model of the NS-Tir dimer (Fig. 3a) consistent with SAXS data (Fig. 3b). AF-2 is an AI-based method that predicts a protein’s 3D structure from its amino acid sequence. It regularly achieves high accuracy competitive with experimental NMR structures54,55. We obtained, with high confidence (pLDDT 93.02 ± 8.92), that the central residues 97–171 form a small β-barrel (SBB) domain56 (Fig. 3c). The predicted features show five strand fragments (β1, residues 109 to 114; β2, residues 120 to 126; β3, residues 129 to 134; β4, residues 153 to 156; β5, residues 168 to 170) which form the SBB, that together with an intercalated helix (αOB, residues 137 to 146) and the N-terminal helix (αN, residues 97 to 105), adopt an oligonucleotide/oligosaccharide-binding (OB)-like fold topology (Fig. 3d) well-encapsulated by its SAXS ab initio reconstruction (Fig. 3e). Moreover, chemical shift analysis57 confirms the secondary structure topology (N-term → C-term: β1-β2-β3–αOB-β4–β5), with the secondary structural elements matching those obtained using the experimental chemical shifts of NS-Tir (Fig. 3f). To reinforce and clarify this finding, we show the expected signature of intramolecular cross-strand NOEs for the SBB domain. In Fig. 3g, the NOE spectrum shows a set of unambiguous long-range NOEs between β1 and β4, β1 and β2, and β2 and β3, which, in combination with their secondary chemical shifts, point to the presence of an antiparallel β-barrel within this region (Fig. 3h). The predicted OB-fold domain is followed by an α-helix (172-187, αC-C) seemingly interacting with the corresponding α-helix from another molecule to assemble an antiparallel coiled-coil homo-dimer (Fig. 3a). To complete the model, we modelled the flanking regions as flexible ensembles with FM (Fig. 3a), obtaining structures that reproduce the experimental scattering profile (Fig. 3b), thus reinforcing the overall architecture of the NS-Tir as an ordered-disordered dimer. We disrupt the dimer by adding a stop codon in the middle of the αC-C (at position 181), producing a stable monomer as seen experimentally by SEC-SAXS, thus implying the involvement of αC-C in dimerization. This construct, encoding for residues 76-180 of Tir, bears the predicted SBB without the αC-C (N-Tir76-180, hereafter SBB-Tir). The synchrotron SEC − SAXS data for SBB-Tir is compatible with a globular protein with an Rg of 17.8 ± 0.8 Å and a Dmax of 70 ± 10 Å, and the molecular weight of a monomer (Supplementary Table 4), in line with the observed gel filtration retention volume. Our results suggest a model of Tir spanning the host plasma membrane with partially disordered N-terminal intracellular domains assembled in an antiparallel dimer bearing an OB-like domain and dangling ELMs.
a Structural ensemble of the NS-Tir antiparallel dimer stabilised by coiled-coil interaction flanked by disordered segments. N-Tir central region encodes for a small β-barrel (SBB) OB-fold domain. b SAXS intensity from NS-Tir (top) and SBB-Tir (bottom), I(s), represented in logarithmic scale as a function of the momentum of transfer, s = (4πsin θ)/λ, where 2π is the scattering angle. The solid lines correspond to the scattering profile calculated from the EOM-selected NS-Tir ensemble (purple line, χ2 = 0.89) and the ab initio model of SBB-Tir (red, χ2 = 1.20), best fitting the experimental data. Point-by-point residuals of fittings and the absolute intensities are shown at the bottom. c Ribbon representation of the best AF2-based model for the SBB domain with colouring according to the predicted local difference test (pLDDT) metric, from high in blue (>90) to yellow (70), orange (50) and low in red (0). d The structural elements (SSEs) are coloured into N-Tir’s SBB model, highlighting an OB-fold topology (N-term → C-term: β1-β2-β3–αOB-β4–β5). e Superposition of the ab initio envelope (transparent red) and the respective AF2 model, with SSEs coloured. f Secondary structure propensity for β-strands (blue) and helices (garnet) calculated from protein backbone chemical shifts using N-Talos57. Highlighted in colour are the SSEs of the AF2-models. Transparency has been added to the 16 residues which could not be assigned. g Strips from a 1H,15N]-NOESY-HSQC spectrum showing cross-strand NOEs (orange dashed lines) between amide and backbone protons compatible with antiparallel beta-sheets. h Cross-strand NOEs (orange dashed lines) are displayed on the SBB domain structure.
Tir bears an intrinsically disordered C-terminal tail
We also evaluated the structural disorder propensity of C-Tir. However, contrary to N-Tir, this region lacks any fixed-structured elements. Analytical SEC in combination with SAXS reveals a monomeric state for C-Tir in solution with an Rg of 38.8 ± 0.2 Å and a Dmax of 128.0 ± 5.0 Å (Supplementary Figure 6, Supplementary Table 5). The Rg-values of IDPs approximately followed a power law of Rg = 2.54N0.522 58,59 as a function of sequence length (N). Using this relationship, the predicted Rg-value is 37.4 Å for the 173-residue C-Tir fused with a StrepTag (8 extra residues). The similarity between the experimental and predicted values suggests that C-Tir adopts IDP-like structures in solution. The SAXS-derived Kratky plot of C-Tir also shows characteristics of disordered or unfolded proteins, monotonically increasing without a well-defined maximum (Fig. 4a). This feature is absent in globular proteins and implies conformational heterogeneity and flexibility for C-Tir. Likewise, the asymmetric pair distance distribution function, P(r), obtained from the scattering data, is compatible with a highly flexible protein sampling pairwise distances far exceeding those expected for a globular protein of the same molecular weight59 (Supplementary Figure 6). The intrinsic disorder of C-Tir is also reflected in its CD and [15N-1HN]-HSQC NMR spectra. C-Tir’s CD profile has negative ellipticity at 200 nm and a shallow band in the 210–230 nm range (Fig. 4b), indicating a high content of random coil with minimal ordered structural elements60. This construct’s 2D-[15N-1HN]-HSQC NMR spectrum displays a typical IDP fingerprint with low chemical shift dispersion61 (Fig. 4c). We observed a similar spectrum for the equivalent region of Tir from EHEC (Fig. 4c), highlighting its structural disorder’s conservation and functional importance for A/E virulence. Overall, the different biophysical measurements are consistent with a highly dynamic protein, unambiguously showing that the C-Tir is an IDR that lacks a well-folded structure. Such inherent flexibility will potentially enable Tir for multiple binding while acting as a scaffold/hub of intracellular host proteins.
a Kratky representation of the SAXS profile measured for C-Tir indicates a lack of compactness. b Far-UV circular dichroism of C-Tir reveals the absence of high-populated structural secondary elements. c C-Tir EPEC O127:H6 (light blue) and C-Tir EHEC O157:H7 (grey) [15N-1HN]-HSQC spectra reveal the narrow 1H chemical shift dispersion characteristic of IDPs with 1H amide backbone resonances clustering between 7.7 and 8.5 ppm. All together provide definite proof of protein disorder.
C-Tir displays non-random structural preferences at phosphorylation sites
We employed NMR to dissect with further detail the structural features of C-Tir. The chemical shift (CS) is the most readily observable in NMR and sensitive to even partial secondary structural elements present in IDPs61. To access this information, we assigned the backbone resonances of C-Tir nuclei using standard triple-resonance spectra at high-field and reverse labelling (Supplementary Figure 9). Reverse labelling of selected amino acids62 proved to be a cost-efficient approach to assign this disordered protein by reducing the spectral ambiguity and should be readily applied to other IDPs. Given the assignment, we calculated the CS deviations (∆δ) to random coil values of intrinsically disordered proteins63. Although C-Tir has properties of a random coil, these so-called NMR secondary shifts allowed us to identify two regions with non-random structural preferences. In IDPs, transient secondary structural elements are often crucial molecular recognition units, playing critical roles in binding64,65. In Fig. 5a, the consecutive positive ΔδCα–ΔδCβ values indicate a tendency for α-helical structure in two distal segments of C-Tir: (I) D420-Q435; II) I507-A515. Both stretches include residues phosphorylated by host kinases, S43466 and Y51132, respectively. Considering all the assigned backbone resonances in a neighbour-corrected sequence structural propensity calculator (nsSPC)-score67 made these local α-helical regions more evident. New stretches also emerged with a putative α-helical propensity, including a sequence contiguous to Y511 and downstream tyrosine 483 (Fig. 5a). We also employed the CheSPI (Chemical shift Secondary structure Population Inference) method68 to provide additional local structure and disorder assignment. CheSPI analysis (Fig. 5b) further confirmed that C-Tir is disordered with small fractions of folded structural elements (i.e., helix, turn) at the residues with higher nsSPC-score. 15N-relaxation measured on C-Tir also supports the existence of less-disordered segments near Y483 and Y511 with relatively elevated 15N-R2/R1. Interestingly, Y483 and Y511 are core residues of Tir’s host-like immunoreceptor tyrosine-based inhibitory motifs (ITIMs) that recruit tyrosine phosphatases SHP-1/2 in an ITIM phosphorylation-dependent manner to inhibit host innate immune responses32,37.
a Secondary chemical shifts (∆δCα-ΔδCβ) (top), structural propensity plot (middle) for C-Tir where the dashed-lines depict the random-coil threshold and stacked bar plot of CheSPI populations (bottom) of extended (blue), helical (red), Turn (green), and non-folded (grey) local structures. ncSPC values above or below this threshold have ɑ-helix or β-sheet propensities, respectively. Positive values (grey regions) show an increased α-helix tendency at segments D420-Q435 (I) and I507-A515 (II), as well as F489-V495 (a) and T543-V547 (b). b 15N–R2/R1 analysis of C-Tir. c ΔPRE-values measured on V405C, S428C, S463C, S509C, and S536C C-Tir mutants as grey-scale heatmaps, from top to bottom, respectively. Color intensity reflects the relative deviations between the experimental PRE and those predicted from a random-coil model (ΔPRE = [Ipara/Idia]RC - [Ipara/Idia]exp). Dashed red lines mark the position of MTSL tags. d Cartoon representations of 3 representative conformers of the C-Tir ensemble refined with PRE + SAXS data with serine- and tyrosine-phosphorylation sites colored orange and green. e Logarithmic-scale of scattering intensity, I(s), as a function of the momentum transfer, s, measured for C-Tir (empty gray circles). The solid line is the averaged back-calculated curve derived from the PRE + SAXS-based ensemble of C-Tir (blue). f Ensemble-averaged contact map normalized to random-coil distances (αij = 〈rij〉ens/〈rij〉RC). Green and orange circles mark phosphorylation sites on C-Tir for tyrosine and serine residues, respectively. C-Tir is bipartite, with its C-terminal half being more compact.
