Abstract
Osteoporosis is a progressive bone disease caused by impaired function of endogenous bone marrow-derived mesenchymal stem cells (BMSCs). Herein, we investigated the mechanism of lncRNA SNHG14 in osteoporosis progression. BMSCs were isolated from BALB/c mice. The osteogenic ability of BMSCs was assessed by Alkaline phosphatase (ALP) and Alizarin Red S Staining (ARS) staining. The interaction between miR-493-5p and SNHG14 or myocyte enhancer factor 2 C (Mef2c) was confirmed by dual-luciferase reporter assay. Bone histomorphometry changes were evaluated to analyze SNHG14’roles in osteoporosis in vivo. Our results illustrated SNHG14 and Mef2c levels were increased in a time-dependent manner in BMSCs, and miR-493-5p expression was decreased. SNHG14 knockdown inhibited osteogenic differentiation of BMSCs, and SNHG14 upregulation had the opposite effect. SNHG14 overexpression elevated bone mineral density and bone trabecular number, and alleviated osteoporosis progression in vivo. Mechanically, miR-493-5p was a target of SNHG14, and miR-493-5p targeted the Mef2c gene directly. SNHG14 overexpression reversed the inhibition of miR-493-5p on the osteogenic ability of BMSCs, and miR-493-5p silencing accelerated BMSCs osteogenesis by activating Mef2c-mediated autophagy to accelerate BMSCs osteogenesis. In short, SNHG14 activated autophagy via regulating miR-493-5p/Mef2c axis to alleviate osteoporosis progression, which might provide a new molecular target for osteoporosis treatment.
Similar content being viewed by others
Introduction
Osteoporosis is a systemic skeletal disease characterized by low bone density and microarchitectural deterioration of bone tissue, leading to enhanced bone fracture risk. With the increase of the aging population, osteoporosis is threatening human health, especially postmenopausal women and the elderly1. Osteoporosis is a growing medical, social, and economic problem because of its prevalence (affecting more than 10% of the population) and its consequences (fractures)2. At present, bone resorption inhibitors or bone formation enhancers are mainly used for treatment, such as bisphosphonate and intermittent parathyroid hormone. However, long-term use of drugs has side effects, and the therapeutic effect is not satisfactory3. Bone marrow mesenchymal stem cells (BMSCs) are multifunctional cells that can be differentiated into osteoblasts, bone cells, and adipocytes4. It is reported that the differentiation capacity of BMSCs is related to osteoporosis. The specific manifestation is that the decrease of osteogenic differentiation of BMSCs will cause the disorder of bone formation and promote the occurrence and development of osteoporosis5. Therefore, promoting osteogenic differentiation of BMSCs in a certain way will help to alleviate osteoporosis.
Long noncoding RNAs (lncRNAs) participate in several biological processes, including control of gene transcription, pre-mRNA processing, regulation of messenger RNA (mRNA) stability, and protein translation6,7. Meanwhile, lncRNA is involved in the regulation of an increasing number of diseases, such as osteosarcoma and Rheumatoid Arthritis, etc.8,9,10. The latest studies have shown that there are abnormal lncRNA in sequence, spatial structure, expression, and protein interaction, which have a vital effect on the pathogenesis of osteoporosis11. Moreover, lncRNAs act as competing endogenous RNAs (ceRNAs) to target and degrade microRNAs (miRNAs)12. Small nucleolar RNA host gene 14 (SNHG14), also known as UBE3A-ATS, is located on chromosome 15q11.213. Du et al. showed that SNHG14 induced osteogenic differentiation of hMSC in vitro by targeting miR-286114. Preliminarily, by applying miRDB software (http://www.mirdb.org/index.html), we found that there was a binding site between SNHG14 and miR-493-5p. However, it was not clear whether SNHG14 participated in the progression of osteoporosis through targeting miR-493-5p.
MiRNA is a noncoding RNA about 22 bp length that binds to the target mRNA 3′ - untranslated region (3′-UTR) to inhibit its levels. Increasing evidence shows that miRNA is released into bone microenvironment and participates in bone remodeling in the process of osteoporosis15. Further analyzing the specific mechanism, it is possible that miRNA may increase bone mineral density by accelerating the osteogenic differentiation of endogenous BMSCs16. Tilde et al. findings showed that overexpression of miR-138 inhibited the differentiation of hBMSCs into osteoblasts6. However, Yuan et al. indicated that miR-92b-5p could improve osteoporosis by promoting osteogenic differentiation of BMSCs via targeting ICAM-117. Recently, researches showed that high glucose inhibited the osteogenic differentiation of BMSCs by regulating miR-493-5p/ZEB2 axis18. Exactly, the roles and specific mechanism of miR-493-5p in osteoblast differentiation remain large elusive.
Autophagy maintains cytoplasmic balance by eliminating macromolecules and cells damaged by hunger or oxidative stress, which is beneficial to cell survival and growth19. The role of autophagy in reducing osteoporosis has been reported. Previous studies showed that in hypoxic microenvironments, autophagy maintained bone remodeling by removing reactive oxygen species, thereby alleviating osteoporosis4. In osteoporosis, oxidative stress induces TP53INP2 downregulation and suppresses osteogenic differentiation of BMSCs during osteoporosis through the autophagy degradation pathway20. Furthermore, autophagy is regulated by a variety of mechanisms, such as transcription factors, miRNA, and other related signaling pathways. Li et al. reported that miR-223 inhibited autophagy by targeting ATG16L1 to promote central nervous system inflammation21. Importantly, miR-15b accelerated the development of osteoporosis by regulating osteocyte differentiation and autophagy22. Moreover, miR-493-5p also had been described as a autophagy-related miRNA in bladder cancer23. However, it is not clear whether miR-493-5p affects autophagy and regulates the osteogenic differentiation of BMSCs and osteoporosis progression.
Strikingly, preliminary bioinformatics analysis found there was a putative binding target of miR-493-5p on the 3′-UTR regions of Mef2c. Previous literature revealed that single nucleotide polymorphism (SNP) of Mef2c gene locus was greatly correlated to adult osteoporosis and osteoporotic fractures24. More importantly, it also observed that enhancement of the p38/Mef2c pathway might be associated with autophagy activation25. Therefore, we hypothesized that SNHG14 regulated the expression of Mef2c and activated autophagy through miR-493-5p, promoting osteogenic differentiation of BMSCs, thus alleviating osteoporosis development.