We further probed C-Tir’s non-random coil preferences using paramagnetic relaxation enhancement (PRE) NMR experiments. To this end, five residues scattered along C-Tir’s sequence were single-point mutated to cysteine to attach a thiol-reactive nitroxide radical probe (i.e., MTSL). The relaxation induced by this paramagnetic tag resulted in substantial peak broadening (Supplementary Figure 10). This effect only affects residues near the MTSL tag in ideal random coils. Yet, if long-range contacts are present within the disordered chain, residues far away in the sequence can be spatially close to the tag and experience the PRE effect. We calculated the deviations between the experimental PRE and those predicted from a random-coil model (i.e., ∆PRE) to detect non-random coil structural preferences. We measured ∆PRE-values for each single-cysteine mutant. Figure 5c shows, in a grayscale, the ∆PRE induced by MTSL at positions 405, 428, 463, 509, and 536, all consistent with long-range compact conformations in the disordered C-Tir. The paramagnetic probe in positions 405 and 428 showed noticeable long-range interactions (∆PRE > 0.2) with distant downstream residues. The MTSL at S463C caused substantial PRE (∆PRE > 0.6) in the residues 497–526, and when placed in position 509, it induced broadening in the region 438-479, including position 463. These complementary long-range interactions observed between residues around positions 463 and 509 are mutually self-consistent with compact conformations. Interestingly, both areas include tyrosine-based ELMs involved in intracellular signalling. To better describe and visualise the non-random preferences of C-Tir, we integrated the PRE data with SAXS data to determine the conformational ensemble of C-Tir (Fig. 5d). The established sub-ensembles agree with the experimental data (Fig. 5e, Supplementary Figure 10), revealing a bias for expanded and compact structures. The ensemble-averaged distances between the Cα atoms for the experimentally selected conformations (〈rij〉ens), normalised by the respective random coil (RC) ensemble (〈rij〉RC), show an N-terminal region more extended relative to the RC. At the same time, the C-terminal half is more compact (Fig. 5f), highlighting a bipartite nature. Compaction is clustered around the host-like ITIMs that interact with host proteins.
C-Tir can interact with the host SHP-1 pre-phosphorylation
The intracellular signalling associated with Tir ITIM-like sequences relies on the phosphorylation of the central tyrosine by Src family protein tyrosine kinases (PTKs), creating a binding site for SH2–containing proteins, which become activated upon recruitment. Nevertheless, unphosphorylated C-Tir can recognize host SH2-domains with low-intermediate affinity. Using the C-terminal SH2 domain (C-SH2) of phosphatase SHP-1, we identified, by NMR, that residues surrounding the unphosphorylated Y511 ITIM-like motif are involved in binding SH2 domains (Fig. 6a). Binding to C-Tir caused a selective loss of the backbone amide cross-peaks intensities, mainly from residues A512LLA515 (Fig. 6b). Visible NMR signals retained low dispersion, indicating that the protein remains mostly disordered and flexible in the complex. We estimated an apparent KD of ca. 68.5 μM for this interaction (Fig. 6c). Thus, besides encoding for less-disordered structures, the sequence surrounding Y511 (i + 4) has a chemical signature for C-SH2 binding pre-phosphorylation. To gain insight into the complex interface, we harvest the capability of AF-2 to accurately model protein-peptide interactions69. In that sense, we employed Alphafold2-multimer39 using the sequence stretch of the unphosphorylated C-Tir mapped by NMR to bind C-SH2, obtaining a complex with high local and inter-chain accuracy (Fig. 6d, Supplementary Figure 11). The SH2 domain shows its typical central β-sheet flanked by two α-helices (αA, αB) (Fig. 6e). The Y511-motif binds perpendicularly to the β-sheet and docks into two abutting recognition sites formed by the β-sheet with each of the α-helices, similar to the complex of C-SH2 with the peptide NKG2A (PDB 2YU7). The Y511 points to the canonically defined “pTyr pocket”, and the SH2 recognizes the hydrophobic Leu residue in position +3 via the “specific pocket”70. Moreover, residue in position −2 interacts with αA, forming a “three-pronged plug” interaction altogether. This binding mode was observed for the SH2 domain of the SLAM-associated protein (SAP). Surprisingly, the SH2 SAP also recognizes unphosphorylated motifs due to the additional binding site, but with a lower affinity than the sequences containing pTyr residues70,71.
a Overlay of [15N-1HN]-HSQC spectra of C-Tir in the absence (blue) and the presence of C-SH2 at 2.0 (pink) and 8.0 (dark grey) equivalents. We show the NMR signal of residues A512LLA515 with an intensity drop due to C-SH2 binding. b Relative [15N-1HN]-peak intensities from the titration analysis. The green circle marks the tyrosine Y511 position. c Global fitting NMR quantification of the binding of C-SH2 to unphosphorylated C-Tir. The data shown are normalized intensities by the initial value. The error bars are the uncertainty from random noise in the ratio of two peak heights. The solid lines correspond to the fitting. d Ribbon representation of the best AF2-based model for the Y511 motif in complex with C-SH2 from SHP-1 with colouring according to the pLDDT metric. e Scheme of the binding interface between an SH2 domain (in pink) and the unphosphorylated Y511 peptide (in blue). Y511 and other interface residues are in stick format. The peptide is numbered by counting Y511 as 0. f Dashed white circles indicate the pY binding pocket and the specificity pocket.
Multi-phosphorylated C-Tir is a disordered multivalent cytosolic tail
To fully assess Tir ability to recruit SH2 domains, we reconstructed C-Tir’s Tyr-phosphorylated state and evaluated its binding to C-SH2. To this end, we incubated C-Tir with the Src family PTK Fyn, and quantitatively monitored its phosphorylation by NMR72. The sensitivity of chemical shifts to changes caused by phosphorylation allowed us to identify four phosphorylated tyrosine sites along the disordered C-Tir modified by Fyn (i.e., Y454, Y474, Y483, Y511). Phosphorylation caused small but noticeable changes in the chemical shifts of the tyrosines and adjacent residues. The [15N-1HN]-HSQC spectrum displayed low amide proton dispersion, a diagnostic that the C-Tir remains disordered upon multisite phosphorylation. Moreover, the secondary chemical shifts of this 4-fold phosphorylated state (pC-Tir) did not reveal substantial changes in local structure propensity due to phosphorylation (Supplementary Figure 12), as similarly reported for other disordered proteins73,74.
To evaluate the interaction of pC-Tir with an SH2 domain, we titrated unlabeled C-SH2 into a 15N-labelled pC-Tir solution and monitored the binding interaction broadening of signals in NMR [15N-1HN]-HSQC spectra (Fig. 7a, b). With the resonance re-assignment of pC-Tir, we identified that upon phosphorylation, all tyrosine sites interact with C-SH2, and not exclusively the Y511-based motif as observed for unphosphorylated C-Tir. The interaction caused a drop in peak intensities around each site (i.e., i = pY454, pY474, pY483, pY511), including roughly residues i + 4 and i-1, even at sub-stoichiometric conditions (Fig. 7b). This suggests a “fuzzy” multivalent binding of C-SH2 to four xYx(x)ϕ motifs, where ϕ is hydrophobic (V/L/I) (Fig. 7c), with multiple pYs on C-Tir interacting with C-SH2 in dynamic equilibrium73,74. Except for these residues, backbone chemical shift changes were relatively small, with spectra displaying sharp line widths and low dispersion, both characteristics of a disordered protein. Thus, pC-Tir binds C-SH2 retaining its high level of disorder. The relative signal intensities at each phospho-tyrosine (pY) and surrounding residues approximately reflect the fraction of unbound sites, allowing us to estimate their apparent local binding affinity (Supplementary Figure 13). Phosphorylation enhanced the binding to Y511 site by ~10 fold and enabled other tyrosine-based motifs to engage C-SH2. Among the four pYs, the resonances around pY454 were less broadened during the titration, suggesting lower local binding. The remaining pYs bind C-SH2 with similar strength (~3-9 μM). Overall, phosphorylation at multiple tyrosine sites provides various SH2 docking sites and reinforces the role of Tir as a scaffolding hub.
a NMR-detected binding of C-SH2 to phosphotyrosine pY454, pY474, pY483, and pY511 before (orange) and after adding 1.0 and 6.0 equivalents of C-SH2 (blue and grey, respectively). b Site-specific NMR signal attenuation on pC-Tir due to C-SH2 binding, with local intensity drop around each phosphorylation site. c Sequence alignment of xYx(x)ϕ motifs of C-Tir interacting with C-SH2 and their respective apparent KD values obtained by global fitting 1:1 model the intensities drop around each phosphorylation site. Darker shades of blue refer to higher sequence identity. The conservation of physicochemical properties in each alignment position is reported in the corresponding bar-plot below the alignment. Green circles denote phosphorylation tyrosine-sites.
Disordered C-Tir binds lipids
Remarkably, Tir ITIMs and cytoplasmic tyrosine-based sequence motifs of host receptors share the ability to adopt α-helical conformations. Indeed, host receptor tyrosine-based motifs can form dynamic/transient α-helices, whether free75,76 or membrane-bound state77. Their dynamic binding to membranes is postulated to regulate tyrosine sites’ access to phosphorylation, providing a switch between functional and non-functional conformations78. The helical conformation in ITIMs/ITAMs is stabilised in vitro in the presence of TFE or through binding to detergent micelles and lipids76,77,79. In the presence of TFE, we found that C-Tir exhibited a substantial increase in secondary structure, with the CD signature of a partial helical protein (Supplementary Figure 14a). This conformational change is followed by chemical shift differences (Supplementary Figure 14b) and a differential decrease in peak intensities of the C-Tir amide proton resonances in increasing amounts of TFE, mainly around Y511 and Y483 and not at S434 (Supplementary Figure 14c). This effect supports an enhanced α-helical conformation propensity around the two tyrosines, and a putative role in membrane-binding, as postulated for some host tyrosine-based motifs that undergo TFE-induced structural changes76.
The ability to form α-helices stabilised by TFE prompted us to investigate lipid binding by C-Tir. We incubated 15N-labelled C-Tir with different bicelles, small planar bilayers of long-chain lipids closed by curved micelle-like walls of short-chain lipids. These disk-like structures are membrane models extensively used to explore protein-membrane interactions, including those involving disordered proteins80. As long-chain lipids, we used 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) or the acidic lipid 1,2-dimyristoyl-sn-glycero-3-phosphorylglycerol (DMPG) mixed with the short-chain lipid 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC), offering the possibility to create bicelles with different lipid charge density.
In the presence of bicelles containing DMPC lipids, we observed a marked residue-specific decrease in the ratio of NMR signals around Y511, yet more subtle, also around Y483 (Fig. 8). This finding shows that C-Tir does interact with non-charged lipid bilayers predominantly via its Y511-based motif, including mostly hydrophobic residues, such as A512LLA515. We further tested the influence of increasing negatively charged lipid head groups on C-Tir membrane association with bicelles containing DMPG lipids. Our NMR data show a more extended NMR signal attenuation in the presence of DMPG/DHPC bicelles, even including the first stretch of residues of this construct, which display some positively charged residues (e.g., R388 and R389) (Fig. 8). In the context of the full-length protein, these residues define the membrane-proximal region (Fig. 1e, Supplementary figure 3). Given its proximity to the cell plasma membrane, it is reasonable to assume its involvement in membrane binding. Aside from Y511 surrounding residues, the sequence around Y483 is also notably affected by anionic lipid content, indicating that lipid-charge content modulates C-Tir membrane binding modes. Contrary, the residues 395-475 did not show any membrane interaction. This region has a net negative charge (pI~4.5), whereas the sequences displaying membrane ability are positive (pI~9.9), thus in line with an electrostatic model. In this residue range, C-Tir bears two experimental confirmed host protein binding sites: a) NCK Src Homology 2 (SH2) domain binding motif30; and b) NPY motif that interacts with the I-BAR domain of host IRSp53/IRTKS81. With both bicelles, the last C-terminal residues of C-Tir also show NMR-signal attenuation indicative of membrane-binding (Fig. 8b). These residues A541PTPGPVRFV550 emerge as part of a potential new lipid-binding region, also affected by TFE (Supplementary Figure 14) and showing pre-existing compaction (Fig. 5). This indicates that C-Tir can undergo multivalent and tunable electrostatic interaction with lipid bilayers via pre-structured elements, suggesting that membrane-protein interplay at the intracellular side might control the activity and interactions of Tir in host cells.
a NMR [15N-1HN]-HSQC spectra of C-Tir in the absence (blue) and the presence of DMPC/DHPC (6% w/v), yellow) or DMPG/DHPC (6% w/v), orange) bicelles. C-Tir’s HSQC data in bicelles superimpose well to free C-Tir, with identical chemical shifts for most visible resonances, pointing to intrinsic disorder. Residues with NMR signal attenuation upon the addition of DMPC/DHPC bicelles are indicated in the spectra (left). The additional residues affected by DMPG/DHPC are shown (right). b, c Lipid-induced NMR attenuation profiles are plotted against C-Tir primary sequence. b Residues comprising the Y483 and Y511 ITIM motifs became less intense with bicelles, reinforcing their inherent ability to bind lipids. (C) With phosphorylation, the NMR signal attenuation around Y483 and Y511 becomes essentially less pronounced. The grey bar highlights the C-terminal residues affected by lipids.
We also tested if phosphorylation of Y483 and Y511 could affect lipid-binding. To this end, we mutated Y454 and Y474 to phenylalanine (Supplementary Table 6) and phosphorylated the remaining two tyrosines. Next, we incubated the phosphorylated and 15N-labelled mutant (pC-Tir454F474F) with both types of bicelles to probe by NMR lipid binding. Our NMR data reveal that the phosphorylation of Y483 and Y511 drastically hampers the membrane association of those sites (Fig. 8c) while partially unaffecting the ability to interact with anion bicelles of the first and last stretch of residues. Our NMR results show that unphosphorylated C-Tir can adopt pre-formed structures potentially relevant to membrane binding and molecular recognition of signalling proteins.
Tir-induced actin polymerization and cell death depend on C-Tir lipid-binding
Tir promotes actin polymerization at the bacterial attachment site, leading to pedestal formation29; moreover, Tir activates pyroptotic cell death in intestinal epithelial cells34. To investigate whether the lipid-binding residues affect the known biological functions of Tir during the infection, we mutated key residues in the C-terminus of Tir (R388, R389, and L513) (Supplementary Table 6). Plasmid-encoded wild-type and mutant Tir were expressed in EPEC-0, which encodes the T3SS and intimin but is missing all the known effector genes, including tir82. These strains were then used to infect the model intestinal epithelial cell line SNU-C534.
We first mutated the positively charged residues R388 and R389, as well as L513 located in the Y511 ITIM motif to the negatively charged glutamic acid (Supplementary Table 7). Similar to EPEC-0-TirWT, EPEC-0-Tir388E389E and EPEC-0-Tir513E triggered the formation of actin pedestals (Fig. 9a) and propidium iodide (PI) uptake as a marker of lytic cell death (Fig. 9b). We then introduced a STOP codon at residue A541, removing the ten residues of the C-terminus tail (A541PTPGPVRFV550), which are also implicated in lipid-binding (Supplementary Table 7), creating EPEC-0-Tir541*. While pedestal formation was unaffected in EPEC-0-Tir541*-infected cells (Fig. 8a), we detected a mild yet significant reduction in PI uptake compared to EPEC-0-TirWT (Fig. 9b) that became more evident over time (Supplementary Figure 15). Accordingly, mutations in all three sites individually did not affect pedestal formation and only resulted in a limited reduction in Tir-induced cell death.
a Immunofluorescence images of SNU-C5 cells infected with EPEC-0 (empty plasmid), EPEC-0-TirWT, EPEC-0-Tir388E389E, EPEC-0-Tir513E, EPEC-0-Tir541*, EPEC-0-Tir388E389E513E or EPEC-0-Tir388E389E513E541* at 2 h post-infection. Enlarged cell-attached EPEC are seen in the white box (right-hand side panels). Shown are representative images from 3 independent biological repeats. Scale bar: 5 µm. Blue: DAPI (DNA); Green: EPEC; Red: phalloidin (actin). b PI uptake into SNU-C5 cells infected with EPEC-0 (empty plasmid), EPEC-0-TirWT, EPEC-0-Tir388E389E, EPEC-0-Tir513E, EPEC-0-Tir541*, EPEC-0-Tir388E389E513E, or EPEC-0-Tir388E389E513E541* at 8 h post-infection. Data shown are mean ± SEM from 3 (EPEC-0-Tir388E389E513E541*) or 6 (all other strains) independent biological repeats. Statistical analysis was performed using 1-way ANOVA with Tukey’s post-test. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns: non-significant.
Next, we generated a triple R388E / R389E / L513E mutant (Supplementary Table 7). The resulting strain EPEC-0-Tir388E389E513E triggered typical actin pedestals (Fig. 9a); however, we observed significantly reduced cell death compared to both EPEC-0-Tir388E389E and EPEC-0-Tir513E (Fig. 9b). Finally, we mutated all four sites simultaneously, including point mutations and the C-terminus ten residue truncation (Supplementary Table 7). The resulting EPEC-0-Tir388E389E513E541* was impaired in actin pedestal formation (Fig. 9a) and was not cytotoxic, similar to the negative control EPEC-0 with an empty plasmid (Fig. 9b). Therefore, while the lipid-binding sites at the C-terminus of Tir appear to be essential for its biological functions, they might be partly redundant as only the quadruple mutant affected Tir-induced cell death and pedestal formation.
Discussion
Although IDPs represent only 5% of typical bacterial proteomes, it has become clear that they control critical aspects of bacterial biology83. Here, we have determined the structural disorder propensity of EPEC O127:H6 sequences and two additional representatives of A/E bacteria: EHEC O157:H7 and CR ICC168. We observed that the three studied prototype strains of A/E pathogens have a structurally diverse repertoire of protein effectors enriched in disorder-prone residues. Some effectors are fully disordered proteins with a high density of host-like short linear motifs, challenging the general trend that prokaryotic proteins are less disordered. Among such disordered effectors emerged Tir, a 56 kDa cell surface receptor able to reshape host cellular behaviour during infection. We examined the structure of Tir from EPEC O127:H6, finding that its intracellular domains (i.e., N-Tir and C-Tir) are flexible with IDR features similar to those found in the cytoplasmic domains of host transmembrane proteins. We unravelled that the N-Tir is highly disordered (~52%) but forms a stable antiparallel dimer. The central region of N-Tir comprises an SBB domain followed by a coiled-coil dimerization helix. The structural and functional implications of an OB-fold in Tir are still unknown. But, many SBB-containing proteins with OD-fold often act as modules that bind to various RNAs, DNAs, phospholipids, and other proteins56. Many OB-folds are domains in proteins that frequently oligomerize. Clustering of Tir triggers host signalling events when binding to intimin. The extracellular intimin binding domain (IBD) of Tir binds intimin as a dimer35 in a reticulating model84. Thus, Tir’s intracellular self-assembly of SBB domains questions whether host signalling activation involves intimin-induced dimerization or structural changes of preformed dimers. It would be interesting to assess the impact of N-Tir oligomerization on the function of Tir. Nevertheless, the N-terminus Tir dimerization embodies a typical feature of cell receptors that Tir is meant to mimic.
Moreover, NMR analysis showed that unbound C-Tir is not exclusively disordered but can adopt partial secondary structural elements and long-range compaction around phosphorylation sites and residues involved in biomolecular interactions (i.e., host protein and lipid-binding). This observation supports the idea of transient pre-structured motifs (PreSMos) as potential pre-existing signatures for binding and function within target-free Tir85. However, we cannot rule out that C-Tir also undergoes structural rearrangement upon binding via induced fit86.
As in disordered cytosolic tails of host receptors, C-Tir displays tyrosine-based motifs, which are phosphorylated by Src family PTKs enabling the interaction with SH2–containing proteins. Interestingly in C-Tir, we found that the motif around Y511 can bind a single C-SH2 without phosphorylation. NMR shows that the residues adjacent to Y511 define a binding site for C-SH2, likely serving as a precursor to a tighter binding upon phosphorylation. Another notable feature of this motif is its α-helical propensity. Although we cannot tell whether a helix is a prerequisite for C-SH2 binding, our results suggest an alternative interaction mode to the “two-pronged plug two-hole socket” model87,88. This atypical SH2 binding may offer an additional mechanism for engaging with host SH2-containing proteins pre-phosphorylation. The Y511 motif also interacts with lipid bilayers. Notably, subsequent multi-site phosphorylation of C-Tir enables the transition from binary to multivalent fuzzy complex with C-SH2. These observations highlight a functional diversity for C-Tir and possible cooperation between host proteins, phosphorylation, and the membrane in signalling processes inside the host14.