Results
SNHG14 gradually enhanced in osteogenic differentiation of BMSCs
BMSCs were firstly isolated from mouse femur and tibia tissues. The results of microscopic morphological observation showed that the P0 generation cells were round or fusiform, and the P3 generation cells were fusiform. The adhesion of P0 generation cells was lower than that of P3 generation cells (Fig. 1a), which accorded with the morphological characteristics of BMSCs26. Flow cytometry analysis indicated that BMSCs surface antigen molecules CD90, CD105, CD44, and CD29 were positive, while CD34 and CD45 were negative (Fig. 1b), these findings suggested that BMSCs was isolated successfully. After the induction of osteogenic differentiation for 21 days, obvious mineralized nodules appeared in BMSCs (Fig. 1c). ALP staining analysis indicated that the ALP activity of BMSCs were significantly increased after the induction of osteogenic differentiation (Fig. 1d). Osteogenic differentiation markers levels, including Runx2, BSP, ALP and OCN, were strikingly increased in BMSCs following the induction of osteogenic differentiation (Fig. 1e). Next, SNHG14 expression was gradually increased during BMSCs osteogenic differentiation (Fig. 1f). In short, these results showed BMSCs were successfully isolated from mice tissue.
a Microscope observed the morphology of isolated BMSCs (100 ×). b Flow cytometry detected BMSCs surface antigen molecules, including CD90, CD105, CD44, CD29, CD34 and CD45. BMSCs were cultured in osteogenic medium for 21 days. c Alizarin Red S (ARS) staining analyzed mineralized nodules of BMSCs. d Representative images of ALP staining. e, f qRT-PCR assessed the level of SNHG14 and osteogenic differentiation markers, including Runx2, BSP, ALP, and OCN. Data shown as Mean ± SD. c–e diagram using students’ t-test, and f diagram using one-way ANOVA with turkey post-test. NM indicates normal growth medium, GM indicates differentiation medium. N = 3, *p < 0.05, **p < 0.01, ***p < 0.001.
SNHG14 knockdown inhibited osteogenic differentiation of BMSCs
To investigate the effect of SNHG14 on osteogenic differentiation of BMSCs, sh-SNHG14 was transfected into BMSCs for SNHG14 knockdown, sh-NC served as the negative control. qRT-PCR analysis showed that SNHG14 expression was significantly down-regulated in BMSCs after transfected with sh-SNHG14 for 48 h (Fig. 2a). Subsequently, transfected BMSCs were further cultured in osteogenic differentiation medium. SNHG14 knockdown blocked the formation of BMSCs mineralized nodules (Fig. 2b). ALP staining analysis displayed that the ALP activity in BMSCs were decreased by SNHG14 knockdown (Fig. 2c). Moreover, qRT-PCR and western blot analysis disclosed that SNHG14 knockdown significantly inhibited the mRNA and protein level of Runx2, BSP, ALP and OCN (Fig. 2d, e). To sum up, SNHG14 knockdown inhibited osteogenic differentiation of BMSCs.
sh-SNHG14 was transfected into BMSCs for SNHG14 knockdown, sh-NC served as the negative control. 48 h later, sh-SNHG14 transfected BMSCs were further cultured in osteogenic differentiation medium for 21 days, sh-NC transfected BMSCs served as control. a qRT-PCR detected SNHG14 mRNA level. b ARS staining analyzed the mineralized nodules of BMSCs. c Representative images of ALP staining. d The mRNA levels of Runx2, BSP, ALP, and OCN were assessed by qRT-PCR. e The protein levels of Runx2, BSP, ALP, and OCN were measured by Western blot. Data shown as Mean ± SD. Figure 2 using students’ t-test. N = 3, *p < 0.05, **p < 0.01, ***p < 0.001.
SNHG14 targeted miR-493-5p to regulate Mef2c expression
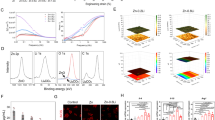
MiRDB software (http://www.mirdb.org/index.html) was applied to search the downstream targets gene of SNHG14. As shown in Fig. 3a, a binding site between SNHG14 and miR-493-5p was found. Subsequently, the direct binding relationship between SNHG14 and miR-493-5p was confirmed by dual-luciferase reporter assay. The results showed that co-transfection of SNHG14-wt plasmid and miR-493-5p mimetic into BMSCs resulted in a decrease in luciferase activity, which was blocked when mutations occurred at the assumed binding site (Fig. 3b). miR-493-5p level was significantly up-regulated in BMSCs after transfected with sh-SNHG14. Inversely, SNHG14 knockdown significantly decreased Mef2c level (Fig. 3c). Subsequently, miRDB software (http://www.mirdb.org/index.html) was used to search the downstream targets gene of miR-493-5p. Importantly, a binding site between miR-493-5p and 3′-UTR of Mef2c was found (Fig. 3d). And the luciferase activity of Mef2c-wt reported plasmids was inhibited by co-transfection of miR-493-5p mimics, but the co-transfection of miR-493-5p mimics did not affect the luciferase activity of Mef2c-mut reported plasmids (Fig. 3e). Furthermore, miR-493-5p inhibition significantly increased Mef2c expression, whereas Mef2c expression was significantly inhibited by miR-493-5p overexpression (Fig. 3f, g). Taken together, these data suggested that SNHG14 targeted miR-493-5p, and Mef2c served as a target of miR-493-5p.
a miRDB software (http://www.mirdb.org/index.html) predicted the binding sites of SNHG14 and miR-493-5p. b SNHG14 and miR-493-5p binding relationship was confirmed by dual-luciferase reporter assay. sh-SNHG14 was transfected into BMSCs for SNHG14 knockdown, sh-NC served as control. c qRT-PCR detected the mRNA levels of miR-493-5p and Mef2c in BMSCs after transfected with sh-SNHG14. d miRDB database (http://www.mirdb.org/index.html) predicted the downstream targets of miR-493-5p and Mef2c. e miR-493-5p and Mef2c binding relationship was confirmed by double luciferase reporter assay. f qRT-PCR detected Mef2c level in BMSCs after transfected with miR-493-5p mimics or inhibitor. g Western blot measured Mef2c protein level in BMSCs after transfected with miR-493-5p mimics or inhibitor. Data shown as Mean ± SD. b, c, e diagram using students’ t-test, and f, g diagram using one-way ANOVA with turkey post-test. N = 3, *p < 0.05, **p < 0.01, ***p < 0.001.