Some T-cell receptor cytoplasmic disordered domains associate with the plasma membrane via tyrosine-based motifs with a helical propensity, and such association regulates phosphorylation and downstream signalling78. Like the CD3ɛ cytoplasmic domain of the T-cell receptor77, C-Tir also has two tyrosines involved in lipid-binding and helical propensity. Our results suggest that the C-terminal half Tir can mimic this regulatory stratagem to fine-tune its ability to interact with human cell components, thereby interfering with normal cellular functions. Further studies are needed to corroborate this initial observation. Nevertheless, we show that the C-Tir can interact with membranes via two ITIMs and its last C-terminal residues. Importantly, our results establish that this membrane affinity is residue-specific and modulated by lipid composition and phosphorylation in a quantitative and site-resolved way, thus suggesting the existence of a regulation layer based on lipid composition and phosphorylation. Our results support that transiently structured lipid-binding regions might bury tyrosine residues rendering them inaccessible, while their phosphorylation perturbs membrane anchoring89. Importantly, we uncover that lipid binding by Tir C-terminus is essential for Tir-induced host cell death and pedestal formation. Interestingly, Tir-induced cell death was previously shown to be dependent on mechanosensitive Ca2+ channels and can be modulated by membrane mechanical stress90. As high density of lipid-binding proteins, such as in the case of intimin-clustered Tir, can potentially produce lateral pressure to bend the lipid membrane91, it remains to be seen whether the lipid-binding ability of Tir directly results in the mechanical changes in the plasma membrane, leading to cell death.
The flexibility and conformational plasticity of C-Tir make it readily accessible for phosphorylation. We found in C-Tir four bona fide tyrosine phosphorylation sites modified by Fyn that render the ability for multivalent/promiscuous binding, supporting the idea of Tir acting as a signalling hub. Even though we only used protein fragments and explored one phosphorylation state, our results support the possibility of four phospho-tyrosines (pY) in the C-Tir sequence coexisting simultaneously. All can bind the C-SH2 in the micromolar range, as reported for other pY-containing motifs92. Interestingly, the pY454 site, which displays the lowest affinity, is part of a conserved NPY binding motif for the I-BAR domain of host IRSp53/IRTKS, linking Tir to the actin polymerization machinery81 in a phosphorylation-independent manner. So, Y454 might act as a binding site for distinct and competing host proteins depending on its phosphorylation state. The superposition of interplaying functional elements observed in C-Tir mimics the mechanics of human intrinsically disordered domains. Future work using high-resolution structural studies targeting the interaction with multiple partners and different C-Tir’s phosphorylation states will provide additional insights into Tir binding specificity and action mode.
In brief, we establish an updated picture of Tir’s intracellular side, supporting that in host cells, Tir dimers can interact with the host plasma membrane via their C-terminus with implications to downstream signalling triggering actin polymerization and cell death. Phosphorylation impacts lipid binding and renders the ability to interact with multiple host SH2 domains. In eukaryotic cells, protein disorder is a frequent feature of protein hubs controlling the complex intracellular networks that influence physiological responses4,93. Notably, the structural disorder of Tir and several other A/E effectors reinforce the idea of a positive evolutionary selection towards disordered proteins in the pool of secreted effectors by bacterial pathogens to target host cellular machinery19. Therefore, ELMs in disordered regions of A/E effectors may play a key structural and signalling role in protein function through interactions with host factors facilitated by their conformational flexibility. Given the ubiquitous presence of IDPs as transcriptional factors94 and, more generally, as hubs in host networks93, bacterial effectors with host-like disordered protein features are an efficient way to subvert host eukaryotic systems and promote infection.
Methods
Disorder prediction and short linear motif analysis
We collected the sequence of A/E pathogen effectors and corresponding reference proteomes from the UniprotKB database95 and a representative set of 20365 human proteins from the human reference proteome (Supplementary Table 1). For A/E bacteria, we assembled three sets of effectors for the following representative strains: CR (strain ICC168), EHEC O157:H7, and EPEC O127:H6 with 28, 39, and 24 sequences, respectively. To compute disorder propensity at the residue level, we used the structural disorder predictors DISOPRED 340 and IUPred 1.0 (set to “long” mode)41. Disordered residues were those with a propensity score equal to or above 0.5. We used this metric to calculate the fraction of disordered residues for each protein. Next, we used the one-sided Mann-Whitney U-test96 to compare the disorder fraction distribution of effector collections and their corresponding proteomes. Only effectors with reference proteomes were considered for disorder fractions comparison and removed from their proteome sets. Based on per-residue scores and aggregated disorder fractions, we classify each protein according to the structural categories adapted from97 (IDP: Intrinsically disordered proteins; PDR: Proteins with intrinsically disordered regions; FRAG: Proteins with fragmented-disorder; NDR: Not disordered proteins; ORD: Ordered Proteins). For this classification (Supplementary Table 2), we changed the original threshold for disordered stretches from 30 amino acids to 22 to be more in line with the shorter average length of prokaryotic proteins compared to eukaryotic ones (320/472 = 0.68)98. We downloaded the list of eukaryotic short linear motifs (ELMs) from the ELM database99 (version 1.4, May 2017). Next, we searched for putative ELMs in effector sequences using the ANCHOR tool100. We calculated the motif density for each effector sequence as the fraction between the amino acids belonging to putative ELMs predicted as disordered and the total number of amino acids predicted as disordered. See all data in Supplementary Table 3.
Protein expression and purification
The genes encoding for N-, NS-, SBB- and C-Tir, (residues 1–233, 61–200, 76–180, and 388–550, respectively; Supplementary Table 4) were amplified by PCR from a synthetic E. coli codon-optimised gene of the full-length protein (Uniprot code B7UM99 | TIR_ECO27). The human SHP-1 C-SH2 domain (residues 101-217; Uniprot code P29350 | PTN6_HUMAN) was PCR-amplified from a vector with the cDNA encoding the tandem SH2 domains of SHP-1 acquired from Addgene (pGEX SHP-1(NC)-SH2; Plasmid #46496). We inserted all PCR products into the IPTG-inducible pHTP8 vector (NZYTech) fused to an N-terminal Trx-His6-tag and a C-terminal Strep-tag. These constructs encoded TEV or HRV-3C protease recognition sites to remove the Trx-His6-tag linked to N-, NS- and SBB-Tir, or C-Tir and C-SH2, respectively. For C-Tir, Y454 and Y474 were mutated to phenylalanine using primers in Supplementary Table 6, resulting in the construct C-TirF454F474.
E. coli BL21 Star (DE3) pLysS were transformed with the desired construct and grown at 37 °C to a mid-log phase (OD600 ~ 0.6–0.8) in LB media containing kanamycin (50 μg/mL) and chloramphenicol (34 μg/mL). Protein expression was induced at 20 °C with 0.1 mM and 1 mM IPTG for SHP-1 C-SH2 domain and Tir constructs, respectively. After 24 h, bacteria were pelleted at 4 °C by centrifugation and stored at −20 °C until purification. For N- and NS-Tir and C-SH2, frozen cell pellets were resuspended in lysis buffer A (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 7.5) supplemented with EDTA-free protease inhibitor cocktail; and then lysed in a French Press. IDPs are known to resist high temperatures. The cell pellets with C-Tir were resuspended in lysis buffer B (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, pH 7.5) and lysed by boiling at 90 °C for 30 min in a water bath101. Total cell lysates were clarified by centrifugation for 40 min, 42000 rpm and 4 °C. For C-Tir, the resulting pellet was washed and resuspended two times in buffer B with 1% Triton X-100 and 0.5 M urea. The supernatants containing the soluble proteins were loaded onto 5 mL Strep-Tactin®XT high-capacity columns (IBA Lifesciences) equilibrated in buffer A. The N-terminal Trx-His6 tag, in C-Tir or C-SH2, was cleaved on-column with HRV-3C protease (1:100 protease:target protein ratio) overnight at 4 °C. We dismissed this step for N-Tir variants since they were expressed in cells containing an auxiliary plasmid, pRK793 (Addgene), which produces MBP-fused TEV protease S219V mutant. This approach enabled in vivo Trx-His6-cleaved N-Tir variants with a C-terminal Strep-tag102. Proteins were eluted with the buffer D (100 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1 mM EDTA, pH = 8.0) containing 50 mM Biotin. Pure fractions were pooled and purified using a Superdex 200 Increase 10/300 (GE Healthcare). Pure C-Tir was eluted in 20 mM sodium phosphate, 150 mM NaCl, 1 mM EDTA pH 6.5; C-SH2 in 20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH = 6.8; and pure N-Tir, NS-Tir and SBB in 20 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM TCEP, pH=6.5.
(un)Labelling for NMR experiments
For producing uniformly isotopic-labelled proteins, cells in LB were transferred before induction to M9 minimal media containing stable isotope precursors sources: 1 g/L of 15N-NH4Cl (U-15N-labelling) or 1 g/L of 15N-NH4Cl together with 2 g/L of 13C D-glucose (U-13C/15N-labelling). We also used reverse labelling to help overcome the signal-overlap problem often found in IDPs spectra. To this end, we produced isotopic-labelled C-Tir with a small set of unlabeled residues by growing cells in M9 media containing 1 g/L of 15N-NH4Cl and 1 g/L of unlabeled amino acids supplemented exogenously 1 hour before induction. We selected unlabeled amino acids with reduced 14N-isotope scrambling (i.e., L-arginine, L-asparagine, L-glutamine, or L-histidine)62 to avoid unwanted reverse labelling. So, only the supplemented unlabeled amino acids were selectively absent from NMR spectra of C-Tir. For paramagnetic NMR experiments, single cysteine variants of C-Tir were produced as described above and labelled with 1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)-methanethiosulfonate (MTSL). A 10-fold excess of MTSL was immediately added to DTT-free samples after elution from PD-10 columns and left reacting overnight at 4 °C in the dark. Excess MTSL was removed using PD-10 columns, and samples were exchanged into the NMR buffer.
NMR spectroscopy
We recorded all NMR experiments on a Bruker Avance II+ spectrometer operating at 18.8 T (800 MHz) and equipped with a helium cold TXI-cryoprobe. Spectra were processed with TopSpin and NMRPipe103 and analysed using CARA104. 1H chemical shifts were referenced directly, and 15N chemical shifts indirectly using 3-trimethylsilyl-1-propanesulfonic acid sodium salt (DSS; methyl 1H signal at 0.00 ppm).
For C-Tir’s backbone resonance assignment, we recorded a set of 3D HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, HN(CO)CACB, together with 2D [15N-1HN]-HSQC experiments at 283 K using 250-950 μM U-13C/15N-labelled samples in 20 mM sodium phosphate, 150 mM NaCl, 1 mM EDTA pH 6.5 with 20 μM DSS and 8% 2H2O (NMR buffer). We also measured 3D [1H,15N]-TOCSY-HSQC and [1H,15N]-NOESY-HSQC experiments using a 480 μM U-15N-labelled sample. In reverse labelling experiments with unlabelled L-arginine, L-asparagine, L-histidine, or L-glutamine residues, we recorded four regular 2D [15N-1HN] HSQC spectra. By comparing 2D [15N-1HN]-HSQC data of the uniformly labelled C-Tir with spectra where only one amino acid type is unlabeled, we could link the missing NMR signals to those respective amino acids. We used this helpful information to efficiently identify C-Tir’s backbone resonances and validate the sequential NMR assignment. This reverse labelling approach helped identify resonances by amino-acid type and simplified the spectral overlap by reducing observed peaks without any disadvantages to the protein expression and should be readily applied to other IDPs. Using the experimentally assigned chemical shifts, we employed the nsSPC67 and CheSPI68 methods to provide local structure information within the disordered C-Tir.