The inhibitory effect of miR-493-5p mimic on osteogenic differentiation was rescued by overexpressing SNHG14
First, we found that the expression of miR-493-5p was gradually reduced during osteoblast differentiation (Fig. 4a). Next, we investigated the role of SNHG14/miR-493-5p axis in osteogenic differentiation of BMSCs. The expression of miR-493-5p or SNHG14 was significantly up-regulated in BMSCs after transfected with miR-493-5p mimics or OE-SNHG14 (Fig. 4b). Then, BMSCs overexpressed with miR-493-5p were further transfected with OE-SNHG14, and transfected BMSCs were further cultured in osteogenic differentiation medium. Alizarin Red S staining results indicated that miR-493-5p overexpression blocked the formation of BMSCs mineralized nodules, while SNHG14 overexpression reversed the effect of miR-493-5p overexpression (Fig. 4c). MiR-493-5p overexpression decreased ALP activity of BMSCs, whereas SNHG14 overexpression reversed the downward trend (Fig. 4d). Furtherly, the mRNA and protein level of osteogenic differentiation marker proteins, including Runx2, BSP, ALP and OCN were down-regulated by miR-493-5p overexpression, but the downward trend was reversed by SNHG14 overexpression (Fig. 4e, f). Overall, the inhibitory effect of miR-493-5p mimic on osteogenic differentiation was rescued by overexpressing SNHG14.
BMSCs were transfected with miR-493-5p mimics and OE-SNHG14 for the overexpression of miR-493-5p and SNHG14. NC mimics and OE-NC vector served as the negative control. a qRT-PCR detected miR-493-5p expression in BMSC by different time induced. b qRT-PCR detected miR-493-5p and SNHG14 mRNA level. c ARS staining analyzed the mineralized nodules of BMSCs. d Representative images of ALP staining. e the mRNA levels of Runx2, BSP, ALP, and OCN were assessed by qRT-PCR. f the protein levels of Runx2, BSP, ALP, and OCN were measured by Western blot. Data shown as Mean ± SD. b diagram using students’ t-test, and a, c–f diagram using one-way ANOVA with turkey post-test. N = 3, *p < 0.05, **p < 0.01, ***p < 0.001.
Inhibition of miR-493-5p activated Mef2c-mediated autophagy to accelerate osteogenic differentiation of BMSCs
Based on previous finding27, we aimed to investigate whether Mef2c is related to the regulation of miR-493-5p in the differentiation of BMSCs. Mef2c expression was gradually increased in the process of osteogenic differentiation of BMSCs (Fig. 5a). Subsequently, we silenced Mef2c by transfecting sh-Mef2c into BMSCs, which was confirmed by RT-qPCR and western blot, showing Mef2c was reduced after sh-Mef2c transfection (Fig. 5b, c). Alizarin Red S and ALP staining demonstrated that miR-493-5p inhibition accelerated the formation of BMSCs mineralized nodules and ALP activity, but these effects were all abolished after Mef2c silencing (Fig. 5d, e). The high levels of osteogenic differentiation markers including Runx2, BSP, ALP, and OCN induced by miR-493-5p inhibitor were remarkably reversed by Mef2c knockdown (Fig. 5f, g). These findings suggested that miR-493-5p regulated osteogenic differentiation of BMSCs by targeting Mef2c. It was reported that autophagy was vital for the survival, differentiation, and function of osteocytes28. P38/Mef2c signaling was also identified to be linked to autophagy activation25. Herein, we further examined the expression of autophagy-related proteins during osteogenic differentiation of BMSCs. Immunofluorescence detected LC3 expression, and the results showed miR-493-5p inhibitor increased LC3 level, while Mef2c silencing revised the effect of miR-493-5p inhibitor (Fig. 5h). The results demonstrated that miR-493-5p inhibition enhanced the ratio of LC3II/I and Beclin-1 expression, but inhibited p62 expression, while these trends were strikingly reversed by Mef2c inhibition (Fig. 5i), suggesting the miR-493-5p/Mef2c axis participates in the osteogenic differentiation process of BMSC by regulating autophagy.
a qRT-PCR detected level of Mef2c after osteogenic induction for 0, 7, 14, and 21 days. sh-Mef2c and sh-NC were transfected into BMSCs, respectively. sh-NC served as control. b, c qRT-PCR, and Western blot detected Mef2c expression. MiR-493-5p inhibitor or sh-Mef2c was co-transfected into BMSCs, and followed by exposed to osteogenic induction medium for 21 days. d ARS staining analyzed mineralized nodules of BMSCs. e Representative images of ALP staining. f, g the mRNA and protein levels of Runx2, BSP, ALP, and OCN were detected by qRT-PCR and Western blot. h Immunofluorescence assay detected the expression of LC3. i the expression of LC3II/I, Beclin-1 and p62 proteins were measured by Western blot. Data shown as Mean ± SD. b, c diagram using students’ t-test, and a, d–i diagram using one-way ANOVA with turkey post-test. N = 3, *p < 0.05, **p < 0.01, ***p < 0.001.