The resonances of backbone nuclei of N-Tir disordered residues were assigned using standard triple-resonance spectra. We collected HNCA, HN(CO)CA, HNCACB, HN(CO)CACB, HNCO, and HN(CA)CO experiments at 278 K using 600-760 μM U-13C/15N-labelled N-Tir samples in 20 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM TCEP, pH=6.5 with 8% 2H2O, and 20 μM DSS. Comparison of the [15N-1HN]-HSQC data of N-Tir to NS-Tir spectra enabled us to assign and corroborate the resonances within the flanking regions. Due to their distinct hydrodynamic properties, variable-temperature NMR experiments often report differently folded and disordered residues. Accordingly, we collected NMR at different temperatures from 5 to 35 °C, exploiting the differential temperature-dependent NMR sensitivity to ordered and disordered regions to investigate the N-Tir protein order-disorder interplay. We have probed mainly the N-Tir’s flexible protein regions with NMR at low temperatures, with ordered residues’ resonances broadened beyond detection due to slower tumbling. The folded residues’ resonances then became observable due to faster tumbling with higher temperatures. In contrast, the exchange of amide protons with the solvent at increased temperatures led to broadening of exposed disordered residues’ resonances, only allowing the detection of folded and less solvent-exposed regions. In addition, protein backbone resonances of NS-Tir were assigned from Best-TROSY-based 3D spectra105 [HNCO, HN(CA)CO, HNCA, iHNCA, HN(CO)CA, HNCACB, HN(CO)CACB, HNHA, H(CCO)NH and CC(CO)NH] all recorded with nonuniform sampling on a 1.23 mM U-[15N,13C]-labelled protein in the NMR buffer at 278 K. We also measured a 3D [1H,15N]-TOCSY-HSQC and [1H,15N]-NOESY-HSQC with a mixing time of 120 ms using a 607 μM U-15N-labelled sample. We computed the secondary structure propensities in terms of α-helix or β-sheet from the assigned chemical shifts using N-TALOS.
Chemical shift perturbations
NMR is highly sensitive to probe changes in the chemical environment. We extensively used this method to detect subtle chemical perturbations due to 2,2,2-Trifluoroethanol (TFE)-induced structural changes, lipid or protein binding, and tyrosine phosphorylation. To monitor the effect of TFE on C-Tir, we prepared protein samples (100 μM) in the NMR buffer with increasing amounts of TFE (1, 2, 5, 10 and 12.5% (v/v)) and acquired a 2D [15N-1HN]-HSQC spectrum for each condition. TFE-induced effects were determined as the ratio of [15N-1HN]-HSQC cross-peak intensity of the samples in the absence and presence of TFE.
15N-R 1 and R 2 experiments
T1 and T2 relaxation measurements were obtained using standard pulse sequences as previously described106 at 283 K. For the T1 relaxation experiments, the time intervals for inversion recovery were set at 20, 60, 100, 200, 400, 600, 800, 1200, 1400, 1600, 1800 and 2000 ms. For T2, the number of loops were set at 1, 2, 4, 8, 10, 12, 14, 16, 20, 24, 30, 40. Regarding the Free Induction Decay (FID) parameters, the data matrix was established with (1H)1024 × (15N) 256 points and 16 scans for all conducted experiments. Finally, the relaxation rates were obtained by applying an exponential function to the peak intensities as implemented in CcpNmr AnalysisAssign107.
Paramagnetic relaxation enhancement experiments (PREs)
We acquired PRE datasets for five paramagnetic centres engineered at different positions on the C-Tir to probe long-range contacts. Those positions were evenly scattered around the C-Tir sequence and preferable at serine residues in the wild-type. 1H-15N-HSQC spectra were acquired for the paramagnetic and diamagnetic (reduced with ascorbic acid) states C-Tir. Peak intensities of the paramagnetic (Ipara) and the diamagnetic (Idia) states were extracted by fitting the peaks with Lorentzian line-shapes. Only well-resolved resonances were used to quantify PRE ratios. Simulated random coil PRE values were calculated from a pool of 10,000 conformers of C-Tir using the average line width in the 1H dimension of the diamagnetic spectrum of each protein. ΔPREs were calculated as the difference between simulated and experimental (Ipara/Idia) data for each residue and smoothed with a 1D Gaussian kernel of window size = 7 and one standard deviation, interpolating the values for prolines or missing signals used to generate the heatmaps108.
Small-angle X-ray scattering (SAXS)
We employed synchrotron SAXS coupled with size exclusion chromatography (SEC) to probe the overall size and conformational properties of Tir intracellular regions and their ab initio shape (N-Tir) or ensemble (NS-Tir and C-Tir) representations. SAXS data for C-Tir were collected on the BM29 beamline (ESRF, Grenoble, France) and for N- and NS-Tir in the B21 beamline (DSL, Didcot, UK)109, exploiting their unique in-line HPLC systems (Shimadzu and Agilent 1200 respectively). We injected 50 μL samples with 10.0–25.0 mg mL−1 of SEC-purified protein into a 2.4 mL Superdex 200 Increase 3.2/300 column (GE) at a flow rate of 0.075 mL min−1 at BM29 and into a 4.6 mL Shodex KW403-4F column at a flow rate of 0.16 mL min−1 at B21. We acquired 1-second and 2-second frames in BM29 and B21, respectively. We did not detect radiation damage or signs of interparticle interference or aggregation. The SEC mobile phase consisted of 20 mM Phosphate pH 6.5, 150 mM NaCl, and 1 mM EDTA, for C-Tir, and 100 mM Tris·HCl at pH 8.0, 150 mM NaCl, 2 mM DTT, and 1 mM EDTA for N-Tir and NS-Tir. The scattering intensities from the respective monomeric (C-Tir) or dimeric elution (N-Tir or NS-Tir) single-peak region were integrated and buffer subtracted to produce the SAXS-profiles using ScÅtter (http://www.bioisis.net) and further processed with ATSAS110. A 50 µL series of samples with 1, 2.5, 5, 7.5, and 10 mg/ml were also run in batch mode. The scattering intensities of the 7.5 and 10 mg/mL injections of C-Tir (10 frames each) were merged to produce a SAXS profile. The Rg values were estimated using the Guinier approximation in the range s < 1.3/Rg. From SEC-SAXS, we created low-resolution ab initio molecular envelopes for N-Tir and NS-Tir, with the program DAMMIF and GASBOR in the ATSAS package110, assuming point symmetry P2. With DAMMIF, twenty independent models were generated, superimposed, and averaged to define the models’ most populated volume and assess modelling robustness. Details are in Supplementary Table 5.
Structural ensembles
We used AlphaFold2-multimer39, a refined version of AlphaFold252 for complex prediction, to predict the structure of the folded part of the NS-Tir dimer. We did not use template structures in the modelling, iterated for up to 48 recycles, followed by energy refinement with AMBER using default settings implemented in LocalColabFold111 and using MMseqs2 for creating multiple sequence alignments112. We assessed the AF-2 models’ confidence by the predicted Local Distance Difference Test (pLDDT) and inter-chain complex Predicted Alignment Error (PAE), i.e., the uncertainty about the interface. pLDDT is closely related to the pre-existing metric lDDT-Cα113 that measures the local accuracy of a prediction by determining the fraction of preserved local distances (higher is better). As a superposition-free method, lDDT is insensitive to relative domain orientation and correctly identifies segments in the full-length model deviating from the reference structure. pLDDT is given on a scale from 0-100. Regions with pLDDT > 90 are expected to have high accuracy. Parts displaying low pLDDT and high disorder propensity that flanked the folded dimer were trimmed from the AF-2 model and modelled as an ensemble. Despite the impressive performance of AF2 algorithms in predicting the native single static lowest energy structure of ordered proteins, they fail to represent disordered regions accurately114. Thus, the disordered regions of NS-Tir (61 to 96 and 187-200) were built on the predicted model using the statistical coil algorithm Flexible-Meccano (FM)53. Ten thousand conformations were individually compared to experimental SAXS data using EOM115 to find those subsets of structures that collectively best fit the SAXS data.
For C-Tir ensemble modelling, we also used the EOM approach to select a subset of conformations from a large starting pool in simultaneous agreement with intramolecular distances from PREs and overall size/shape distribution encoded by the SAXS data. In brief, we created 50000 structures with FM53, followed by side-chain modelling116, energy-refinement and in-silico incorporation of multiple spin-label dispositions at single-cysteines to interpret the PRE data117,118. We simulated the SAXS pattern for each conformer using CRYSOL110 and back-calculated the intramolecular 1H-15N-PRE-PRE rates assuming the Solomon-Bloembergen approximation117. We used multiple states derived from a 100 ns Molecular Dynamics (MD) simulation118. We extracted a frame every 100 ps to create a library of 1000 different MTSL conformations used to in-silico label each single-cysteine residue C-Tir conformer with multiple sterically allowed spin-label dispositions. This MTSL ensemble representation enabled the estimation of the order parameters associated with the dipolar proton-electron interaction vector motion119. With this strategy, we independently calculated the PRE rates for V405C, S428C, S463C, S509C, and S536C. We next searched for subsets of 200 models that best reproduce the five PRE rates datasets and the experimental SAXS profile using a genetic algorithm115.
AlphaFold-2 modelling of unphosphorylated C-Tir bound to C-SH2
Again, we used AlphaFold-Multimer to predict the binding interface as described above for NS-Tir. As a first stage, we used the sequences of C-SH2 and C-Tir as input to predict the 3D structure of C-Tir bound to C-SH2. Next, we run independent predictions replacing the full-length C-Tir with the ITIM sequence of C-Tir mapped to bind C-SH2, comprising the G506IQSTYALLANSGG519 peptide. As before, model confidence was assessed by the pLDDT and PAE metrics. PAE is not an inter-residue distance map or a contact map but the expected distance error in Å. It indicates the expected positional error at residue x if the predicted and actual structures are aligned on residue y (using the C, N and C atoms). PAEs are measured in Å and capped at 30 Å. PAE helps to assess the confidence in the relative position and orientation of the model parts (e.g., two domains or chains). For residues x and y in two chains, if the PAE values (x, y) are low, AF-2 predicts the chains to have well-defined relative positions and orientations120.