SNHG14 overexpression activated Mef2c-mediated autophagy to accelerate osteogenic differentiation of BMSCs
We furtherly explore whether SNHG14 regulated osteogenic differentiation of BMSCs by regulating Mef2c-mediated autophagy. Alizarin Red S and ALP staining demonstrated that SNHG14 overexpression accelerated the formation of BMSCs mineralized nodules and ALP activity, but these effects were all abolished after Mef2c silencing (Supplementary Fig. 1a, b). The high levels of osteogenic differentiation markers including Runx2, BSP, ALP, and OCN induced by SNHG14 overexpression were remarkably reversed by Mef2c knockdown (Supplementary Fig. 1c, d). Besides, the level of LC3 in BMSCs was detected by immunofluorescence assay. Results indicated that SNHG14 overexpression enhanced LC3 expression, while this trend was strikingly reversed by Mef2c inhibition (Supplementary Fig. 1e). Western blot analysis also discovered that SNHG14 overexpression significantly elevated the levels of autophagy-associated proteins, including LC3II/I and Beclin-1, but reduced p62 level, while Mef2c knockdown reversed these trends (Supplementary Fig. 1f). These findings indicated that SNHG14 overexpression activated Mef2c-mediated autophagy to accelerate osteogenic differentiation of BMSCs.
SNHG14 overexpression promoted bone formation and alleviated osteoporosis in vivo
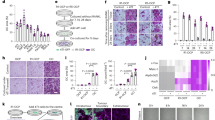
For further explore the role of SNHG14 in bone formation in vivo, the osteoporotic mouse model. A statistically significant decrease in BMD and bone volume/total volume ratio (BV/TV) of OVX mice was detected, but OE-SNHG14 impeded the trends (Fig. 6a, b). Quantitative analysis of tissue morphology showed that BFR/BS, Ob.S/BS, Oc.S/BS, and N.Oc/B.Pm of OVX mice were all statistically increased compared to sham group, after treatment by OE-SNHG14, the above trends were all alleviated (Fig. 6c–f). Similarly, H&E staining showed that the number of bone trabeculae decreased in OVX mice compared with sham mice, but OE-SNHG14 reversed this trend (Fig. 6g). qRT-PCR analysis indicated that SNHG14 and Mef2c expression was decreased, but miR-493-5p level was up-regulated in femurs and tibias tissues of OVX mice, while these effects were reversed by OE-SNHG14 treatment (Fig. 6h). Subsequently, BMSCs and osteoclasts were isolated from mouse bone marrow. qRT-PCR was performed to detected the level of SNHG14 in BMSCs and osteoclasts. Results discovered that compared to the sham group, the level of SNHG14 was significantly reduced after OVX treatment, while overexpression of SNHG14 had the opposite effects (Fig. 6i, j). Besides, Western blot analysis also discovered that the levels of osteogenic differentiation markers, including Runx2, BSP, ALP, and OCN, and autophagy-associated proteins, including LC3II/I and Beclin-1 were significantly elevated, but p62 level was reduced by OE-SNHG14 treatment compared to OVX group (Fig. 6k, l). These findings suggested that SNHG14 overexpression promoted bone information and alleviated osteoporosis, which might be correlated to Mef2c-mediated autophagy activation.
The mice were randomly divided into 4 groups (n = 5/each group): Shame group, OVX group, OVX + OE-NC, OVX + OE-SNHG14 group. a PIXImus densitometer was used to measure BMD of femurs. b Micro-CT measured BV/TV of structural parameters. c Bone formation parameter (BFR/BS) in distal mouse femur. d Osteoblast parameter (Ob.S/BS) was measured by histomorphometric analysis. e, f Bone resorption parameters (Oc.S/BS and N.Oc/BS) in distal mouse femur. g H&E staining was used to detect the pathological change. h qRT-PCR assessed the levels of SNHG14, miR-493-5p and Mef2c genes. i, j qRT-PCR measured the expression of SNHG14 in BMSCs and osteoclast. k Western blot detected the levels of Runx2, BSP, ALP, and OCN. l Western blot measured the proteins levels of LC3II/I, Beclin-1, and p62. Data shown as Mean ± SD. Figure 6 using one-way ANOVA with turkey post-test. N = 5, *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
With the increase of the elderly population, osteoporosis has become a worldwide disease, which will increase the risk of fracture and affect human health. The decrease of osteogenic differentiation of BMSCs leads to the decrease of bone formation and accompanied by osteoporosis29. In the present work, we isolated BMSCs from femurs and tibias tissues of mice to establish an osteogenic differentiation model in vitro. Moreover, we also conducted an osteoporotic mouse model in vivo. The finally data showed that SNHG14 targeted miR-493-5p/Mef2c axis to regulate autophagy activation, resulting the promotion effect in osteogenic differentiation of BMSCs, suggesting SNHG14 might be an effective target for osteoporosis treatment.
LncRNAs are a type of universal genes with multiple biological functions. In this paper, our findings suggest that SNHG14 was a sponge of miR-493-5p. There is growing evidence that the lncRNA/miRNA axis is involved in regulating various physiological and pathological processes in BMSCs, including osteogenic differentiation. Liu et al. showed that SNHG14 promoted the osteogenic differentiation of BMSCs by regulating the miR-185-5p/WISP2 axis30. Similarly, our findings indicated that SNHG14 knockdown inhibited osteogenic differentiation of BMSCs. Mechanically, we found that there was a binding site between SNHG14 and miR-493-5p.
MiRNAs have been shown to be widely involved in various biological processes. In recent years, the abnormal expression of miRNAs in osteoporosis has received widespread attention11. As previous mentioned, kynurenine accumulation led to an oxidative stress environment, resulting in miRNA profile changes in BMSCs, including the increased let-7f-5p, miR-493-5p, and the decreased miR-1281, miR-330-3p, miR-210-3p, which were the key factor to modulate BMSCs proliferation and differentiation31. Importantly, it reported that high glucose regulated the osteogenic differentiation of BMSCs by regulating miR-493-5p18. In our present work, we have found that miR-493-5p was up-regulated in osteoporotic mice and reduced gradually during BMSCs osteogenic differentiation. Subsequently, we also observed that miR-493-5p functioned as a negative regulator of osteoblast differentiation of BMSCs to decrease the formation of mineralized nodules, ALP activity, and osteogenic markers, indicating miR-493-5p plays a negative role in osteogenic differentiation of BMSCs. What’s more, we found that miR-493-5p served as a target of SNHG14, and the inhibition of osteogenic differentiation of BMSCs induced by miR-493-5p overexpression was relieved by SNHG14 overexpression. These findings suggested that SNHG14 via targeting miR-493-5p to regulate osteogenic differentiation of BMSCs. Furthermore, SNHG14 participates in the regulation of autophagy through targeted regulation of miRNAs. Deng et al. reported that SNHG14 regulates mouse hippocampal neurons through miR-182-5p/BINP3 axis32. SNHG14 regulated autophagy through the miR-223-3p/Foxo3a axis to alleviate acute lung injury induced by lipopolysaccharide33. Notably, we also observed the autophagy activation in miR-493-5p silenced BMSCs, presenting the increase of LC3II/I, Beclin-1, and the reduction of p62. Similarly, Moreover, a great number of evidences was widely confirmed that autophagy activation was a critical driven factor in osteogenic differentiation34,35,36. Thus, it could be inferred that the anti-osteogenic differentiation of miR-493-5p might be correlated to the inactivation of autophagy, which deserves further verification in the future.
Mef2c is a transcription factor associated with various cells’ development, including bone cells37. However, the biological role of Mef2c in osteoporosis and related bone formation is multidirectional. On the hand, it has been showed that the double deficiency of Mef2c and Mef2d will lead to cartilage angiogenesis and ossification in mice38. Likewise, Takayuki et al. revealed that Mef2c play as a positive regulator of osteoclast differentiation by facilitating c-FOS and NFATc137. On the other hand, Mef2c was negatively targeted by miR-23 in hBMSCs, and exhibited a promoting role in osteogenic differentiation by activating MAPK signaling39. In addition, Ni et al. investigation observed that enforced Mef2c expression could enhance p38, Osx and Runx2 and multiple osteogenesis‑associated genes in mouse MSCs40. From these observations, we believe that more experiments are needed to verify the exact biological functions and mechanisms of Mef2c. In this work, Mef2c was found gradually up-regulated during osteogenic induction of BMSCs, and the depletion of Mef2c led to an inhibitory role in osteogenic differentiation of BMSCs, which strikingly diminished miR-493-5p knockdown. Studies have shown that Mef2c is involved in the regulation of autophagy. For example, rapamycin treatment enhanced Mef2c expression, suggesting autistic behavior in rats can be improved by enhancing autophagy-activated p38/Mef2c signal pathway25. Importantly, we also observed that Mef2c silencing inhibited autophagy-related LC3II/I and Beclin-1 expression, but increased p62 accumulation, which consistent to Luo et al. findings25. More importantly, we have confirmed the targeting relationship between miR-493-5p and Mef2c. Therefore, we considered that Mef2c functioned as a promoting factor in osteogenic differentiation to involve in the regulation of miR-493-5p on BMSCs differentiation.
In summary, our findings firstly elucidated the roles and exact mechanism of SNHG14/miR-493-5p/Mef2c axis in osteogenic differentiation of BMSCs. It has demonstrated that SNHG14 by regulating miR-493-5p/Mef2c-mediated autophagy activation, leading to the promotion effect on osteogenic differentiation of BMSCs, thereby improving osteoporosis progression. In conclusion, SNHG14 activated autophagy via regulating miR-493-5p/Mef2c axis to alleviate osteoporosis progression. Our research provided more evidence for further understanding of the pathological mechanism of osteoporosis.
Materials and methods
Isolation and identification of BMSCs
Balb/c female mice were purchased from CasGene Biotech company (Beijing, China, 20 g to 25 g). The 8-week-old mice were sacrificed according to Animal Care and Use Guidelines of the First Affiliated Hospital, Department of Spine Surgery, Hengyang Medical School, University of South China (NO. 2021ll0518003). In detail, isoflurane anesthetized mice were subjected to 15 min in CO2 and were euthanized. Subsequently, the femur and tibia of mice were immediately removed for BMSCs isolation, according to previously description5. BMSCs cells were washed with syringe needle system and cultured in DMEM medium (Invitrogen, Carlsbad, CA, USA) containing 10% FBS (Solarbio, Beijing, China). For osteogenesis, BMSCs (4 × 105 cells/cm2) were cultured in α-MEM plus 10% FBS added with 50 μg/mL ascorbate-2, 10 mM β-glycerol phosphate and 0.1 μM dexamethasone for 21 d. Finally, the morphology of BMSCs was observed by microscope.
Flow cytometric analysis of BMSCs
The BMSCs cells were isolated with trypsin and washed twice with PBS. The cell suspension containing 1 × 105 cells was fixed in precooled 2% frozen formaldehyde. Subsequently, BMSCs were incubated with anti-CD90 (Abcam, ab3105, 1:50), anti-CD105 (Abcam, ab2529, 1:50), anti-CD44 (Abcam, ab112178, 1:100), anti-CD29 (Abcam, ab36219, 1:50), anti-CD34 (Abcam, ab81289, 1: 50) and anti-CD45 (Abcam, ab10558, 1:50) for 30 min. Finally, FACS Calibur system was used for flow cytometry, and cellquest graphics software was used to collect and analyze data.
Alizarin red S staining
After osteogenic induction of BMSCs, calcification was analyzed by Alizarin Red S staining, according to previous reports41. In detail, PBS was used to wash the BMSCs cultured in osteogenic differentiation medium for 15 days. Subsequently, BMSCs was fixed for 30 min with 10% formalin. After washing with PBS for 3 times, the cells were stained with alizarin red for 3 h at room temperature. Finally, wash the BMSCs with distilled water and take pictures under a microscope (OLYMPUS, Tokyo, Japan).
ALP staining
ALP staining was performed according to previous reports42. BMSCs was fixed with 4% formaldehyde, incubated with BCIP/NBT solution (Beyotime, Nanjing, China) under dark conditions, and washed with PBS three times to stop the reaction. Subsequently, the coloring was observed with a microscope, and the results were statistically analyzed.