CD spectroscopy
We performed Far-UV CD spectroscopy in a J-815 spectrophotometer (Jasco), using a 1-mm optical pathlength cuvette for high performance (QS) (Hellma). The protein concentration was 6, 10, or 11.9 μM for N-Tir, NS-Tir, or C-Tir samples. N-Tir and NS-Tir were in 0.4 mM Tris-HCl pH 8, 15.0 mM NaF, and 0.1 mM TCEP. C-Tir was in 2 mM phosphate pH 6.5, 15 mM NaF and 0.1 mM EDTA supplemented with 0, 5, 10, 20, and 50% TFE. CD spectra are averages of 5-10 accumulations collected in the 190 nm to 260 nm range using a 0.1 nm data pitch and 2 nm bandwidth with a data integration time (DIT) of 8 sec at a scan speed of 50 nm/min at 10 °C. The thermal denaturation of NS-Tir was followed by monitoring temperature-dependent stepwise changes in spectral features ranging from 5 to 95 °C in 5 °C steps. We also recorded the CD signal at 222 nm probing for α-helix secondary structure by applying a temperature ramp of 1 °C min−1.
Tyrosine phosphorylation
Fully phosphorylation of four tyrosine residues in C-Tir was achieved by incubating the protein with human recombinant Fyn Kinase (Thermo Fisher Scientific)75. To this end, uniformly 13C/15N-labelled C-Tir (1.1 mM) was incubated with Fyn (1.93 μM) overnight at 25 °C in 20 mM HEPES pH 7.5 150 mM NaCl, 12 mM MgCl2 and 5.5 mM ATP. The amount of Fyn kinase used took into account its optimal specific activity at 25 °C and pH=7.5, and its capacity to completely phosphorylate four potential phosphorylation sites. C-Tir’s full phosphorylation was assessed by comparing 2D [15N-1HN]-HSQC data of pC-Tir to non-phosphorylated protein (969 μM) in the same buffer. Several aliquots of an equivalent 15N-labelled C-Tir were collected at different time points (2, 4, 8, 16, 24, 32, 48, and 64 minutes) upon adding Fyn to monitor the reaction. These sample aliquots were incubated at 95 °C for 5 min to stop the phosphorylation reaction. A 2D [15N-1HN]-HSQC spectrum of non-phosphorylated C-Tir was acquired as a control. Spectral dimensions were Ω1(15N) × Ω2(1H) = 23.5 ppm × 10.99 ppm, and the acquisition time was 68 ms in the Ω1 dimension and 92 ms in the Ω2 dimension. 2D [15N-1HN]-HSQC spectra were acquired for the different samples (50 μM each) using the same acquisition parameters as the control samples. The stubble changes in phosphorylated C-Tir (pC-Tir) backbone resonances were checked and re-assigned by HNCACB and HNCO experiments measured for the pC-Tir.
Phosphorylated C-Tir and binding to SHP-1 C-SH2 domain
Once C-Tir tyrosine phosphorylation was tested and implemented, we produced a 15N-labelled pC-Tir sample to reconstruct and investigate C-Tir interaction with the SHP-1 C-SH2 domain. As such, pC-Tir (95 μM) was titrated with 0.25, 0.5, 0.75, 1, 2, 4 and 6 equivalents of unlabeled C-SH2. 2D [15N-1HN]-HSQC spectra were acquired using the same parameters described. Intensity ratio perturbation (\({I}_{{obs}}^{i}\)) of the NMR signals from pC-Tir were plotted against the molar ratio of C-SH2/C-Tir. Peaks adjacent to each phosphorylation site exhibited a similar intensity drop, so we clustered them by site and globally fitted them to a 1:1 binding model using the equation:
where \({I}_{\max }^{i}\) is the maximal relative intensity of protein in free-state and \({I}_{\min }^{i}\) is the minimal relative intensity of bound protein under saturation conditions for residue i. \({K}_{D}\) is the apparent dissociation constant for each site. \([{CTir}]\) is the total protein concentration of C-Tir, and \([{CSH}2]\) is the concentration of the C-SH2. \({K}_{D}\) and \({I}_{\min }^{i}\) were defined as adjustable parameters. We employed the same approach to estimate the affinity of unphosphorylated C-Tir to C-SH2, by collectively fitting Eq. 1 to the NMR attenuation of residues A512LLA515.
Bicelles preparation
Bicelles with different charges were prepared based on the protocols described in refs. 80,121. In brief, we prepared 12.5% (w/w) dispersions by mixing long-chain (DMPC or DMPG) and short-chain (DHPC) phospholipids (Avanti Polar Lipids) in chloroform or chloroform:methanol 65:35, evaporating the solvent under a nitrogen flow and rehydrating the resulting lipid film in 20 mM sodium phosphate, 150 mM NaCl, 1 mM EDTA pH 6.5. The mixtures underwent 35 freeze-thaw cycles with vortexing until the lipid suspension was clear and transparent and then checked by dynamic light scattering. The molar ratio of lipids in bicelle samples was 1.0 DHPC:0.8 DMPG (q = 0.8, 44.4% DMPG molar ratio) and 1.0 DHPC:0.8 DMPC (q = 0.8, 44.4% DMPC molar ratio). The q-ratio (DMPC or DMPG)/DHPC) of 0.8 provides isotropic fast tumbling particles. To investigate C-Tir:bicelle interaction, 15N-labelled protein was added to the bicelles to a final concentration of 115 μM protein and 6% (w/v) of lipids. Lipid-binding effects were determined as the ratio of [15N-1HN]-HSQC cross peak intensity of the samples in the absence and presence of bicelles.
Tir lipid-binding site mutations
The pSA10-TirWT plasmid was used as a template for mutations. Primer sequences are listed in Supplementary Table 6. R388 and R389 were mutated to glutamic acid using primer Tir_388E389E_Fwd and Tir_388E389E_Rvs. L513 were mutated to glutamic acid using primer Tir_513E_Fwd and Tir_513E_Rvs. STOP codon was introduced at A541 using primer Tir_541*_Fwd and Tir_541*_Rvs. The resulting plasmids were transformed into E. coli DH5α, isolated and sequenced by Eurofins using primer Tir_CT_seq before the next round of mutation. To create pSA10-Tir388E389E513E, a second round of mutation was performed using primer Tir_513E_Fwd and Tir_513E_Rvs to introduce the 513E mutation into pSA10-Tir388E389E. To create pSA10-Tir388E389E513E541*, a third round of mutation was performed using primer Tir_541*_Fwd and Tir_541*_Rvs to introduce the 541* STOP codon into pSA10-Tir388E389E513E. The resulting plasmids were also transformed into E. coli DH5α, isolated and sequenced using primer Tir_CT_seq. All sequencing-verified plasmids (Supplementary Table 7) were electroporated into EPEC-0.
EPEC infection
SNU-C5 cells were seeded at 1105 on glass coverslips in each well of a 24-well plate (immunofluorescence) or 5104 in each well of a black clear bottom 96-well plate (PI uptake) in Park Memorial Institute (RPMI) medium (Sigma-Aldrich) with 10% (v/v) foetal bovine serum (FBS) (Gibco), 2 mM Glutamax (Gibco), 1 mM sodium pyruvate, 10 mM HEPES, and 2,5 μg/ml glucose. SNU-C5 cells were primed by IFNγ at 10 µg/ml 24 h before infection. EPEC E2348/69 strains were cultured at 37 °C 180 rpm shaking overnight in Luria Bertani (LB) medium and sub-cultured 1:50 in unsupplemented Dulbecco’s Modified Eagle Medium (DMEM) (low glucose) for 3 h at 37 °C with 5% CO2 static before infection. IPTG at 0.5 mM was added to the bacterial culture to induce protein expression from the pSA10 plasmid 30 min before infection. Infection was performed at a multiplicity of infection of 50:1. Infected cells were centrifuged at 700 g for 10 min and incubated at 37 °C with 5% CO2 static for required time.
Immunofluorescence imaging
Infection was performed as described above. At 2 h post-infection, infected cells were fixed by 4% paraformaldehyde for 15 min, washed by PBS for 3 times, permeabilized by 0.2% Triton X-100 in PBS for 4 min, washed again by PBS and blocked by 1% bovine serum albumin (BSA) for 10 min. Samples was incubated with rabbit anti-EPEC serum (VLA) (1:1000) in 1% BSA for 45 min, washed by PBS, re-blocked and incubated with the donkey anti-rabbit IgG-Alexa 488 antibody (Jackson Immunoresearch) (1:200), phalloidin-TRITC (Sigma-Aldrich) (1:200) and DAPI (Sigma-Aldrich) (1:1000) in 1% BSA for another 30 min and washed by PBS afterwards. Coverslips were mounted on glass slides (VWR) with Gold-Pro-Long-Anti-fade (Invitrogen). Imaging was performed using a Zeiss AxioImager Z1 microscope (Carl Zeiss) equipped with 100 lens.
Propidium iodide uptake assay
Infection was performed as described above. Cells were incubated with 5 µg/ml propidium iodide (PI) (Invitrogen) from 1 h before infection in phenol red-free serum-free RPMI medium. Positive control was performed by treating uninfected cells incubated in 0.05% Triton X-100 in PI-supplemented RPMI medium. Uninfected cells were used as negative control. No cell medium-only wells were used as blank. Time-course measurements were carried out in a FLUOstar Omega Microplate Reader (BMG Labtech) from 10 min to 8 h post-infection with 10-min intervals, measuring 620-nm emission with 520-nm excitation. After 2 h post-infection, 200 µg/ml gentamicin (Sigma-Aldrich) was added to each well. All PI uptake data were blank-corrected and normalised to negative control. Increase in percentage PI uptake was then calculated using the normalised PI uptake value of the infected sample divided by the normalised PI uptake value of the positive control. Statistical analysis of PI uptake assay was performed using GraphPad Prism 5.1.
Statistics and reproducibility
We compared disorder fraction distributions with the Mann-Whitney U-test. All biochemical experiments were performed at least three times. SAXS experiments were performed in duplicates at independent facilities (ESRF-BM29 and DLS-B21 Bio-SAXS beamlines) using different protein samples. All NMR samples were freshly prepared, followed by immediate recording of NMR spectra with multiple scans to ensure a good signal-to-noise ratio. NMR spectra were reproduced on different samples and also using technical replicates. Cell-based assays were performed with at least 3 biological replicates. Statistical analysis was performed using 1-way ANOVA with Tukey’s post-test. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns: non-significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The NMR chemical shifts of C-Tir, pC-Tir and NS-Tir are available at the BMRB with accession codes 50758 (C-Tir), 50759 (pC-Tir) and 52064 (NS-Tir). The SEC-SAXS data and models are available at SASBDB)122 under the project “SAXS studies on the intracellular region of the translocated intimin receptor”. The accession codes are detailed in Supplementary Table 5. Proteomes and effector collections, disorder predictions in Fig. 1 are available for download at https://osf.io/3mka9/ and supplementary material. A PRE/SAXS-based structural sub-ensemble of 200 structures for C-Tir is in the open-access Protein Ensemble Database (PED)123 with the identifier code PED00210. Numerical source data for graphs in Figs. 2 to 9 can be found in the supplementary data 1 file.