BMSCs transfection
The shRNAs targeting SNHG14 or Mef2c, miR-493-5p inhibitor, miR-493-5p mimics, and their negative control (shNC, inhibitor NC, mimics NC) were synthesized by GeneChem (Shanghai, China). And cells were transfected with above plasmids by liposome 2000 reagent (Invitrogen, CA, USA). Then, lentiviral carried with overexpression vectors (OE-SNHG14) were co-transfected into human embryonic kidney 293 T (HEK293T) cells, and the three packaging plasmids included pHBLVTM, psPAX2, and pMD2.G. Lentiviral particles were obtained after 48 h of transfection via cell harvest and concentration with ultracentrifugation at 72,000 g for 2 h. Following titer determination, BMSCs were transfected at a multiplicity of infection (MOI) of 50 for 24 h.
qRT-PCR
The total RNA of the femur was isolated using an EASYspin Plus Bone Tissue RNA Kit (RN54, Aidlab Biotechnologies Co., Ltd., Beijing, China) according to the manufacturer’s instructions. The concentration and purity of the total RNA was calculated by measuring the absorbance at 260 nm and 280 nm. Total RNA was extracted from cells using a High Pure RNA isolation kit (BioTeke, China). cDNA was reversely transcribed from total RNA using M-MLV reverse transcriptase and random primers (Invitrogen, CA, USA). miR-493-5p level was detected by Taqman microRNA assay kit (Takara). Data analysis used U6 as endogenous control. A one-step SYBR Prime Script Plus RT-PCR kit (Takara) detected SNHG14, OCN, ALP, BSP, Runx2, and Mef2c levels. Standard procedure for two-step PCR amplification: stage 1, pre-denaturation, 95 °C 30 s. Stage 2, PCR reaction, 95 °C 5 s, 60 °C 30 s (40 cycles). Finally, extend 72 °C for 5 min. GAPDH served as endogenous control. The 2−ΔΔCt method calculated the relative expression level of genes. Primers were shown in Table 1.
Western blot
After the femurs were crushed in liquid nitrogen, then bone tissue protein extraction kit (BestBio, Shanghai, China) was used to extract the protein in bone tissues. BMSCs were gently lysed by incubation in ice-cold RIPA buffer (Beyotime, China) containing protease inhibitors (Beyotime, China) for 30 min followed by centrifugation at 12,000 × g for 15 min; then, the supernatant was collected. The total protein concentration was determined using a BCA kit (Beyotime, Nanjing, China). Protein (20 μg) was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroprinted on polyvinylidene fluoride membranes produced by Invitrogen in California, USA. The PVDF membrane was incubated with anti-OCN (Abcam, ab93876, 1:1000), anti-ALP (Abcam, ab229126, 1:1000), anti-BSP (GeneTex, GTX12155, 1:2000), anti-RUNX2 (Abcam, ab236639, 1:1000), anti-Mef2c (Abcam, ab211493, 1:1000), anti-LC3 (Abcam, ab128025, 1:1000), anti-p62 (Abcam, ab109012, 1:10000) and anti-Beclin-1 (Abcam, ab207612, 1:2000) overnight at 4 °C. Subsequently, the second strain of anti-rabbit IgG antibody was conjugated to horseradish peroxidase (Abcam, ab150077, 1:2000) on the membrane for 2 h at room temperature, and the bands were detected with an immobilized Western chemiluminescence meter (AutoLumo A2000Plus, Autobio, Zhengzhou, China). Finally, the strength of the bands was quantified using ImageJ software. Anti-β-actin (Abcam, ab8226, 1:1000) acted as internal reference. All western blot images have been cropped according to the protein molecular weight recommended by the manufacturer’s instructions.
Dual-luciferase reporter assay
miRDB software (http://www.mirdb.org/index.html) to predict the potential binding site between SNHG14, miR-493-5p and Mef2c 3′-UTR. Subsequently, the physical interaction between SNHG14, miR-493-5p, and Mef2c was confirmed by dual-luciferase reporter assay. In detail, the wild type (wt) or mutation type (mut) 3′-UTR fragments of Mef2c or SNHG14 were amplified by PCR and followed by inserted into pmirGLO vector (Promega, Fitchburg, WI, USA) to construct recombinant luciferase vector (Mef2c-wt and Mef2c-mut). The Mef2c-wt/mut or SNHG14-wt/mut vectors were co-transfected with miR-493-5p mimics or mimics NC into BMSCs by lipofectamine 2000 reagent (Invitrogen, CA, USA). Luciferase activity was assessed 48 h after transfection.
Osteoporotic mouse model
CasGene Biotech company (Beijing, China, 20 g to 25 g) provided 8-week-old Balb/c female mice. The mice were kept in standard cages with a room temperature of about 25 °C, a humidity of about 60%, a light-dark cycle of 12 h, and adequate food and water every day. All experimental produce was approved by the Ethics Committee of the First Affiliated Hospital, Department of Spine Surgery, Hengyang Medical School, University of South China (NO. 2021ll0518003). The mice were randomly divided into 4 groups (n = 5/each group): Sham group, OVX group, OVX + vector, OVX + Overexpressed (OE)-SNHG14. After anesthesia with pentobarbital sodium (50 mg/kg, intraperitoneal injection), the mice were subjected to sham operation or surgical ovariectomy (OVX), as described earlier43. Ovariectomy was performed in the OVX group. The bilateral ovaries of the sham group were identified and placed back into the abdominal cavity. Two months after ovariectomy, OVX mice were tail vein injected with lentivirus-packaged OE-NC or lentivirus-packaged OE-SNHG14 (10 nmol/per mouse). Once a week, four weeks after continuous injection, the mouse was euthanized by using CO2.
Micro-CT analysis
Microcomputed tomography (micro-CT) was applied to determine bone mass. After anesthesia with 1% pentobarbital, the mice were secured on the table in the prone position. Then, femurs were imaged by microcomputed tomography (PerkinElmer, USA) at 55 kV at an 8 μm voxel size. After the image was reconstructed, the area of interest (ROI) of trabecular bone was analyzed within the 0.3–0.8 mm distal metaphysis from the growth plate. Analyze V12.0 software was used to analyze the data and quantify the parameters, including bone mineral density (BMD), trabecular bone thickness (Tb. Th), trabecular bone number (Tb. N), bone volume per tissue volume (BV/TV), and trabecular separation (Tb. Sp).
Osteoclast culture
BMSCs were harvested from the femora and tibiae of 8-week-old mice. Then, the bone marrow cells (0.5 × 106) were differentiated into bone marrow-derived macrophages in α-MEM supplemented with 50 ng/mL recombination macrophage colony stimulating factor Glutamax, 10% heat-inactivated FBS, and 1% PBS in a 12-well plate for 16 h in a 37 °C incubator with 5% CO2. The non-adherent cells were then maintained in the presence of 50 ng/mL receptor activator for nuclear factor-κB ligand (RANKL) and 50 ng/mL M-CSF for an additional 5 days. For the coculture assay, osteoclasts were cocultured with BMSCs at a 1:3 ratio for 5 d in a Transwell system. Tartrate resistant acid phosphatase (TRAP) staining was performed using a kit (Solarbio, China) following the manufacturer’s protocols.
Histomorphometric analysis
Balb/c female mice were intraperitoneally injected with 20 mg/kg calcein (Sigma–Aldrich, USA), dissolved in PBS at a concentration of 2 mg/ml with 1 mg/ml NaHCO3 (Sigma–Aldrich, USA) at 16 d and 2 d before sacrifice. After sacrifice, the left femur was obtained, fixed in 4% formaldehyde, and embedded in methyl methacrylate. The specimen was cut into 30 mm slices by using a microtome (SP1600; Leica, Germany) under the condition of avoiding light. Then, cortical endosteum surfaces were evaluated using a fluorescence microscope (STP6000; Leica, Germany) with an excitation wavelength of 488 nm. Bone formation rate/bone surface area (BFR/BS), osteoblast surface area/osteoblast surface area (Ob.S/BS), osteoclast surface /bone surface area (Oc. S/BS) and number of osteoclast per bone surface (N.Oc/B.Pm) were used for quantitative analysis using ImageJ 1.47 software.
Hematoxylin-eosin (H&E) staining
The femur was decalcified by 10% EDTA and then embedded in paraffin. Five-micrometer sagittal sections of the metaphysis were prepared, stained with Toluidine blue, TRAP, hematoxylin-eosin (HE), and observed by light microscopy (Zeiss, Germany).
Statistics and reproducibility
Mean ± standard deviation (SD) represents data from at least three independent trials. All data were statistically analyzed using GraphPad Prism 5.0 Software (GraphPad Software, Inc.). Student’s t-test was used for comparison between the two groups, and One-way ANOVA with turkey post-test were used for multi-group comparison. The difference between the two groups was statistically significant (P < 0.05).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data that have been generated or analyzed during this study are included in the published article and the associated supplementary files. Where cropped western blot images have been used in the results figures, uncropped images have also been provided (Supplementary Fig. 7); raw data for graphs are presented in Supplementary Data l.
References
You, L., Pan, L., Chen, L., Gu, W. & Chen, J. MiR-27a is essential for the shift from osteogenic differentiation to adipogenic differentiation of mesenchymal stem cells in postmenopausal osteoporosis. Cell. Physiol. Biochem. 39, 253–265 (2016).
Pilipović, N. [Osteoporosis]. Med. Pregl. 54, 5–9 (2001).
Liang, X. et al. Icariin promotes osteogenic differentiation of bone marrow stromal cells and prevents bone loss in OVX mice via activating autophagy. J. Cell. Biochem. 120, 13121–13132 (2019).
Zhang, P. et al. Insulin impedes osteogenesis of BMSCs by inhibiting autophagy and promoting premature senescence via the TGF-β1 pathway. Aging 12, 2084–2100 (2020).
Li, Z. H. et al. MiR-291a-3p regulates the BMSCs differentiation via targeting DKK1 in dexamethasone-induced osteoporosis. Kaohsiung J. Med. Sci. 36, 35–42 (2020).
Eskildsen, T. et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl Acad. Sci. USA 108, 6139–6144 (2011).
Lan, T. et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 18, 186 (2019).
Han, J. J., Wang, X. Q. & Zhang, X. A. Functional interactions between lncRNAs/circRNAs and miRNAs: insights into rheumatoid arthritis. Front. Immunol. 13, 810317 (2022).
Ghafouri-Fard, S., Shirvani-Farsani, Z., Hussen, B. M. & Taheri, M. The critical roles of lncRNAs in the development of osteosarcoma. Biomed. Pharmacother. 135, 111217 (2021).
Ghafouri-Fard, S. et al. Contribution of miRNAs and lncRNAs in osteogenesis and related disorders. Biomed. Pharmacother. 142, 111942 (2021).
Yang, Y. et al. The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biol. Res. 53, 40 (2020).
Ma, N., Tie, C., Yu, B., Zhang, W. & Wan, J. Identifying lncRNA-miRNA-mRNA networks to investigate Alzheimer’s disease pathogenesis and therapy strategy. Aging 12, 2897–2920 (2020).
Dong, H. et al. Exosome-mediated transfer of lncRNA‑SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int. J. Oncol. 53, 1013–1026 (2018).
Du, M. et al. SNHG14 induces osteogenic differentiation of human stromal (mesenchymal) stem cells in vitro by downregulating miR-2861. BMC Musculoskelet. Disord. 21, 525 (2020).
Li, X. et al. miR-384-5p targets Gli2 and negatively regulates age-related osteogenic differentiation of rat bone marrow mesenchymal stem cells. Stem Cells Dev. 28, 791–798 (2019).
Wang, J., Liu, S., Li, J., Zhao, S. & Yi, Z. Roles for miRNAs in osteogenic differentiation of bone marrow mesenchymal stem cells. Stem Cell Res. Ther. 10, 197 (2019).
Chen, J. et al. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J. Exp. Clin. Cancer Res. 38, 398 (2019).
Zhai, Z. et al. High glucose inhibits osteogenic differentiation of bone marrow mesenchymal stem cells via regulating miR-493-5p/ZEB2 signalling. J. Biochem. 167, 613–621 (2020).
Kondrikov, D. et al. Kynurenine inhibits autophagy and promotes senescence in aged bone marrow mesenchymal stem cells through the aryl hydrocarbon receptor pathway. Exp. Gerontol. 130, 110805 (2020).
Yang, Y. et al. Oxidative stress induces downregulation of TP53INP2 and suppresses osteogenic differentiation of BMSCs during osteoporosis through the autophagy degradation pathway. Free Radic. Biol. Med. 166, 226–237 (2021).