Code availability
The associated code is available at https://osf.io/cxkjf/.
References
Peng, Z. et al. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell. Mol. Life Sci. 72, 137–151 (2015).
Babu, M. M., Kriwacki, R. W. & Pappu, R. V. Structural biology. Versatility protein Disord. Sci. 337, 1460–1461 (2012).
Uversky, V. N. Intrinsically disordered proteins and their “mysterious” (meta)physics. Front. Phys. 7, (2019).
Wright, P. E. & Dyson, H. J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015).
Galea, C. A., Wang, Y., Sivakolundu, S. G. & Kriwacki, R. W. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry 47, 7598–7609 (2008).
Pelham, J. F., Dunlap, J. C. & Hurley, J. M. Intrinsic disorder is an essential characteristic of components in the conserved circadian circuit. Cell Commun. Signal. 18, 181 (2020).
Bah, A. & Forman-Kay, J. D. Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 291, 6696–6705 (2016).
van der Lee, R. et al. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 8, 1832–1844 (2014).
Uversky, V. N. Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front. Mol. Biosci. 1, 6 (2014).
Davey, N. E. et al. Attributes of short linear motifs. Mol. Biosyst. 8, 268–281 (2012).
Van Roey, K. et al. Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 114, 6733–6778 (2014).
Csizmok, V., Follis, A. V., Kriwacki, R. W. & Forman-Kay, J. D. Dynamic protein interaction networks and new structural paradigms in signaling. Chem. Rev. 116, 6424–6462 (2016).
Jespersen, N. & Barbar, E. Emerging features of linear motif-binding hub proteins. Trends Biochem. Sci. 45, 375–384 (2020).
Cornish, J., Chamberlain, S. G., Owen, D. & Mott, H. R. Intrinsically disordered proteins and membranes: a marriage of convenience for cell signalling? Biochem. Soc. Trans. 48, 2669–2689 (2020).
Deryusheva, E. et al. Does intrinsic disorder in proteins favor their interaction with lipids? Proteomics 19, e1800098 (2019).
Kjaergaard, M. & Kragelund, B. B. Functions of intrinsic disorder in transmembrane proteins. Cell. Mol. Life Sci. 74, 3205–3224 (2017).
Basile, W., Salvatore, M., Bassot, C. & Elofsson, A. Why do eukaryotic proteins contain more intrinsically disordered regions? PLoS Comput. Biol. 15, e1007186 (2019).
Xue, B., Dunker, A. K. & Uversky, V. N. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 30, 137–149 (2012).
Marín, M., Uversky, V. N. & Ott, T. Intrinsic disorder in pathogen effectors: protein flexibility as an evolutionary hallmark in a molecular arms race. Plant Cell 25, 3153–3157 (2013).
Hayashi, T. et al. Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA. Cell Host Microbe 12, 20–33 (2012).
O’Brien, D. P. et al. Calmodulin fishing with a structurally disordered bait triggers CyaA catalysis. PLoS Biol. 15, e2004486 (2017).
Aitio, O. et al. Enterohaemorrhagic Escherichia coli exploits a tryptophan switch to hijack host f-actin assembly. Structure 20, 1692–1703 (2012).
Hamada, D., Hamaguchi, M., Suzuki, K. N., Sakata, I. & Yanagihara, I. Cytoskeleton-modulating effectors of enteropathogenic and enterohemorrhagic Escherichia coli: a case for EspB as an intrinsically less-ordered effector. FEBS J. 277, 2409–2415 (2010).
Hazen, T. H. et al. Genomic diversity of EPEC associated with clinical presentations of differing severity. Nat. Microbiol. 1, 15014 (2016).
Pennington, H. Escherichia coli O157. Lancet 376, 1428–1435 (2010).
Tobe, T. et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl Acad. Sci. USA 103, 14941–14946 (2006).
Mills, E., Baruch, K., Charpentier, X., Kobi, S. & Rosenshine, I. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3, 104–113 (2008).
Kenny, B. et al. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520 (1997).
Kalman, D. et al. Enteropathogenic E. coli acts through WASP and Arp2/3 complex to form actin pedestals. Nat. Cell Biol. 1, 389–391 (1999).
Gruenheid, S. et al. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3, 856–859 (2001).
Xue, Y., Du, M., Sheng, H., Hovde, C. J. & Zhu, M.-J. Escherichia coli O157:H7 suppresses host autophagy and promotes epithelial adhesion via Tir-mediated and cAMP-independent activation of protein kinase A. Cell Death Discov. 3, 17055 (2017).
Yan, D., Wang, X., Luo, L., Cao, X. & Ge, B. Inhibition of TLR signaling by a bacterial protein containing immunoreceptor tyrosine-based inhibitory motifs. Nat. Immunol. 13, 1063–1071 (2012).
Goddard, P. J. et al. Enteropathogenic Escherichia coli stimulates effector-driven rapid caspase-4 activation in human macrophages. Cell Rep. 27, 1008–1017.e6 (2019).
Zhong, Q. et al. Clustering of Tir during enteropathogenic E. coli infection triggers calcium influx-dependent pyroptosis in intestinal epithelial cells. PLoS Biol. 18, e3000986 (2020).
Luo, Y. et al. Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405, 1073–1077 (2000).
Blasche, S. et al. The EHEC-host interactome reveals novel targets for the translocated intimin receptor. Sci. Rep. 4, 7531 (2014).
Yan, D. et al. Enteropathogenic Escherichia coli Tir recruits cellular SHP-2 through ITIM motifs to suppress host immune response. Cell. Signal. 25, 1887–1894 (2013).
Zhou, R. et al. Enterohemorrhagic Escherichia coli Tir inhibits TAK1 activation and mediates immune evasion. Emerg. Microbes Infect. 8, 734–748 (2019).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. BioRxiv (2021) https://doi.org/10.1101/2021.10.04.463034.
Jones, D. T. & Cozzetto, D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857–863 (2015).
Dosztányi, Z., Csizmok, V., Tompa, P. & Simon, I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 (2005).
Grishin, A. M. et al. NleH defines a new family of bacterial effector kinases. Structure 22, 250–259 (2014).
Holmes, A., Mühlen, S., Roe, A. J. & Dean, P. The EspF effector, a bacterial pathogen’s Swiss army knife. Infect. Immun. 78, 4445–4453 (2010).
Sallee, N. A. et al. The pathogen protein EspF(U) hijacks actin polymerization using mimicry and multivalency. Nature 454, 1005–1008 (2008).
Via, A., Uyar, B., Brun, C. & Zanzoni, A. How pathogens use linear motifs to perturb host cell networks. Trends Biochem. Sci. 40, 36–48 (2015).
Davey, N. E., Travé, G. & Gibson, T. J. How viruses hijack cell regulation. Trends Biochem. Sci. 36, 159–169 (2011).
Bommarius, B. et al. Enteropathogenic Escherichia coli Tir is an SH2/3 ligand that recruits and activates tyrosine kinases required for pedestal formation. Mol. Microbiol. 63, 1748–1768 (2007).
Tompa, P., Davey, N. E., Gibson, T. J. & Babu, M. M. A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169 (2014).
Campen, A. et al. TOP-IDP-scale: a new amino acid scale measuring propensity for intrinsic disorder. Protein Pept. Lett. 15, 956–963 (2008).
Patel, A. et al. Host protein interactions with enteropathogenic Escherichia coli (EPEC): 14-3-3tau binds Tir and has a role in EPEC-induced actin polymerization. Cell. Microbiol. 8, 55–71 (2006).
Rat, C., Heiby, J. C., Bunz, J. P. & Neuweiler, H. Two-step self-assembly of a spider silk molecular clamp. Nat. Commun. 9, 4779 (2018).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Ozenne, V. et al. Flexible-meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics 28, 1463–1470 (2012).
Fowler, N. J. & Williamson, M. P. The accuracy of protein structures in solution determined by AlphaFold and NMR. Structure 30, 925–933.e2 (2022).
Li, E. H. et al. Blind assessment of monomeric AlphaFold2 protein structure models with experimental NMR data. J. Magn. Reson. 352, 107481 (2023).
Youkharibache, P. et al. The small β-barrel domain: a survey-based structural analysis. Structure 27, 6–26 (2019).
Shen, Y. & Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013).
Bernadó, P. & Blackledge, M. A self-consistent description of the conformational behavior of chemically denatured proteins from NMR and small angle scattering. Biophys. J. 97, 2839–2845 (2009).
Cordeiro, T. N. et al. Structural characterization of highly flexible proteins by small-angle scattering. Adv. Exp. Med. Biol. 1009, 107–129 (2017).
Chemes, L. B., Alonso, L. G., Noval, M. G. & de Prat-Gay, G. Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains. Methods Mol. Biol. 895, 387–404 (2012).
Delaforge, E., Cordeiro, T. N., Bernadó, P. & Sibille, N. Conformational characterization of intrinsically disordered proteins and its biological significance. in Modern Magnetic Resonance (ed. Webb, G. A.) 381–399 (Springer International Publishing, 2018). https://doi.org/10.1007/978-3-319-28388-3_52.
Prasanna, C., Dubey, A. & Atreya, H. S. Amino acid selective unlabeling in protein NMR spectroscopy. Meth. Enzymol. 565, 167–189 (2015).
Tamiola, K., Acar, B. & Mulder, F. A. A. Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J. Am. Chem. Soc. 132, 18000–18003 (2010).
Fuxreiter, M., Simon, I., Friedrich, P. & Tompa, P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 338, 1015–1026 (2004).
De Biasio, A. et al. p15PAF is an intrinsically disordered protein with nonrandom structural preferences at sites of interaction with other proteins. Biophys. J. 106, 865–874 (2014).
Backert, S., Kenny, B., Gerhard, R., Tegtmeyer, N. & Brandt, S. PKA-mediated phosphorylation of EPEC-Tir at serine residues 434 and 463: A novel pathway in regulating Rac1 GTPase function. Gut Microbes 1, 94–99 (2010).
Tamiola, K. & Mulder, F. A. A. Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem. Soc. Trans. 40, 1014–1020 (2012).
Nielsen, J. T. & Mulder, F. A. A. CheSPI: chemical shift secondary structure population inference. J. Biomol. NMR 75, 273–291 (2021).
Tsaban, T. et al. Harnessing protein folding neural networks for peptide-protein docking. Nat. Commun. 13, 176 (2022).
Jaber Chehayeb, R. & Boggon, T. J. SH2 domain binding: diverse flvrs of partnership. Front Endocrinol. (Lausanne) 11, 575220 (2020).
Poy, F. et al. Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition. Mol. Cell 4, 555–561 (1999).
Theillet, F.-X. et al. Site-specific NMR mapping and time-resolved monitoring of serine and threonine phosphorylation in reconstituted kinase reactions and mammalian cell extracts. Nat. Protoc. 8, 1416–1432 (2013).
Mittag, T. et al. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl Acad. Sci. USA 105, 17772–17777 (2008).
Turner, A. L. et al. Highly disordered histone H1-DNA model complexes and their condensates. Proc. Natl Acad. Sci. USA 115, 11964–11969 (2018).
Rosenlöw, J., Isaksson, L., Mayzel, M., Lengqvist, J. & Orekhov, V. Y. Tyrosine phosphorylation within the intrinsically disordered cytosolic domains of the B-cell receptor: an NMR-based structural analysis. PLoS ONE 9, e96199 (2014).
Cheng, H. et al. Conformational Changes in the Cytoplasmic Region of KIR3DL1 upon Interaction with SHP-2. Structure 27, 639–650.e2 (2019).
Xu, C. et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell 135, 702–713 (2008).
Xu, X., Li, H. & Xu, C. Structural understanding of T cell receptor triggering. Cell. Mol. Immunol. 17, 193–202 (2020).
Duchardt, E., Sigalov, A. B., Aivazian, D., Stern, L. J. & Schwalbe, H. Structure induction of the T-cell receptor zeta-chain upon lipid binding investigated by NMR spectroscopy. Chembiochem 8, 820–827 (2007).
Pérez, Y. et al. Lipid binding by the Unique and SH3 domains of c-Src suggests a new regulatory mechanism. Sci. Rep. 3, 1295 (2013).
de Groot, J. C. et al. Structural basis for complex formation between human IRSp53 and the translocated intimin receptor Tir of enterohemorrhagic E. coli. Structure 19, 1294–1306 (2011).
Cepeda-Molero, M. et al. Attaching and effacing (A/E) lesion formation by enteropathogenic E. coli on human intestinal mucosa is dependent on non-LEE effectors. PLoS Pathog. 13, e1006706 (2017).
Cohan, M. C. & Pappu, R. V. Making the case for disordered proteins and biomolecular condensates in bacteria. Trends Biochem. Sci. 45, 668–680 (2020).
Touzé, T., Hayward, R. D., Eswaran, J., Leong, J. M. & Koronakis, V. Self-association of EPEC intimin mediated by the beta-barrel-containing anchor domain: a role in clustering of the Tir receptor. Mol. Microbiol. 51, 73–87 (2004).
Kim, D.-H. & Han, K.-H. PreSMo target-binding signatures in intrinsically disordered proteins. Mol. Cells 41, 889–899 (2018).
Mollica, L. et al. Binding mechanisms of intrinsically disordered proteins: theory, simulation, and experiment. Front. Mol. Biosci. 3, 52 (2016).
Nieto, L. et al. Estrogen receptor folding modulates cSrc kinase SH2 interaction via a helical binding mode. ACS Chem. Biol. 10, 2624–2632 (2015).
Nachman, J. et al. Conformational determinants of phosphotyrosine peptides complexed with the Src SH2 domain. PLoS ONE 5, e11215 (2010).
Arbesú, M., Iruela, G., Fuentes, H., Teixeira, J. M. C. & Pons, M. Intramolecular fuzzy interactions involving intrinsically disordered domains. Front. Mol. Biosci. 5, 39 (2018).
Zhong, Q., Chatterjee, S., Choudhary, J. S. & Frankel, G. EPEC-induced activation of the Ca2+ transporter TRPV2 leads to pyroptotic cell death. Mol. Microbiol. 117, 480–492 (2022).
Stachowiak, J. C. et al. Membrane bending by protein-protein crowding. Nat. Cell Biol. 14, 944–949 (2012).
Wagner, M. J., Stacey, M. M., Liu, B. A. & Pawson, T. Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb. Perspect. Biol. 5, a008987 (2013).
Haynes, C. et al. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2, e100 (2006).
Staby, L. et al. Eukaryotic transcription factors: paradigms of protein intrinsic disorder. Biochem. J. 474, 2509–2532 (2017).
UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 (2021).
Mann, H. B. & Whitney, D. R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18, 50–60 (1947).
Giansanti, A., Deiana, A., Forcelloni, S. & Porrello, A. New classification of intrinsic disorder in the human proteome. BioRxiv (2018) https://doi.org/10.1101/446351.
Tiessen, A., Pérez-Rodríguez, P. & Delaye-Arredondo, L. J. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res. Notes 5, 85 (2012).
Dinkel, H. et al. ELM 2016-data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 44, D294–D300 (2016).
Mészáros, B., Simon, I. & Dosztányi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 5, e1000376 (2009).
Livernois, A. M., Hnatchuk, D. J., Findlater, E. E. & Graether, S. P. Obtaining highly purified intrinsically disordered protein by boiling lysis and single step ion exchange. Anal. Biochem. 392, 70–76 (2009).
Kapust, R. B. & Waugh, D. S. Controlled intracellular processing of fusion proteins by TEV protease. Protein Expr. Purif. 19, 312–318 (2000).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Bartels, C., Xia, T. H., Billeter, M., Güntert, P. & Wüthrich, K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6, 1–10 (1995).
Solyom, Z. et al. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR 55, 311–321 (2013).
Gomes, T. et al. Conformational landscape of multidomain SMAD proteins. Comput. Struct. Biotechnol. J. 19, 5210–5224 (2021).
Skinner, S. P. et al. CcpNmr AnalysisAssign: a flexible platform for integrated NMR analysis. J. Biomol. NMR 66, 111–124 (2016).
Arbesú, M. et al. The unique domain forms a fuzzy intramolecular complex in src family kinases. Structure 25, 630–640.e4 (2017).
Cowieson, N. P. et al. Beamline B21: high-throughput small-angle X-ray scattering at diamond light source. J. Synchrotron Radiat. 27, 1438–1446 (2020).
Manalastas-Cantos, K. et al. ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 54, 343–355 (2021).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Mariani, V., Biasini, M., Barbato, A. & Schwede, T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 29, 2722–2728 (2013).
Ruff, K. M. & Pappu, R. V. Alphafold and implications for intrinsically disordered proteins. J. Mol. Biol. 433, 167208 (2021).
Bernadó, P., Mylonas, E., Petoukhov, M. V., Blackledge, M. & Svergun, D. I. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 (2007).
Eyal, E., Najmanovich, R., McConkey, B. J., Edelman, M. & Sobolev, V. Importance of solvent accessibility and contact surfaces in modeling side-chain conformations in proteins. J. Comput. Chem. 25, 712–724 (2004).
Salmon, L. et al. NMR characterization of long-range order in intrinsically disordered proteins. J. Am. Chem. Soc. 132, 8407–8418 (2010).
Cordeiro, T. N. et al. Interplay of protein disorder in retinoic acid receptor heterodimer and its corepressor regulates gene expression. Structure 27, 1270–1285.e6 (2019).
Chen, Y., Campbell, S. L. & Dokholyan, N. V. Deciphering protein dynamics from NMR data using explicit structure sampling and selection. Biophys. J. 93, 2300–2306 (2007).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Maffei, M. et al. Lipid binding by disordered proteins. Protoc. exch. (2013) https://doi.org/10.1038/protex.2013.094.
Valentini, E., Kikhney, A. G., Previtali, G., Jeffries, C. M. & Svergun, D. I. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res. 43, D357–D363 (2015).
Lazar, T. et al. PED in 2021: a major update of the protein ensemble database for intrinsically disordered proteins. Nucleic Acids Res. 49, D404–D411 (2021).
Elazar, A., Weinstein, J. J., Prilusky, J. & Fleishman, S. J. Interplay between hydrophobicity and the positive-inside rule in determining membrane-protein topology. Proc. Natl Acad. Sci. USA 113, 10340–10345 (2016).
Acknowledgements
We acknowledge using the ESRF-BM29 (MX-2085-BAG) and DLS-B21 Bio-SAXS beamlines (MX20161-1, SM21035-177). We thank Miquel Pons, from UB, Lígia Martins, and Adriano Henriques from ITQB-NOVA, for many helpful discussions. We thank the staff of ITQB-NOVA’s Research Facilities for their technical assistance. MA thanks Prof. Hartmut Oschkinat and FMP-Berlin for the financial support. AZ thanks Fabrice Lopez and Aurélie Bergon (TAGC, Marseille) for technical assistance. We thank Luis Angel Fernandes for sharing EPEC-0. This work was supported by National funds through FCT, Project MOSTMICRO-ITQB (UIDB/04612/2020, UIDP/04612/2020), FEDER Funds through COMPETE 2020 (0145-FEDER-007660), and SR&TD project (PTDC/BIA-BFS/0391/2021). Financial support was provided by European EC Horizon2020 TIMB3 (Project 810856). NMR data were acquired at CERMAX, ITQB-NOVA, Oeiras, Portugal, with equipment funded by FCT, project AAC 01/SAICT/2016. MFMV is funded by a MolBioS FCT PhD program fellowship (PD/BD/135482/2018). GH thanks the PT-NMR FCT PhD Program (PD/BD/147227/2019) for financial support. A MOSTMICRO FCT PhD scholarship supports MLM (UI/BD/154576/2022). An Imperial College president’s scholarship supports QZ. AZ acknowledges the financial support of the JPI HDHL-INTIMIC action co-funded by the Agence Nationale de la Recherche (ANR-17-HDIM-0001). TNC is the recipient of the grant CEECIND/01443/2017.
Author information
Authors and Affiliations
Contributions
MFMV carried out the biochemical, SAXS, and NMR experiments, performed data analysis and protein assignment, and interpreted results related to C-Tir. GH conducted the biochemical, SAXS, and NMR experiments and data analysis for N-Tir constructs. MFMV and GH contributed equally to the work. QZ collected the cell-based data and microscopy fluorescence images. GF supervised the cell-based assays and fluorescence imaging and contributed with analysis tools. MA performed the bioinformatic analysis of disordered predictions. AZ was responsible for motif bioinformatic analysis. MA and AZ contributed to the conceptualization of this project. CF conceived and designed experiments. MLM performed protein purification and sample preparation. TV was responsible for C-Tir phosphorylation. TG assisted with NMR experiments, data analysis, and interpretation. HM assisted with modeling. TNC carried out modeling and protein assignments. All authors interpreted the findings. TNC wrote the paper with input from all authors. TNC supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Peer review
Peer review information
Communications Biology thanks Sulakshana Mukherjee and Neel Sarovar Bhavesh for their contribution to the peer review of this work. Primary Handling Editors: Manidipa Banerjee, Anam Akhtar, and Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vieira, M.F.M., Hernandez, G., Zhong, Q. et al. The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection. Commun Biol 7, 179 (2024). https://doi.org/10.1038/s42003-024-05856-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-05856-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.