Li, Y. et al. Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy 15, 478–492 (2019).
Lu, X., Zhang, Y., Zheng, Y. & Chen, B. The miRNA-15b/USP7/KDM6B axis engages in the initiation of osteoporosis by modulating osteoblast differentiation and autophagy. J. Cell. Mol. Med. 25, 2069–2081 (2021).
Guo, C. et al. Long noncoding RNA SNHG1 activates autophagy and promotes cell invasion in bladder cancer. Front. Oncol. 11, 660551 (2021).
Velazquez-Cruz, R. et al. Analysis of association of MEF2C, SOST and JAG1 genes with bone mineral density in Mexican-Mestizo postmenopausal women. BMC Musculoskelet. Disord. 15, 400 (2014).
Luo, Y. et al. [Enhanced autophagy activates p38/MEF2C pathway to regulate the expression of synapse-associated proteins and improve the symptoms of autistic rats]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 35, 236–242 (2019).
Mohamed-Ahmed, S. et al. Adipose-derived and bone marrow mesenchymal stem cells: a donor-matched comparison. Stem Cell Res. Ther. 9, 168 (2018).
Tang, M. et al. [Expression of myocardial specificity markers MEF-2C and Cx43 in rat bone marrow-derived mesenchymal stem cells induced by electrical stimulation in vitro]. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 32, 629–634 (2015).
Yin, X. et al. Autophagy in bone homeostasis and the onset of osteoporosis. Bone Res. 7, 28 (2019).
Qadir, A. et al. Senile osteoporosis: the involvement of differentiation and senescence of bone marrow stromal cells. Int. J. Mol. Sci. 21, 349 (2020).
Liu, Z. H. et al. LncRNA SNHG14 promotes osteogenic differentiation of human bone marrow-derived mesenchymal stem cells via regulating miR-185-5p/WISP2 axis. J. Biol. Regul. Homeost. Agents 35, 605–615 (2021).
Dalton, S. et al. Accumulation of kynurenine elevates oxidative stress and alters microRNA profile in human bone marrow stromal cells. Exp. Gerontol. 130, 110800 (2020).
Deng, Z. et al. LncRNA SNHG14 promotes OGD/R-induced neuron injury by inducing excessive mitophagy via miR-182-5p/BINP3 axis in HT22 mouse hippocampal neuronal cells. Biol. Res. 53, 38 (2020).
Hong, J. et al. lncRNA-SNHG14 plays a role in acute lung injury induced by lipopolysaccharide through regulating autophagy via miR-223-3p/Foxo3a. Mediat. Inflamm. 2021, 7890288 (2021).
Yang, X., Jiang, T., Wang, Y. & Guo, L. The role and mechanism of SIRT1 in resveratrol-regulated osteoblast autophagy in osteoporosis rats. Sci. Rep. 9, 18424 (2019).
Wang, S. et al. The role of autophagy and mitophagy in bone metabolic disorders. Int. J. Biol. Sci. 16, 2675–2691 (2020).
Vidoni, C. et al. Autophagy drives osteogenic differentiation of human gingival mesenchymal stem cells. Cell Commun. Signal. 17, 98 (2019).
Fujii, T. et al. MEF2C regulates osteoclastogenesis and pathologic bone resorption via c-FOS. Bone Res. 9, 4 (2021).
Kramer, I., Baertschi, S., Halleux, C., Keller, H. & Kneissel, M. Mef2c deletion in osteocytes results in increased bone mass. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 27, 360–373 (2012).
Jiang, K., Teng, G. D. & Chen, Y. Q. MicroRNA-23 suppresses osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting the MEF2C-mediated MAPK signaling pathway. J. Gene Med. 22, e3216 (2020).
Ni, S., Xiong, X. B. & Ni, X. Y. MgCl2 promotes mouse mesenchymal stem cell osteogenic differentiation by activating the p38/Osx/Runx2 signaling pathway. Mol. Med. Rep. 22, 3904–3910 (2020).
Liu, F. et al. LRRc17 controls BMSC senescence via mitophagy and inhibits the therapeutic effect of BMSCs on ovariectomy-induced bone loss. Redox Biol. 43, 101963 (2021).
Shen, G. et al. Foxf1 knockdown promotes BMSC osteogenesis in part by activating the Wnt/β-catenin signalling pathway and prevents ovariectomy-induced bone loss. EBioMedicine 52, 102626 (2020).
Guo, H. L. et al. LINC00472 promotes osteogenic differentiation and alleviates osteoporosis by sponging miR-300 to upregulate the expression of FGFR2. Eur. Rev. Med. Pharmacol. Sci. 24, 4652–4664 (2020).
Acknowledgements
This work was supported by Scientific Research Project of Hunan Provincial Health Commission (No. 202104070077) and Natural Science Foundation of Hunan Province (No. 2023JJ30551).
Author information
Authors and Affiliations
Contributions
J.X: Conceptualization, Methodology, Investigation, Validation, Visualization, Data curation, Writing- Original draft preparation, Supervision, Writing- Reviewing and Editing. L.L: Data curation, Software. H.L: Data curation. Z.L: Validation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The 8-week-old mice were sacrificed according to Animal Care and Use Guidelines of the First Affiliated Hospital, Department of Spine Surgery, Hengyang Medical School, University of South China. All experimental produce was approved by the Ethics Committee of the First Affiliated Hospital, Department of Spine Surgery, Hengyang Medical School, University of South China (NO. 2021ll0518003).
Peer review
Peer review information
Communications Biology thanks Hiroaki Saito and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Martina Rauner and Joao Valente.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xue, J., Liu, L., Liu, H. et al. LncRNA SNHG14 activates autophagy via regulating miR-493-5p/Mef2c axis to alleviate osteoporosis progression. Commun Biol 6, 1120 (2023). https://doi.org/10.1038/s42003-023-05493-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-05493-8
This article is cited by
-
Exploration and bioinformatic prediction for profile of mRNA bound to circular RNA BTBD7_hsa_circ_0000563 in coronary artery disease
BMC Cardiovascular Disorders (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.